Abstract
A wide variety of enveloped viruses infects cells by taking advantage of the low pH in the endocytic pathway to trigger virus-membrane fusion. For alphaviruses such as Semliki Forest virus (SFV), acidic pH initiates a series of conformational changes in the heterodimeric virus envelope proteins E1 and E2. Low pH dissociates the E2/E1 dimer, releasing the membrane fusion protein E1. E1 inserts into the target membrane and refolds to a trimeric hairpin conformation, thus driving the fusion reaction. The means by which E1 senses and responds to low pH is unclear, and protonation of conserved E1 histidine residues has been proposed as a possible mechanism. We tested the role of four conserved histidines by mutagenesis of the wild-type (wt) SFV infectious clone to create virus mutants with E1 H3A, H125A, H331A, and H331A/H333A mutations. The H125A, H331A, and H331A/H333A mutants had growth properties similar to those of wt SFV and showed modest change or no change in the pH dependence of virus-membrane fusion. By contrast, the E1 H3A mutation produced impaired virus growth and a markedly more acidic pH requirement for virus-membrane fusion. The dissociation of the H3A heterodimer and the membrane insertion of the mutant E1 protein were comparable to those of the wt in efficiency and pH dependence. However, the formation of the H3A homotrimer required a much lower pH and showed reduced efficiency. Together, these results and the location of H3 suggest that this residue acts to regulate the low-pH-dependent refolding of E1 during membrane fusion.
Enveloped viruses infect cells by fusing their membrane with that of the target cell through the action of transmembrane proteins in the virus envelope (15, 47). These membrane fusion proteins, although differing in structure among different enveloped viruses, nonetheless act through a common mechanism. Following an initial triggering event, the fusion protein interacts with the target membrane via a hydrophobic fusion peptide(s) and refolds into a hairpin-like conformation with the transmembrane domain and fusion peptide at the same end of the molecule. To date, the postfusion structures of all virus fusion proteins are trimeric hairpins. The triggering events for virus membrane fusion include virus-receptor/coreceptor interactions, exposure to a mildly acidic pH, and a combination of these processes. The critical triggering events can occur at the plasma membrane or within the low-pH environment of the endocytic pathway. While there has been remarkable progress in our understanding of the structures of virus membrane fusion proteins, the mechanism of triggering and the process of conversion from the prefusion conformation to the postfusion conformation are not understood.
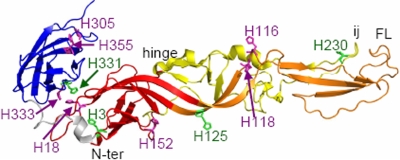
Semliki Forest virus (SFV) is a member of the alphaviruses, a genus of small, enveloped, plus-strand RNA viruses (25). SFV infects cells through a low-pH-triggered fusion reaction mediated by the E1 transmembrane protein (19). E1 is an elongated molecule containing three domains (DI, DII, and DIII) composed primarily of ß-sheets (Fig. 1) (26, 36). The central DI begins at the E1 N terminus. DII is comprised of two long extensions that connect to DI through a flexible hinge region. One extension contains the hydrophobic fusion peptide loop, and the other contains the ij loop, closely associated with the fusion loop at the DII tip. The other side of DI is joined to DIII via a linker region. DIII has an immunoglobulin-like fold and connects to the stem region and TM domain in the full-length E1 protein. On the alphavirus particle, E1 is located tangential to the virus membrane, with the fusion loop covered by E1's heterodimeric interaction with the partner protein E2 (12, 36, 49). The T=4 icosahedral symmetry of the virus particle is stabilized by E1-E1 interactions (26, 33, 36).
FIG. 1.
Locations of conserved histidine residues on the SFV E1 protein. The prefusion structure of the SFV E1 ectodomain is shown (PDB accession number 2ALA) (26, 36), with the nomenclature of ß-strands and other structural features taken from reference 26 and 36. The locations and residue numbers of all E1 histidine residues are indicated, with the conserved histidines shown in green. DI is shown in red, DII is shown in orange (the extension containing the hydrophobic fusion peptide loop) and yellow (the extension containing the ij loop), the DI-DIII linker is shown in gray, and DIII is shown in blue. The positions of the internal fusion peptide loop (FL), the DII ij loop, and the DI-DII hinge region are indicated. The E1 stem and TM regions are missing from this ectodomain structure. This figure was prepared using the PyMOL program (8). N-ter, N terminus.
Exposure to a pH of <6.2 in the endosome triggers a series of conformational changes in SFV (reviewed in reference 22). The E2/E1 dimer dissociates, exposing the E1 fusion loop. The fusion loop inserts into the target membrane, and three E1 molecules trimerize and refold into a stable hairpin in which DIII and the stem region fold back and pack against the central trimer composed of DI and DII (13). This rearrangement from the prefusion heterodimer to the stable target membrane-inserted E1 homotrimer drives the fusion reaction. SFV fusion and infection are inhibited by neutralization of endosomal low pH (16, 23), by mutations that prevent E2/E1 dimer dissociation, E1 membrane insertion, or E1 trimerization (14, 21, 37), and by the presence of exogenous DIII proteins, which bind E1 and prevent hairpin formation (28).
While it is clear that low pH is the trigger for the fusion of SFV with target membranes in vitro and in vivo, the mechanism of low-pH triggering is not understood. Low-pH-induced dissociation of the E2/E1 dimer has been shown to be controlled by the maturation state of E2 (37, 50) and by residues on E2 that affect the heterodimer interaction (14, 51). In addition, however, the monomeric E1 ectodomain has a threshold of pH ∼6 for membrane insertion and trimerization (14), indicating that E1 independently responds to low pH in the absence of E2. The pH 6 threshold suggested a possible role for E1 histidine residues. At neutral pH, histidine is relatively uncharged, while at pH ∼6, it becomes positively charged by the addition of an extra proton.
Comparison of alphavirus E1 protein sequences showed that four histidine residues in the SFV E1 fusion protein are highly conserved: H3, H125, H230, and H331 (Fig. 1) (36). While H230 in the ij loop was previously shown to be critical for SFV's fusion activity, it is not involved in the pH dependence of E1's conformational changes (5, 6). However, the remaining highly conserved histidine residues are positioned within regions of E1 that are known to move or rearrange during trimerization. H331 is located on DIII in a region that interacts with DI in the prefusion conformation and with DII in the trimer. The DIII-DI interface region in SFV contains two additional less-conserved histidines (DI H18 and DIII H333) (Fig. 1). All alphaviruses have at least one histidine residue in this interface (36), suggesting that protonation of these histidine residues could release DIII to permit folding back and hairpin formation. H125 is located in the DI/DII hinge region. Mutations that affect the pH dependence of the structurally related flavivirus fusion protein are frequently located in this flexible hinge (34). Interestingly, in the alphavirus particle, H125 is located at the quasi-twofold axis at a distance of approximately 5 to 6 Å from H125 on the interacting E1 (36). Thus, protonation of H125 could also be involved in the initial release of virus particle E1-E1 interactions by low pH (36). H3 is located on DI. During formation of the trimeric hairpin, H3 would be positioned close to the DI-DIII linker, which takes on a highly extended conformation. Thus, H3 could be involved in potential pH effects on this linker extension step.
In this study, we addressed the role of the histidine residues at E1 positions 3, 125, 331, and 333 by replacing each with alanine in the SFV infectious clone. While H331A, H333A, and H125A mutations did not have significant effects on virus fusion and infection, the H3A mutation decreased virus growth and strongly shifted the fusion threshold to a more acidic pH. This phenotype was due to a change in the pH dependence of E1 trimerization.
(Most of the data in this paper are from a thesis submitted by Z.Q. in partial fulfillment of the requirements for the degree of Doctor of Philosophy from the Second Military Medical University, Shanghai, People's Republic of China.)
MATERIALS AND METHODS
Cells.
BHK-21 cells were maintained in complete BHK medium (Dulbecco's minimal essential medium containing 5% fetal calf serum, 10% tryptose phosphate broth, 100 μg streptomycin/ml, and 100 U penicillin/ml) at 37 or 28°C.
Generation of mutant SFV clones and infectious RNAs.
Mutagenesis was performed as previously described (5, 29), using the subgenomic DG-1 plasmid as the template. The H331A/H333A double mutant was constructed using DG-1 containing the H331A mutation as the template. Mutations were introduced into DG-1 by circular mutagenesis using Pfu Turbo DNA polymerase (Stratagene, Inc., La Jolla, CA). The mutated NsiI/SpeI fragments were subcloned into the pSP6-SFV4 infectious clone (30). Two independent clones for each mutant were constructed from two different mutagenic PCRs to confirm the phenotype. Sequence analysis of mutant infectious clones (Genewiz Inc., North Brunswick, NJ) confirmed the sequence of the mutagenized region of E1. RNAs from the wild-type (wt) and mutant infectious clones were transcribed in vitro and electroporated into BHK-21 cells to produce virus infection (30).
Plaque assay and infectious-center assay.
To compare the growth kinetics of mutant and wt SFV, the amounts of infectious virus particles in the media at various times post-RNA electroporation were determined by plaque titration on BHK-21 cells. For slow-growing mutants, an “infectious-center” (referring to infected cells) assay was used instead of the plaque assay. Serial dilutions of virus were incubated with BHK-21 cells at 37°C for 60 min. The cells were incubated at 28°C overnight in the presence of 20 mM NH4Cl to prevent secondary infection, and infectious centers were quantitated by immunofluorescence using a polyclonal antibody to the SFV envelope proteins (29).
Virus assembly assay.
Virus assembly was evaluated by pulse-chase assays at 37°C as previously described (5, 29). BHK-21 cells were electroporated with wt or mutant RNA, plated in 35-mm dishes, and incubated at 37°C for 6 h. The infected cells were then pulse labeled with [35S]methionine-cysteine at 37°C for 30 min and chased at 37°C in media without label. At 0 h, 2 h, or 3 h, the chase media and cell lysates were harvested and analyzed by immunoprecipitation using a rabbit polyclonal antibody to the SFV E1 and E2 proteins (29). Cell lysate samples were precipitated in the presence of detergent. Medium samples were precipitated in the absence of detergent to permit recovery of intact, capsid-containing virus particles from the chase medium. Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Cell-cell fusion assay.
The fusion activity of viral proteins expressed on the surfaces of infected cells was tested by cell-cell fusion assays as previously described (5, 29). In brief, BHK-21 cells electroporated with virus RNA were diluted 1:20 with nonelectroporated cells and incubated at 37°C for 2 h to allow cell attachment to coverslips. The cells were cultured for ∼16 h at 28°C in the presence of 20 mM NH4Cl to prevent secondary infection and then treated with medium at the indicated pH for 1 min at 37°C to trigger cell-cell fusion. Cells were incubated an additional 3 h at 28°C to permit polykaryons to express the viral proteins and stained to detect virus membrane proteins and cell nuclei. The numbers of nuclei per envelope protein-positive cell were evaluated by fluorescence microscopy, and at least 200 nuclei per coverslip were counted. The fusion index was calculated as 1 − (number of cells/number of nuclei).
Fusion-infection assay.
wt and mutant virus stocks were prepared by electroporation of BHK-21 cells and incubation for 10 h. Tests of plaque size confirmed the absence of revertants within this incubation time. Appropriate dilutions of these virus stocks were incubated with BHK-21 cells on ice with shaking for 90 min. The cells with bound virus were then treated at the indicated pH for 1 min at 37°C to trigger virus fusion with plasma membrane and cultured for ∼16 h at 28°C in medium containing 20 mM NH4Cl to prevent secondary infection. The cells infected due to low-pH-triggered fusion were quantitated by immunofluorescence as described previously (28).
Preparation of radiolabeled virus.
To prepare 35S-labeled virus, BHK-21 cells were electroporated with wt or H3A mutant RNA, plated at 37°C for 6 h in complete BHK medium, and then cultured for 12 to 14 h at 37°C in methionine- and cysteine-deficient minimal essential medium containing 100 μCi of [35S]methionine-cysteine per ml (Promix; Amersham Life Sciences). The virus was purified by banding on a Pfefferkorn gradient (5). Tests of plaque size confirmed the absence of revertants within these incubation times.
Virus-liposome association assay.
Liposomes were prepared by freeze-thaw and extrusion as previously described (5) using a 1:1:1:1.5 molar ratio of phosphatidylcholine, phosphatidylethanolamine, sphingomyelin, and cholesterol (Avanti Polar Lipids, AL). Radiolabeled virus was mixed with liposomes (final concentration, 0.3 mM lipid), and the mixture was treated at the indicated pH for 5 min at 37°C. The samples were then adjusted to pH 8.0 and 40% sucrose in a final volume of 0.45 ml and layered over a 60% sucrose cushion, followed by the addition of 1.1 ml of 25% sucrose and 0.3 ml of 5% sucrose. Sucrose solutions were wt/vol in 50 mM Tris (pH 8.0)-100 mM NaCl. Gradients were centrifuged in a TLS-55 rotor for 2 h at 50,000 rpm at 4°C and fractionated into seven fractions. The percentages of virus radioactivity in the liposome-containing top three fractions were determined. Recovery of virus radioactivity from the gradients ranged from 72% to 98%.
Assays of E2/E1 dimer dissociation and E1 homotrimer formation.
35S-labeled wt or H3A mutant viruses were mixed with liposomes (final concentration, 0.8 mM lipid), treated at the indicated pH for 5 min at 20°C, and adjusted to a neutral pH. To detect E2/E1 dimer dissociation, samples were solubilized in 1% TX-100 and digested with 100 μg trypsin/ml for 10 min on ice (9, 20). To detect the trypsin-resistant E1 homotrimer, samples were solubilized in 1% TX-100 and digested with 100 μg trypsin/ml for 10 min at 37°C (5). Trypsin digestion was terminated by the addition of soybean trypsin inhibitor. To detect the SDS-resistant E1 homotrimer, samples were solubilized in SDS sample buffer for 3 min at 30°C and the proportions of E1 migrating in the monomer and trimer positions were determined (5). All samples were analyzed by SDS-PAGE and fluorometry. Quantitation of 35S-labeled viral proteins was performed by PhosphorImager analysis using Image Quant version 1.2 software (Molecular Dynamics, Sunnyvale, CA).
RESULTS
Generation and initial characterization of SFV E1 histidine mutants.
We tested the role of the highly conserved histidine residues at SFV E1 positions 3, 125, and 331 (Fig. 1). Given the close proximity of H331 and H333 on DIII (Fig. 1), we evaluated the role of both of these residues in the release of DIII during E1 refolding. Mutagenesis of the SFV infectious clone was used to change each histidine to the small nonpolar amino acid alanine, producing the H3A, H125A, and H331A single mutants and the H331A/H333A double mutant. Viral RNAs were transcribed in vitro from the mutant infectious clones, and the phenotypes of the mutants were characterized by electroporation of RNA into BHK-21 cells.
Initial characterization of electroporated cells by immunofluorescence confirmed that all four mutant RNAs caused primary infections comparable to those caused by wt RNA and that the mutant E1 and E2 proteins were efficiently expressed at the cell surface (data not shown). We tested the ability of the mutants to mediate secondary infection by coculturing electroporated and nonelectroporated cells overnight at 28 or 37°C (5). The H125A, H331A, and H331A/H333A mutants produced high levels of secondary infection at both temperatures, while secondary infection by the H3A mutant at either temperature was greatly reduced compared to the level of infection by wt SFV (data not shown).
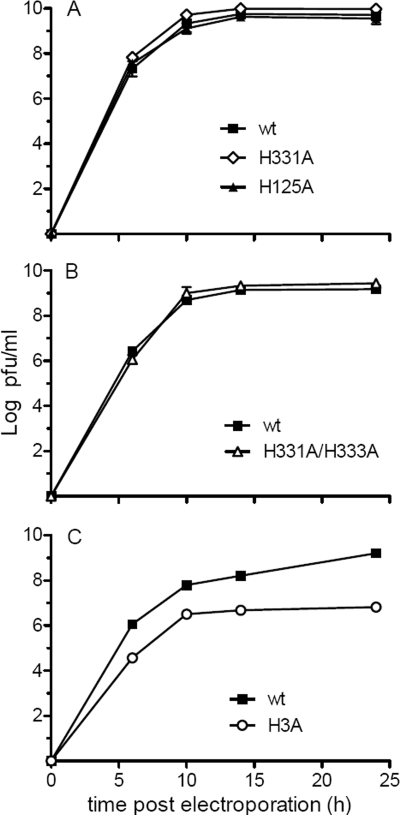
To quantitatively evaluate the growth kinetics of mutant and wt SFV, viral RNAs were electroporated into BHK-21 cells and progeny virus in the culture medium was measured after incubation of the infected cells at 37°C. Plaque assays on indicator BHK-21 cells showed that the growth kinetics of the H125A, H331A, and H331A/H333A mutants were comparable to those of wt SFV, with efficient virus production observed 6 h after electroporation and maximal virus titers observed by 14 to 24 h (Fig. 2A and B). The H3A mutant produced very small plaques that were difficult to count, so progeny virus was quantitated by using an infectious-center assay. While H3A mutant virus-infected cells produced virus that mediated secondary infection, the titer was about two logs lower than that of wt SFV at each time point (Fig. 2C), suggesting a decrease in virus assembly and/or infection efficiency.
FIG. 2.
Growth properties of wt and mutant SFV. BHK-21 cells were electroporated with wt or mutant virus RNA and incubated at 37°C. The cell media were collected at the indicated times after electroporation, and virus in the media was quantitated by plaque assay (A and B) or infectious-center assay (C) on BHK-21 cells. Data are the averages of the results from two independent experiments, and ranges are indicated by error bars.
Assembly of the H3A mutant.
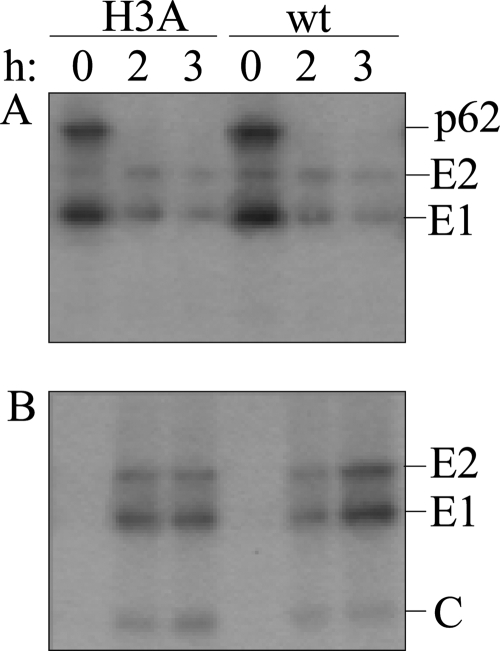
Virus assembly was evaluated by pulse-chase assays of cells electroporated with wt or H3A RNA. The infected cells were pulse-labeled with [35S]methionine-cysteine and chased at 37°C for 0 to 3 h, and the viral envelope proteins in the cell lysates and media were analyzed by immunoprecipitation. Both wt- and mutant-infected cells expressed p62 and E1 and showed efficient processing of the p62 precursor to the mature E2 protein during the chase time (Fig. 3A). Thus, the biosynthesis, transport, and processing of the envelope proteins were not significantly affected by the H3A mutation. Analysis of the chase media showed that wt- and H3A mutant-infected cells released virus particles with similar efficiencies (Fig. 3B). A number of previously described SFV mutants with assembly defects produce a proteolytically cleaved form of E1 termed E1s (5, 31), but H3A mutant-infected cells did not release significant amounts of E1s (Fig. 3B). Thus, these results suggested that the impaired growth of the H3A mutant is due to a decrease in infection efficiency rather than a defect in particle production.
FIG. 3.
Assembly properties of wt and mutant SFV. BHK-21 cells were electroporated with wt or mutant virus RNA, incubated at 37°C for 6 h, pulse labeled with [35S]methionine-cysteine, and chased for the indicated times at 37°C. The cell lysates (A) and media (B) were immunoprecipitated using a polyclonal antibody to the envelope proteins and analyzed by SDS-PAGE. Medium samples were immunoprecipitated in the absence of detergent to allow recovery of intact virus particles containing the viral nucleocapsid. The positions of the envelope (E1 and E2) and capsid (C) proteins are indicated on the right. Shown is an example representative of three experiments.
Membrane fusion activity of wt and mutant SFV.
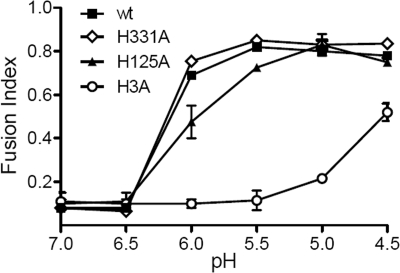
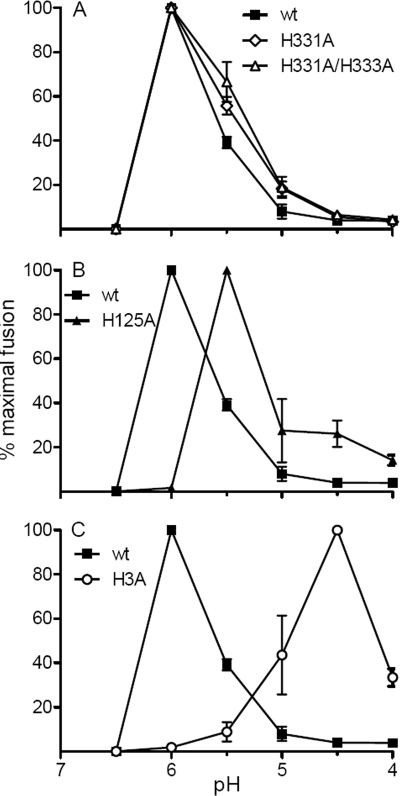
The effect of the histidine-to-alanine mutations on E1's fusion activity was tested by two different assays. Cells were electroporated with wt or mutant RNA and cultured overnight to produce high levels of expression of the E1 and E2 proteins at the cell surface. Cell-cell fusion was then triggered by treating the cells for 1 min at 37°C with buffers of the indicated pH. Polykaryon formation by the virus-infected cells was quantitated and expressed as a fusion index (Fig. 4). wt- or H331A mutant-infected cells showed efficient cell-cell fusion after treatment at pH 6.0, and fusion was maximal at a pH of 5.5. The H125A mutant-infected cells fused at pH 6.0, but the fusion index was somewhat lower than that of wt SFV at pH 6.0. Maximal fusion of H125A occurred at a pH of 5.0. The H3A mutant-infected cells showed no fusion until treatment at a pH of 5.0, and the fusion index was much lower than that of wt SFV even after treatment at a pH as low as 4.5. The fusion index is an average value and does not differentiate among individual polykaryons with different numbers of nuclei. We therefore also compared the fusion activities of wt and mutant SFV by counting the number of nuclei in each infected cell and calculating the distribution of nuclei per infected cell (data not shown). The results showed that there was no significant difference between the sizes of the syncytia generated by the H125A mutant and wt SFV after treatment at either pH 5.5 or pH 5.0. In contrast, the syncytia induced by the H3A mutant after treatment at pH 5.0 or pH 4.5 contained significantly fewer nuclei than those of wt SFV. At either pH, only 2 to 4% of the H3A mutant-infected cells contained three or more nuclei, and about 80% had not fused. In contrast, after treatment at a pH of 5.0 to 4.5, 37% to 34% of the wt-infected cells contained three or more nuclei, and more than 50% of the expressing cells had fused.
FIG. 4.
Cell-cell fusion activities of wt and mutant SFV. BHK-21 cells were electroporated with wt or mutant virus RNA, diluted 1:20 with nonelectroporated cells, and cultured overnight at 28°C. The cells were treated with media of the indicated pHs at 37°C for 1 min and incubated at 28°C for 3 h. The number of nuclei per envelope protein-expressing cell was evaluated, and the fusion index was calculated as described in the Materials and Methods. Data are the averages of the results from two independent experiments, and ranges are indicated by error bars.
We also tested the fusion activity of the wt and mutant virus particles with BHK-21 cells by using a fusion-infection assay. Virus stocks were bound to BHK-21 cells in the cold and treated at low pH to trigger fusion with the plasma membrane. The resulting infected cells were quantitated by immunofluorescence (Fig. 5). The fusion activities of the H331A mutant and the H331A/H333A double mutant were similar to that of wt SFV at each pH point, with maximal fusion at pH 6.0. The H125A mutant showed maximal fusion at pH 5.5. The H3A mutant showed a dramatically shifted pH dependence, with maximal fusion not occurring until treatment at pH 4.5. At pH values below the maximal fusion pH, all viruses showed decreased fusion, reflecting acid inactivation. Acid inactivation of the H3A mutant did not occur before treatment at a pH of <4.5. Thus, of the four E1 histidine residues tested for a role in SFV fusion, only mutation of H3 produced a strong phenotype in both virus infectivity and the pH dependence of virus fusion.
FIG. 5.
pH dependence of wt and mutant virus-membrane fusion. Virus stocks were incubated with BHK-21 cells on ice for 90 min to permit virus-cell binding and then treated with media of the indicated pHs for 1 min at 37°C to trigger virus fusion with plasma membrane. The cells infected due to low-pH-induced fusion were quantitated by immunofluorescence, and results are shown as percentages of maximal fusion for each virus. Data are the averages of the results from two independent experiments, and ranges are indicated by error bars.
Dimer dissociation and E1 membrane insertion.
In order to determine the mechanism for the striking change in low-pH triggering of the H3A mutant, we used purified 35S-labeled viruses to analyze the defined series of conformational changes induced upon exposure of SFV to an acidic pH. These conformational changes include dissociation of the E2/E1 heterodimer, insertion of the hydrophobic E1 fusion loop into the target membrane, and refolding of E1 to the stable homotrimer (reviewed in reference 19).
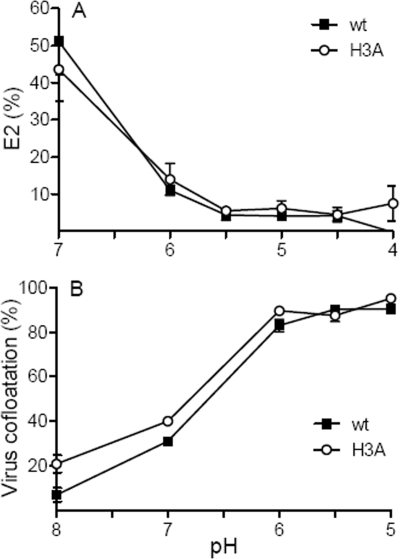
To compare the pH dependence of E2/E1 dissociation in wt SFV and the H3A mutant, radiolabeled virus was mixed with liposomes, incubated in buffers of defined pH, returned to neutral pH, and solubilized in Triton X-100. Under these conditions, the release of the heterodimer interaction upon exposure to low pH can be followed by gradient sedimentation or coimmunoprecipitation (45) or by monitoring the increased trypsin sensitivity of monomeric E2 in comparison to that of E2 heterodimers (9). As shown in Fig. 6A, E2 in wt SFV incubated at pH 7.0 is relatively resistant to trypsin digestion at 4°C but becomes markedly more susceptible after dissociation of the heterodimer by treatment at pH 6.0 and below. The digestions of E2 were similar for wt and H3A virus, indicating that the pH dependences and efficiencies of dimer dissociation were comparable.
FIG. 6.
pH dependence of wt and H3A mutant dimer dissociation and E1-membrane interaction. (A) E2/E1 dimer dissociation. Purified 35S-labeled wt or H3A virus was mixed with liposomes, treated at the indicated pHs for 5 min at 20°C, and adjusted to pH 7.0. Samples were solubilized in 1% Triton X-100, digested with trypsin for 10 min on ice, and analyzed by SDS-PAGE. The E2 protein in each sample was quantitated by phosphorimaging and expressed as a percent of the total E2 protein in control samples incubated with premixed trypsin and soybean trypsin inhibitor. Data in panel A are the averages of the results from three independent experiments, and standard deviations are shown as error bars. (B) Virus-liposome association. Purified 35S-labeled wt or H3A mutant SFV was mixed with liposomes, treated at the indicated pHs for 30 s at 37°C, and adjusted to pH 8.0. Virus-liposome association was analyzed by cofloatation assay on discontinuous sucrose gradients and expressed as a percentage of the total virus in the gradient. Data in panel B are the averages of the results from two independent experiments, and the ranges are indicated by error bars.
To monitor E1 membrane insertion, radiolabeled wt and H3A mutant viruses were mixed with liposomes and incubated at the indicated pH, and the liposome-bound virus was separated from unbound virus on sucrose floatation gradients (Fig. 6B). In keeping with the results of previous studies (3, 21), efficient cofloatation of wt virus and target liposomes was observed after incubation at pH 6.0 and below. Cofloatation of the H3A mutant was similar to that of wt SFV. This result confirms the comparable heterodimer dissociations of the wt and the H3A mutant and demonstrates that the membrane insertion of the H3A mutant E1 protein had wt efficiency and pH dependence. Thus, the initial response of H3A mutant virus to low pH was similar to that of wt virus.
H3A mutant homotrimer formation.
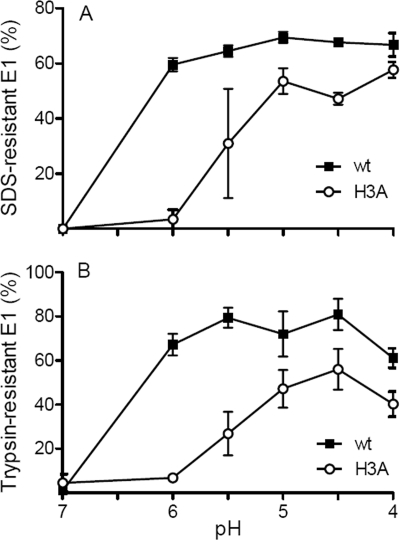
Following the release of E1 from its heterodimeric association with E2 and the insertion of the E1 fusion loop into the target membrane, the virus fusion reaction is driven by the refolding of E1 into the final trimeric hairpin conformation. This postfusion form of E1 is resistant to SDS treatment at 30°C (46), resistant to trypsin digestion at 37°C (11), and reactive with several acid-conformation-specific monoclonal antibodies (1). These assays were used to quantitate E1 homotrimer formation by radiolabeled wt and H3A mutant virus. The wt E1 conformational change was efficiently triggered by low pH, with 60 to 70% of the total wt E1 protein forming SDS-resistant trimers after treatment at pH 6.0 or below (Fig. 7A). By contrast, SDS-resistant trimer formation by the H3A mutant had a pH threshold of 5.5, was maximal only after treatment at pH 5.0, and showed decreased efficiency compared to wt SFV (Fig. 7A). Quantitation of trypsin-resistant E1 trimers showed that 70 to 80% of wt E1 trimerized after treatment at pH 6.0 or below (Fig. 7B). Maximal H3A mutant trimerization was not observed until treatment at pH 4.5, and the final extent of trimerization was ∼50%. Similar reductions in pH threshold and efficiency were observed when H3A mutant reactivity with an acid-conformation-specific monoclonal antibody was compared to that of wt SFV (data not shown).
FIG. 7.
pH dependence of E1 homotrimer formation for wt and H3A mutant SFV. Purified 35S-labeled wt or H3A virus was mixed with liposomes, treated at the indicated pHs for 5 min at 20°C, and adjusted to pH 7.0. (A) Aliquots of the samples were solubilized in SDS sample buffer at 30°C, and the SDS-resistant E1 homotrimer bands were quantitated by SDS-PAGE and phosphorimaging. (B) Aliquots of the samples were digested with trypsin for 10 min at 37°C and the trypsin-resistant E1 homotrimers were quantitated by SDS-PAGE and phosphorimaging. Data shown are averages of the results from two independent experiments, with ranges indicated by error bars (A), or three independent experiments, with standard deviations shown as error bars (B).
DISCUSSION
The available data suggest that low-pH triggering of the alphavirus fusion reaction occurs through several sequential changes in the viral envelope proteins. The first of these is the release of the dimeric interaction of E2 and E1, a step required to free E1 for subsequent conformational changes that mediate virus-membrane fusion (3, 45). Based on biochemical assays, E2/E1 dimer dissociation begins at pH ∼7.0, a somewhat more basic pH threshold than those for E1 conformational changes and membrane fusion (45, 50, 52). E2 is synthesized as a p62 precursor that is processed by furin late in the exocytic pathway (7, 50). The release of the p62/E1 heterodimer interaction has a markedly more acidic pH threshold (pH ∼5.0) than that of E2/E1 (45, 50), and association with p62 is believed to protect E1 from premature fusion during transport to the plasma membrane. In spite of these strong influences of p62/E2 on the pH dependence of alphavirus fusion, however, several findings argue that E1 is also independently triggered by low pH. Even when the dimer interaction is completely eliminated by isolation of E1 ectodomain monomers, these E1 proteins require exposure to low pHs to trigger membrane interaction and trimerization (24). Although the hydrophobic fusion loop is solvent exposed in E1 ectodomains, its stable insertion into target membranes remains low-pH dependent (12, 38). The pH threshold of isolated E1 ectodomains is unaffected by the pH dependence of the original heterodimer (14).
Although considerable evidence supports the concept of direct effects of low pH on the E1 protein, the mechanism of E1's response to low pH is not known. We evaluated the role of conserved E1 histidine residues in this process by substituting alanine for H3, H125, H331, and H333 in the SFV infectious clone. In E1's prefusion conformation, H331 and H333 are located in the interface of DIII with DI, and in the postfusion homotrimer, both residues are on the side of DIII that faces the trimer core. In spite of their location in these interface regions, alanine substitution of H331 and H333 did not affect E1's low-pH-dependent fusion activity. Thus, protonation of H331 and H333 is not required to “release” DIII during E1 refolding at low pHs. In keeping with this result, truncated E1 proteins containing only DI and DII retain a low-pH requirement for membrane insertion and trimerization even though they lack DIII (38). H125 is located within the flexible hinge region between DI and DII. On the virus particle, the H125 residues of E1 proteins at the quasi-twofold axes are within range to be involved in electrostatic repulsion (36). However, our results with the H125A mutant suggest that protonation of H125 does not play a significant role in SFV fusion and infection. Thus, if virus fusion requires the quasi-twofold E1 interactions to be disrupted, other mechanisms must be involved.
In contrast to the other histidine mutants tested here, the SFV E1 H3A mutant showed a significant decrease in virus growth, a markedly more acidic fusion pH threshold, and an overall decrease in fusion efficiency in comparison to the wt. Virus assembly was comparable to that of wt SFV. H3 is located close to the N terminus of E1 on the B0 strand of DI, an E1 region that does not interact with E2 in the heterodimer or in the virus particle (36). In keeping with this location, E2/E1 dimer dissociation and E1-liposome binding showed comparable efficiencies and low-pH dependences in wt and H3A mutant viruses. However, the subsequent formation of the H3A E1 homotrimer had a lower pH threshold and reduced efficiency. Once formed at a low pH, the H3A E1 homotrimer was resistant to protease and SDS, so by these rough criteria the overall stability of the H3A E1 homotrimer was unaffected. We therefore hypothesize that H3A mediates its effects by directly modulating the low-pH-dependent refolding of E1 into the trimer conformation, thus explaining the striking decrease in the H3A mutant's fusion activity.
A recent paper suggested that protonation of histidine residues could serve as a general triggering mechanism for pH-dependent virus membrane fusion proteins (18). This model for triggering proposes that in the prefusion conformation, a critical histidine(s) would be located close to a positively charged residue, possibly interacting via hydrogen bonds. At low pH, histidine becomes positively charged, disrupting any existing hydrogen bonds and leading to electrostatic repulsion. Upon fusion protein refolding, histidine would form a salt bridge with a negatively charged residue, thus stabilizing the postfusion conformation. This interaction could also raise the effective pKa of the histidine, contributing to irreversibility of the conformational change.
Both this model and a number of papers on the structures and functions of low-pH-triggered virus fusion proteins proposed histidines as critical components of the pH sensor. For example, histidine residues have been implicated in the low-pH-dependent fusion reactions of viruses with class I fusion proteins (39, 44) and class III fusion proteins (4, 17, 35). Recent mutagenesis studies of the tick-borne encephalitis (TBE) virus fusion protein have provided important direct evidence for the role of histidine as an initial pH sensor (10). TBE virus is a flavivirus whose class II membrane fusion E protein is structurally highly related to the alphavirus protein E1 (2, 22, 34, 42). Similar to the SFV E1 protein, at low pH, the flavivirus protein E converts to a target membrane-inserted homotrimer in which DIII folds back against the trimer core. Refolding into this trimeric hairpin conformation drives the membrane fusion reaction (28, 43). It is thus quite informative to consider the similarities and differences in the low-pH control of these two fusion proteins.
Similar to alphaviruses, the flavivirus fusion protein E is synthesized together with a chaperone/companion protein, prM, which acts to protect the E protein from low pH during transport to the plasma membrane. prM is processed by furin, but this processing, unlike that of alphavirus p62, requires a low-pH-dependent conformational change to permit furin access (27, 40, 48). The mature flavivirus particle contains E protein homodimers that are organized in a head-to-tail fashion with the fusion loop hidden by interaction with DI and DIII of the dimeric partner (34). As is the case with the alphavirus E2/E1 heterodimer, the flavivirus E homodimer must dissociate to allow fusion loop insertion into the target membrane and subsequent membrane fusion (41). Treatment of TBE virus at pH 10 dissociates the dimer and allows fusion loop insertion into target membranes but without permitting homotrimer formation and fusion (43). It thus appears that TBE virus fusion loop insertion is not itself low-pH dependent. The recent TBE study tested the role of five conserved E protein histidine residues (10). Mutation of H323 on DIII blocked low-pH-triggered dimer dissociation, fusion loop exposure, and fusion, and thus, this residue acts as an initial pH sensor in the TBE fusion pathway. The results also suggest that protonation of H323 is required to release DIII from its association with DI and permit folding back to the mature trimer. This finding could explain the previous block in TBE fusion at pH 10, as under these conditions, the dimer would dissociate and the fusion loop would insert but DIII would not be released from its interaction with DI. In contrast, our data on the H331A/H333A mutants do not support a similar mechanism of, or requirement for, low-pH-triggered release of alphavirus DIII.
Is it possible that, similar to the case in the TBE model, insertion of the alphavirus fusion loop does not itself require a low pH but occurs simply as a consequence of its release from the E2 dimer interaction? Stable membrane insertion of the fusion loop in the monomeric E1 ectodomain is clearly low-pH dependent (12, 24). However, structural studies show that, unlike the situation when E1 is associated with E2 in the virus particle, the fusion loop in isolated E1 appears to fold back to shield hydrophobic groups from solvent (36). Thus, it is difficult to know whether the pH dependence of the E1-membrane interaction is influenced by the conformation of the fusion loop in isolated E1 and whether more-sensitive assays of fusion loop-membrane interaction might reveal membrane insertion at a neutral pH, similar to the TBE results.
Based on their sequence conservation and proximity to charged residues in the prefusion and postfusion structures, the alphavirus E1 residues H125 and H3 were suggested as candidates to control low-pH triggering (18). H125 does not play a major role in alphavirus pH dependence. Our data and those from the studies of flaviviruses thus suggest that inspection of the structures alone may not reliably identify residues critical in pH dependence (10, 32). Our results indicate that H3 is indeed involved in the triggering or refolding of E1 into the postfusion form. Inspection of the SFV E1 prefusion structure did not reveal any positively charged side chains within 5 Å of H3, so initial protonation of H3 at low pH appears unlikely to produce significant electrostatic repulsion. In the postfusion form, H3 is involved in a variety of interactions both within and between E1 molecules. The DI-DIII linker takes on a highly extended conformation to allow the folding back of DIII toward the trimer core, and H3 is positioned to affect linker extension. H3 also appears to mediate intermolecular interactions that could promote trimerization. Further mutagenesis studies based on the trimer structure and careful analysis of H3A revertants will define the relative importance of such H3 interactions during fusion.
Acknowledgments
We thank all of the members of our laboratory for helpful discussions and experimental suggestions, Anuja Ogirala for technical assistance, and Félix Rey and the members of our laboratory for critical comments on the manuscript. Z.Q. thanks her thesis mentor Z.T. Qi for his encouragement.
This work was supported by a grant to M.K. from the National Institute of Allergy and Infectious Diseases (R01 AI075647) and by Cancer Center Core Support Grant NIH/NCI P30-CA13330. Z.Q. was supported in part by a visiting student grant from the China Scholarship Council.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Ahn, A., D. L. Gibbons, and M. Kielian. 2002. The fusion peptide of Semliki Forest virus associates with sterol-rich membrane domains. J. Virol. 763267-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bron, R., J. M. Wahlberg, H. Garoff, and J. Wilschut. 1993. Membrane fusion of Semliki Forest virus in a model system: correlation between fusion kinetics and structural changes in the envelope glycoprotein. EMBO J. 12693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carneiro, F. A., F. Stauffer, C. S. Lima, M. A. Juliano, L. Juliano, and A. T. Da Poian. 2003. Membrane fusion induced by vesicular stomatitis virus depends on histidine protonation. J. Biol. Chem. 27813789-13794. [DOI] [PubMed] [Google Scholar]
- 5.Chanel-Vos, C., and M. Kielian. 2004. A conserved histidine in the ij loop of the Semliki Forest virus E1 protein plays an important role in membrane fusion. J. Virol. 7813543-13552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chanel-Vos, C., and M. Kielian. 2006. Second-site revertants of a Semliki Forest virus fusion-block mutation reveal the dynamics of a class II membrane fusion protein. J. Virol. 806115-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Curtis, I., and K. Simons. 1988. Dissection of Semliki Forest virus glycoprotein delivery from the trans-Golgi network to the cell surface in permeabilized BHK cells. Proc. Natl. Acad. Sci. USA 858052-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLano, W. L. 2002. The PyMOL user's manual. DeLano Scientific, San Carlos, CA.
- 9.Duffus, W. A., P. Levy-Mintz, M. R. Klimjack, and M. Kielian. 1995. Mutations in the putative fusion peptide of Semliki Forest virus affect spike protein oligomerization and virus assembly. J. Virol. 692471-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritz, R., K. Stiasny, and F. X. Heinz. 2008. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J. Cell Biol. 183353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbons, D. L., A. Ahn, P. K. Chatterjee, and M. Kielian. 2000. Formation and characterization of the trimeric form of the fusion protein of Semliki Forest virus. J. Virol. 747772-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbons, D. L., A. Ahn, M. Liao, L. Hammar, R. H. Cheng, and M. Kielian. 2004. Multistep regulation of membrane insertion of the fusion peptide of Semliki Forest virus. J. Virol. 783312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbons, D. L., M.-C. Vaney, A. Roussel, A. Vigouroux, B. Reilly, J. Lepault, M. Kielian, and F. A. Rey. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427320-325. [DOI] [PubMed] [Google Scholar]
- 14.Glomb-Reinmund, S., and M. Kielian. 1998. fus-1, a pH-shift mutant of Semliki Forest virus, acts by altering spike subunit interactions via a mutation in the E2 subunit. J. Virol. 724281-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison, S. C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15690-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helenius, A., M. Marsh, and J. White. 1982. Inhibition of Semliki Forest virus penetration by lysosomotropic weak bases. J. Gen. Virol. 5847-61. [DOI] [PubMed] [Google Scholar]
- 17.Kadlec, J., S. Loureiro, N. G. Abrescia, D. I. Stuart, and I. M. Jones. 2008. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 151024-1030. [DOI] [PubMed] [Google Scholar]
- 18.Kampmann, T., D. S. Mueller, A. E. Mark, P. R. Young, and B. Kobe. 2006. The role of histidine residues in low-pH-mediated viral membrane fusion. Structure 141481-1487. [DOI] [PubMed] [Google Scholar]
- 19.Kielian, M. 2006. Class II virus membrane fusion proteins. Virology 34438-47. [DOI] [PubMed] [Google Scholar]
- 20.Kielian, M., and A. Helenius. 1985. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J. Cell Biol. 1012284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kielian, M., M. R. Klimjack, S. Ghosh, and W. A. Duffus. 1996. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J. Cell Biol. 134863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kielian, M., and F. A. Rey. 2006. Virus membrane fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 467-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kielian, M. C., S. Keränen, L. Kääriäinen, and A. Helenius. 1984. Membrane fusion mutants of Semliki Forest virus. J. Cell Biol. 98139-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klimjack, M. R., S. Jeffrey, and M. Kielian. 1994. Membrane and protein interactions of a soluble form of the Semliki Forest virus fusion protein. J. Virol. 686940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn, R. J. 2007. Togaviridae: the viruses and their replication, p. 1001-1022. In D. M. Knipe (ed.), Fields virology, 5th ed., vol. 1. Lippincott, Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 26.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105137-148. [DOI] [PubMed] [Google Scholar]
- 27.Li, L., S. M. Lok, I. M. Yu, Y. Zhang, R. J. Kuhn, J. Chen, and M. G. Rossmann. 2008. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 3191830-1834. [DOI] [PubMed] [Google Scholar]
- 28.Liao, M., and M. Kielian. 2005. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus-membrane fusion. J. Cell Biol. 171111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao, M., and M. Kielian. 2006. Functions of the stem region of the Semliki Forest virus fusion protein during virus fusion and assembly. J. Virol. 8011362-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liljeström, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 654107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, Y. E., C. H. Eng, S. G. Shome, and M. Kielian. 2001. In vivo generation and characterization of a soluble form of the Semliki Forest virus fusion protein. J. Virol. 758329-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mueller, D. S., T. Kampmann, R. Yennamalli, P. R. Young, B. Kobe, and A. E. Mark. 2008. Histidine protonation and the activation of viral fusion proteins. Biochem. Soc. Trans. 3643-45. [DOI] [PubMed] [Google Scholar]
- 33.Pletnev, S. V., W. Zhang, S. Mukhopadhyay, B. R. Fisher, R. Hernandez, D. T. Brown, T. S. Baker, M. G. Rossmann, and R. J. Kuhn. 2001. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2A resolution. Nature 375291-298. [DOI] [PubMed] [Google Scholar]
- 35.Roche, S., F. A. Rey, Y. Gaudin, and S. Bressanelli. 2007. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 315843-848. [DOI] [PubMed] [Google Scholar]
- 36.Roussel, A., J. Lescar, M.-C. Vaney, G. Wengler, G. Wengler, and F. A. Rey. 2006. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure 1475-86. [DOI] [PubMed] [Google Scholar]
- 37.Salminen, A., J. M. Wahlberg, M. Lobigs, P. Liljeström, and H. Garoff. 1992. Membrane fusion process of Semliki Forest virus II: cleavage-dependent reorganization of the spike protein complex controls virus entry. J. Cell Biol. 116349-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez San Martin, C., H. Sosa, and M. Kielian. 2008. A stable prefusion intermediate of the alphavirus fusion protein reveals critical features of class II membrane fusion. Cell Host Microbe 4600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schowalter, R. M., A. Chang, J. G. Robach, U. J. Buchholz, and R. E. Dutch. 26 November 2008. Low pH triggering of human metapneumovirus fusion: essential residues and importance in entry. J. Virol. doi: 10.1128/JVI.01381-08. [DOI] [PMC free article] [PubMed]
- 40.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 718475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stiasny, K., S. L. Allison, J. Schalich, and F. X. Heinz. 2002. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 763784-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stiasny, K., and F. X. Heinz. 2006. Flavivirus membrane fusion. J. Gen. Virol. 872755-2766. [DOI] [PubMed] [Google Scholar]
- 43.Stiasny, K., C. Kossl, J. Lepault, F. A. Rey, and F. X. Heinz. 2007. Characterization of a structural intermediate of flavivirus membrane fusion. PLoS Pathog. 3e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thoennes, S., Z. N. Li, B. J. Lee, W. A. Langley, J. J. Skehel, R. J. Russell, and D. A. Steinhauer. 2008. Analysis of residues near the fusion peptide in the influenza hemagglutinin structure for roles in triggering membrane fusion. Virology 370403-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahlberg, J. M., W. A. M. Boere, and H. Garoff. 1989. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J. Virol. 634991-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wahlberg, J. M., and H. Garoff. 1992. Membrane fusion process of Semliki Forest virus I: low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J. Cell Biol. 116339-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White, J. M., S. E. Delos, M. Brecher, and K. Schornberg. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43189-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, I. M., W. Zhang, H. A. Holdaway, L. Li, V. A. Kostyuchenko, P. R. Chipman, R. J. Kuhn, M. G. Rossmann, and J. Chen. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 3191834-1837. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, W., S. Mukhopadhyay, S. V. Pletnev, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2002. Placement of the structural proteins in Sindbis virus. J. Virol. 7611645-11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, X., M. Fugere, R. Day, and M. Kielian. 2003. Furin processing and proteolytic activation of Semliki Forest virus. J. Virol. 772981-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, X., and M. Kielian. 2005. An interaction site of the envelope proteins of Semliki Forest virus that is preserved after proteolytic activation. Virology 337344-352. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, X., and M. Kielian. 2004. Mutations that promote furin-independent growth of Semliki Forest virus affect p62-E1 interactions and membrane fusion. Virology 327287-296. [DOI] [PubMed] [Google Scholar]