Abstract
Adenovirus infection induces a cellular DNA damage response that can inhibit viral DNA replication and ligate viral genomes into concatemers. It is not clear if the input virus is sufficient to trigger this response or if viral DNA replication is required. Adenovirus has evolved two mechanisms that target the Mre11-Rad50-Nbs1 (MRN) complex to inhibit the DNA damage response. These include E4-ORF3-dependent relocalization of MRN proteins and E4-ORF6/E1B-55K-dependent degradation of MRN components. The literature suggests that degradation of the MRN complex due to E4-ORF6/E1B-55K does not occur until after viral DNA replication has begun. We show that, by the time viral DNA accumulates, the MRN complex is inactivated by either of the E4-induced mechanisms and that, with E4-ORF6/E1B-55K, this inactivation is due to MRN degradation. Our data are consistent with the conclusion that input viral DNA is sufficient to induce the DNA damage response. Further, we demonstrate that when the DNA damage response is active in E4 mutant virus infections, the covalently attached terminal protein is not cleaved from viral DNAs, and the viral origins of replication are not detectably degraded at a time corresponding to the onset of viral replication. The sequences of concatemeric junctions of viral DNAs were determined, which supports the conclusion that nonhomologous end joining mediates viral DNA ligation. Large deletions were found at these junctions, demonstrating nucleolytic procession of the viral DNA; however, the lack of terminal protein cleavage and terminus degradation at earlier times shows that viral genome deletion and concatenation are late effects.
Adenovirus (Ad) has a linear, double-stranded DNA genome with inverted terminal repeats (ITRs) at each extremity that contain the origins of replication. Several viral proteins have been found to be key components in supporting viral DNA replication, and of particular importance here, this includes the E4-ORF3 and E4-ORF6 products. With mutant viruses that lack E4-ORF3 and E4-ORF6, early viral transcription and gene expression are normal; however, there is a significant delay and reduction in viral DNA replication, and virus yield is reduced by ∼1,000-fold (20, 49). Either the E4-ORF3 or E4-ORF6 protein is individually sufficient to complement the DNA replication defect and virus growth (7, 24); thus, these proteins are considered functionally redundant.
An important function of these two Ad E4 proteins is the inhibition of the Mre11-Rad50-Nbs1 (MRN) complex (30, 50). The MRN complex is involved in double-strand break repair (DSBR) and is regarded as the sensor of double-strand breaks (DSBs) (9, 12, 46). When a DSB occurs in the cellular genome due to a multitude of causes, ranging from ionizing radiation (33) to VDJ recombination (10), the MRN complex recognizes the lesion and recruits the protein kinases ataxia-telangiectasia mutated (ATM) and ATM-Rad3 related (ATR) to the site of the break to initiate the process of nonhomologous end joining (NHEJ) (46). These two kinases are central players in activation of the cellular DNA damage response. Through intermolecular autophosphorylation, ATM is phosphorylated on S1981, resulting in the dissociation of dimers into monomers and enzymatic activation (3). ATM subsequently phosphorylates downstream effectors involved in checkpoint signaling, such as Nbs1 (17, 31) and H2AX (8, 41), and recruits other proteins involved in repairing the lesion. Large foci form at the site of the DNA break due to the accumulation of γH2AX, the phosphorylated form of the histone variant H2AX. γH2AX recruits, among other proteins, Mdc1 (mediator of DNA damage checkpoint 1), which serves as a bridge to sustain protein-protein interactions at the DNA lesion (45).
Mre11 has both single-stranded endonuclease and 3′-5′ exonuclease activity and can process the ends of the DNA lesion to yield regions of microhomology that are between 1 and 5 nucleotides (nt) in length (38). Rad50 is proposed to be involved in holding the two ends of DNA together by dimerization through the coiled-coil domains (36). Rad50 has ATPase activity that is important for regulating DNA binding and Mre11 nuclease activity (5, 13, 23). Nbs1 is important in directing the localization of the MRN complex. In cells that lack Nbs1, which contains a nuclear localization signal, Mre11 and Rad50 remain cytoplasmic (14). Also, the forkhead-associated and BRCA1 C-terminal domains of Nbs1 are involved in binding to γH2AX and retaining the MRN complex at the site of the lesion (27). DNA-PK and DNA ligase IV/XRCC4 are involved in ligating the DNA ends together to repair the DSB (2).
The role of the MRN complex in NHEJ is relevant to an Ad infection due to the fact that MRN will perceive the linear, double-stranded Ad DNA genomes as DSBs. Following infection with a mutant virus that lacks both E4-ORF3 and E4-ORF6, ATM is phosphorylated, checkpoint signaling occurs, and the genomes are eventually ligated together to form large concatemers (reviewed in references 30 and 50). These concatemers are too large to be packaged into virus particles and would be inefficiently replicated due to the fact that the internal genomes lack a free terminus. The junctions of the concatemers also have deletions (48), suggesting that the ends of the Ad genome are degraded. Since the origins of replication are located at the termini of the viral genome, this would inhibit viral DNA replication.
For this reason, Ad has evolved several mechanisms to counteract the detrimental effects of NHEJ, two of which specifically target the MRN complex. The E4-ORF3 protein of group C Ad causes the redistribution of the MRN complex from the nucleoplasm to the E4-ORF3-dependent PML tracks (16, 43, 44). This effectively sequesters the MRN complex away from the viral genomes, which are located at the replication centers and do not colocalize with redistributed MRN proteins (16, 44). The E4-ORF6 protein interacts with another viral protein, E1B-55K, to form an E3 ubiquitin ligase complex with the cellular proteins Rbx1, Cullin 5 (CUL5), and elongins B and C (21, 39). This complex targets specific proteins, such as p53, Mre11, and DNA ligase IV, for proteasome-dependent degradation (2, 21, 39). E1B-55K and E4-ORF3 also are involved in relocalizing Mre11 to cytoplasmic aggresomes (1, 32). With either a virus that lacks E4-ORF6 or E4-ORF3 alone, checkpoint signaling and concatemer formation are inhibited, but the deletion of both of these Ad gene products results in checkpoint signaling induction, concatemer formation, and a significant decrease of viral DNA replication (30, 50).
It is not clear if the input Ad genome is sufficient to trigger the DSBR response or if viral DNA replication is required. The E4-ORF3 protein causes the redistribution of the MRN complex by 6 h postinfection (hpi), before viral DNA replication begins (16). The sequestration of MRN by E4-ORF3 is associated with inhibition of the DSBR response. However, in the absence of the E4-ORF3 protein, when MRN activity is solely inhibited by E4-ORF6/E1B-55K-induced degradation, MRN protein levels are not detectably reduced until later after infection (32, 43). Here, we show that, when looking at individual cells, the MRN complex is effectively inactivated by either E4-induced mechanism by the time Ad DNA replication is evident and that the degradation of the MRN complex by E4-ORF6 and E1B-55K occurs prior to viral DNA accumulation.
Ad terminal protein (TP) is covalently linked to the 5′ end of the viral genome and is involved in priming the viral DNA replication reaction. It is possible that TP can prevent the recognition of the viral genome as a DSB during the early phase of infection. It is thought that the nucleolytic activity of Mre11 may be involved in cleaving TP from the genome, as it has a similar function in cleaving Spo11 off the Saccharomyces cerevisiae meiotic-specific DSBs (35). This led to the speculation that it is the cleavage of TP by Mre11 that allows for full recognition of the Ad termini as DSBs (43, 50). Here, we show that Ad genomes show no signs of TP cleavage or significant degradation of the Ad genomic termini at the onset of viral DNA replication in cells infected with mutant viruses that lack E4-ORF3 and E4-ORF6 activities. However, later during such an infection, large deletions were found at the junctions of Ad concatemers. The concatemeric junctions had variably sized deletions, with little to no sequence homology, supporting the role of NHEJ in Ad genome end joining. These results uncouple the role of the MRN complex in the inhibition of Ad DNA replication from the ligation of viral genomes into concatemers.
MATERIALS AND METHODS
Cells, viruses, and infections.
A549 cells and ATCC HeLa cells were grown in Dulbecco's modified Eagle medium supplemented with 10% calf serum at 37°C in 5% CO2. The viruses used, which were purified on CsCl equilibrium gradients, are dl309 (phenotypically wild-type Ad5) (26, 35), dl355 (E4-ORF6 mutant) (24), inORF3 (E4-ORF3 mutant) (24), dl1520 (E1B-55K mutant) (4), dl355/inORF3 (E4-ORF6 and E4-ORF3 double mutant) (24), dl1520/dl341 (E1B-55K and E4-ORF3 double mutant) (42), dl366 (E4 deletion mutant) (20), and dl808 (E4 deletion mutant) (49). Infections were performed at 200 particles per cell for 1 h, followed by replacement with media and incubation at 37°C in 5% CO2.
Viral DNA replication assay.
A549 cells were infected at various time points, and lysates were subsequently prepared by resuspending infected cells in isotonic buffer (150 mM NaCl, 10 mM Tris [pH 7.4], 1.5 mM MgCl2) containing 0.6% NP-40. The samples were incubated on ice for 10 min, followed by centrifugation at 800 × g for 5 min to pellet nuclei. Nuclei were then resuspended in sodium dodecyl sulfate (SDS) lysis buffer (0.5% SDS, 200 mM Tris [pH 8.0], 50 mM EDTA, 0.1 mg/ml proteinase K) and incubated at 55°C overnight. The samples were subsequently subjected to phenol-chloroform extraction, and the DNA was ethanol precipitated and resuspended in TE (10 mM Tris [pH 8.0], 1 mM EDTA) overnight at 4°C. Samples were standardized by measuring absorbance at 260 nm and diluted as follows in TE: at 4 to 12 hpi, samples were diluted at 1:10; at 15 hpi, 1:100; at 18 hpi, 1:200; and at 24 hpi, 1:300. The DNA was denatured in 0.3 N NaOH for 1 h at 65°C and then neutralized in 2 M ammonium acetate. The samples were then applied to a Hybond-N+ membrane (Amersham) with a slot blot apparatus. The DNA was cross-linked using a Stratalinker (Stratagene) and subjected to Southern hybridization with a 32P-labeled probe made by random priming with the whole Ad5 genome (15). The DNA was visualized on an ABI Storm 860 Phosphorimager and quantified using ImageQuant 1.1 software (Molecular Dynamics).
Immunoblots.
Whole-cell extracts were prepared by resuspending infected cells in SDS lysis buffer (1.2% SDS, 150 mM Tris [pH 6.8], 30% glycerol) and boiling them for 10 min. The samples were centrifuged at 16,100 × g for 30 min, supernatants were collected, and the total protein concentration was determined using the Pierce bicinchoninic acid method. Standardized amounts of protein were subjected to 12.5% SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and blocked in 1% casein in Tris-buffered saline. Each membrane was probed with an anti-γ-tubulin rabbit polyclonal antibody (Sigma) at a dilution of 1:5,000 and one of the following antibodies at 1:1,000: anti-Mre11 mouse monoclonal (Genetex), anti-Rad50 mouse monoclonal (Novus), or anti-Nbs1 mouse monoclonal (Genetex). The secondary antibodies IR680 goat anti-mouse and IR800 goat anti-rabbit (Li-Cor) were used, and the proteins were detected and quantified using the Odyssey infrared imaging system (Li-Cor).
Immunofluorescence.
A549 and HeLa cells were seeded on glass coverslips and transfected according to the manufacturer's instructions using FuGene 6 (Roche) and/or infected as described above. In some cases, the cells were then exposed to 1 Gy of ionizing radiation from Cs at 10 hpi. At the times indicated above, the cells were washed with phosphate-buffered saline (PBS), fixed with −20°C methanol for 5 min, and washed again with PBS. The cells were blocked with 10% goat serum diluted in PBS and then incubated with primary antibodies diluted in 10% goat serum. The antibodies used were anti-DNA binding protein (DBP) mouse monoclonal (40) at a dilution of 1:50, anti-Mre11 rabbit polyclonal (Novus) at 1:600, anti-Rad50 rabbit polyclonal (Novus) at 1:600, anti-Nbs1 rabbit polyclonal (Novus) at 1:1,200, and anti-phospho-ATM (pATM) S1981 rabbit polyclonal (Rockland) at 1:300. The cells were washed and incubated with Alexa350-conjugated goat anti-mouse immunoglobulin G (Molecular Probes) and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin G (Zymed). The cells were washed a final time and mounted on slides with Shandon Immu-Mount (Thermo Scientific). The microscope used was a Zeiss Axiovert 200 M digital deconvolution microscope fitted with a Chroma filter set and an ApoTome system, and images were captured with a Peltier-cooled charge-coupled-device AxioCam HRm camera and analyzed with AxioVision 4.5 software.
TP cleavage assay.
A549 cells were infected, and nuclei were isolated using isotonic buffer containing NP-40, as described above. Viral DNA was then isolated using the procedure of Hirt (22); however, only half of the sample was treated with proteinase K. DNA was purified on a Qiagen PCR MinElute column, digested with BglII, separated on a 1% agarose gel, and analyzed by Southern hybridization with a 32P-labeled Ad5 total genome probe made by random primer labeling (15).
Isolating individual concatemer junctions.
A549 cells were infected with dl366 or dl808 at a multiplicity of infection of 10 infectious units/cell using crude virus lysates. After 4 days, intracellular DNA was isolated using a variation of the Hirt extraction procedure (47). Approximately 1 μg of the DNA was digested with ClaI and BglII. These enzymes have recognition sites close to the ends of the Ad5 genome at nt 917 (ClaI), nt 30616, and nt 3328 (BglII). Concatemers formed between genomes oriented in a left-end to right-end fashion, and where deletions did not extend beyond the enzyme sites, generated fragments containing ClaI and BglII termini. Genomes oriented in a left-end to left-end fashion with deletions extending beyond the ClaI site at nt 914 at one terminus also generated fragments with ClaI and BglII termini. Right-end to right-end genomes will not be sampled using this strategy. The digested DNA was then ligated to DNA of a plasmid derived from pSL180, which had been digested with ClaI and BamHI and transformed into the SURE strain of Escherichia coli (Stratagene). Resulting colonies were replica plated to two nitrocellulose filters, and the DNA was prepared for hybridization with two separate probes specific for either the left-hand end or the right-hand end of the Ad5 genome. Colonies whose DNA tested positive for both probes were amplified, DNA was extracted, and the DNA was sequenced. Note that three clones isolated using this procedure, which should identify left-end to right-end junctions, proved to have left-end to left-end junctions and, thus, were incorrectly classified in the original screen.
RESULTS
Time course of MRN degradation compared to that of viral DNA replication during Ad infection.
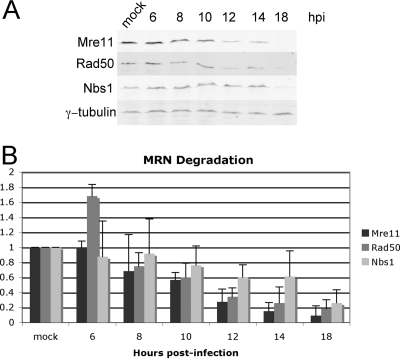
In the literature, the earliest decrease in the levels of the MRN complex during an Ad infection is at 12 to 16 hpi (32, 43). To examine this in A549 cells, a time course was performed in which cell lysates were prepared from mock-infected cells and inORF3-infected cells at 6, 8, 10, 12, 14, and 18 hpi, and MRN levels were measured by quantitative Western blot analysis using antibodies against Mre11, Rad50, and Nbs1. γ-Tubulin levels were measured to normalize each sample (Fig. 1A). Mutant inORF3 (lacking E4-ORF3) was used for this analysis so that only the E4-ORF6/E1B-55K complex contributes to MRN regulation. At 6 and 8 hpi, the levels of the components of the MRN complex were similar to or slightly greater than the levels of mock-infected cells (Fig. 1B). By 10 and 12 hpi, however, the Mre11 and Rad50 signals were diminished by about half. Significant degradation of these proteins was observed by 18 hpi. As previously described, Nbs1 protein levels decreased at a reduced rate (32, 43).
FIG. 1.
MRN degradation does not occur until late during Ad infection. Cell lysates from mock-infected A549 cells and mutant inORF3-infected (E4-ORF3 mutant) A549 cells were harvested at 6, 8, 10, 12, 14, and 18 hpi. (A) MRN degradation was visualized by immunoblot analysis of each component of the MRN complex, along with γ-tubulin as a loading control. (B) Each band was quantified and normalized to γ-tubulin. The values for mock-infected samples are set at 1. Relative levels of Mre11, Rad50, and Nbs1 in infected cell lysates are shown at the indicated time points. The results represent the average of three independent experiments.
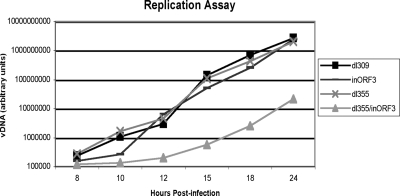
Since it is not known if it is important for the MRN complex to be inhibited prior to the onset of viral DNA accumulation, we wished to determine an accurate time point for the beginning of viral DNA replication in Ad-infected A549 cells. We analyzed viral DNA levels during a time course from cells infected with phenotypically wild-type Ad5 (dl309) and mutant viruses deleted for E4-ORF6, E4-ORF3, or both proteins (dl355, inORF3, or dl355/inORF3, respectively) starting at 4 hpi and until 24 hpi (Fig. 2). Wild-type and dl355 viral DNA began to accumulate starting at 10 hpi. Mutant inORF3 grew with similar kinetics but showed a slight lag at the onset of replication. The double mutant dl355/inORF3, which has a viral DNA replication defect (24), was significantly delayed for viral DNA replication. We conclude that significant levels of degradation for the MRN complex do not occur prior to the accumulation of viral DNA when looking at a population of infected cells.
FIG. 2.
The onset of viral DNA replication occurs at 10 to 12 hpi with viruses that inactivate MRN. Cells were infected with dl309 (phenotypically wild-type Ad5), dl355 (E4-ORF6 mutant), inORF3, and dl355/inORF3 (E4-ORF6/E4-ORF3 mutant). High-molecular-weight DNA was prepared from infected cell lysates at 4, 6, 8, 10, 12, 15, 18, and 24 hpi. Levels of DNA in each sample were quantified by slot blot analysis and plotted on the graph. Equivalent levels of viral DNA (vDNA) were observed with the 4 and 6 hpi samples compared to the level of the 8 hpi sample. The results represent the average of three independent experiments.
The MRN complex is functionally inhibited by E4-ORF3 and E4-ORF6/E1B-55K at the onset of replication.
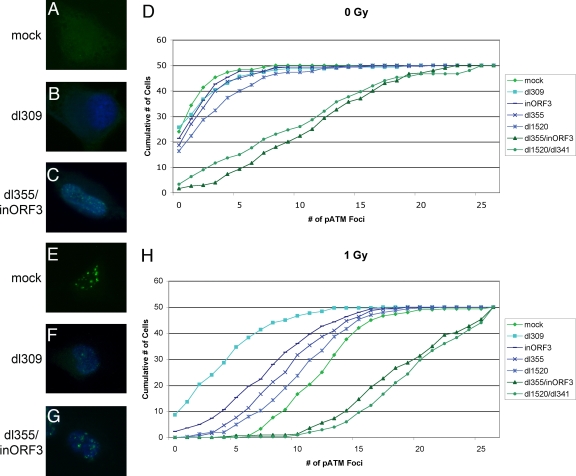
To examine MRN degradation on a single-cell basis in Ad-infected A549 cells, we used checkpoint signaling, as seen by pATM focus formation, as an indication of MRN function. MRN activity is required for the induction of pATM foci following the induction of DNA damage (29). We costained Ad-infected cells at 10 hpi for the Ad DBP and ATM phosphorylated at S1981 (pATM) and counted the number of pATM-S1981 foci that formed in DBP-positive cells (Fig. 3). DBP is an early viral protein and served as a marker for infected cells.
FIG. 3.
The MRN complex is inactivated by 10 hpi in wild-type and single-mutant infections. A549 cells were mock infected (A, E) or infected with dl309 (B, F); dl355, inORF3, dl1520 (E1B-55K mutant), and dl355/inORF3 (C, G); or dl1520/dl341 (E1B-55K/E4-ORF3 mutant) for 10 h. The cells were then fixed (A to D) or treated with 1 Gy of ionizing radiation and fixed 1 h postirradiation (E to H). The cells were stained with antibodies against DBP and pATM-S1981 in Alexa Fluor 350 (blue) or FITC (green), respectively. (A to C, E, and F) Representative immunofluorescence images. (D, H) The number of pATM foci were counted in DBP-expressing cells for 50 cells infected with each virus in triplicate, and the average number was plotted in the graphs. The results represent the cumulative value of pATM foci observed: i.e., 0 represents the number of cells with 0 pATM foci, 1 represents the number of cells with 0 and 1 pATM foci, 2 represents the number of cells with 0, 1 and 2 pATM foci, etc.
With mock-infected cells, very few pATM foci were evident (Fig. 3A and D). Similarly, in cells infected with dl309, dl355, inORF3, or dl1520 (an E1B-55K mutant), very few pATM foci were observed, suggesting that there was significant protection of the viral genomes from detection by the MRN complex and subsequent checkpoint signaling by 10 hpi (Fig. 3B and D). In contrast, the two double mutants, dl355/inORF3 and dl1520/dl341 (an E1B-55K E4-ORF3 double mutant), had a significant number of pATM foci, with an average of ∼10 per cell (Fig. 3C and D). Taken together, these results show that when either E4-ORF3 or the combination of E1B-55K and E4-ORF6 is present, they functionally inhibit the MRN complex and impair checkpoint signaling elicited by the recognition of viral genomes. The increase in pATM foci observed in cells infected with the replication-defective double mutants compared to the amount of those observed in mock-infected cells strongly indicates that the input viral genomes from the initial infection are sufficient to induce MRN signaling.
Infected cells were also irradiated with 1 Gy of ionizing radiation at 10 hpi, fixed at 1 h postirradiation, and stained for DBP and pATM. In mock-infected, irradiated cells, there was an average of ∼12 pATM foci per cell (Fig. 3E and H). Wild-type virus (dl309) was able to strongly inhibit checkpoint signaling with only ∼3 foci per cell, even after exogenously introduced DSBs were formed (Fig. 3F and H). Each of the single mutants (dl355, inORF3, and dl1520) was able to prevent a full checkpoint signaling response with ∼7 to 10 foci per cell (Fig. 3H). Both double mutants (dl355/inORF3 and dl1520/dl341) were unable to inhibit checkpoint signaling, and ∼18 pATM foci formed per cell (Fig. 3G and H). These results demonstrate that by 10 hpi, checkpoint signaling is inhibited not only for recognition of viral genomes in wild-type and single-mutant infections but also for DSBs in cellular DNA induced by ionizing radiation. Once again, the comparison of the results using the mock versus double mutant virus infections supports the conclusion that the input viral genomes are sufficient to induce a DSBR response.
Degradation of the MRN complex occurs before the accumulation of viral DNA.
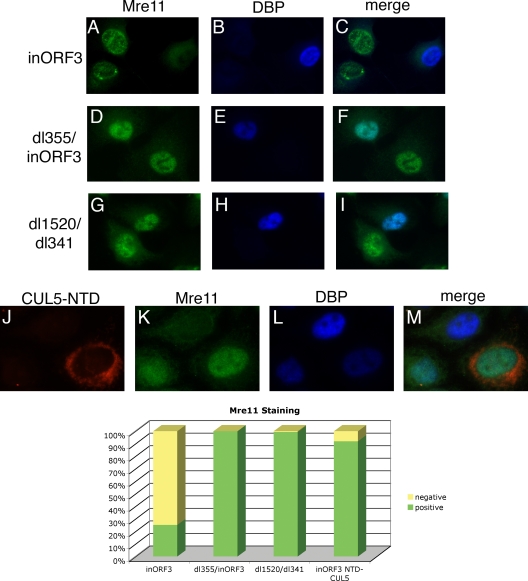
Immunofluorescence experiments were also performed to visualize the localization of the MRN complex in infected cells. An E4 mutant was used that does not express a functional E4-ORF3 protein to prevent MRN relocalization and functional inactivation in order to directly assess MRN regulation by E4-ORF6/E1B-55K. Cells were infected with mutant inORF3 and then immunostained at 8 hpi with antibodies against DBP and Mre11, Rad50, or Nbs1. In about 75% of DBP-positive cells at 8 hpi, the signal for each of the components of the MRN complex disappeared (Fig. 4A to C and bottom and data not shown). The disappearance of the signal was dependent on both E1B-55K and E4-ORF6, as seen by a return of the signal in nearly 100% of cells infected with viruses that lacked E4-ORF6 and E4-ORF3 (dl355/inORF3) or E1B-55K and E4-ORF3 (dl1520/dl341) (Fig. 4D to I and bottom and data not shown). Since the loss of the MRN signal was observed at 8 hpi, whereas viral DNA replication in mutant inORF3-infected cells was evident only between 10 and 12 hpi (Fig. 2), we conclude that MRN is degraded in Ad-infected cells prior to viral DNA accumulation.
FIG. 4.
Immunofluorescence shows a disappearance of Mre11 signal by 8 hpi that is due to degradation by E4-ORF6 and E1B-55K. A549 cells were infected with inORF3 (A to C), dl355/inORF3 (D to F), or dl1520/dl341 (G to I) or cotransfected with CUL5-NTD and DsRed2-Mito expression plasmids overnight and infected with inORF3 (J to M) for 8 h. The cells were then stained with antibodies for Mre11 (green, FITC) and DBP (blue, Alexa Fluor 350). (Bottom) Cells were scored positive or negative for Mre11 for staining, and the percentages of 50 cells in triplicate were plotted on the graph.
If the disappearance of the MRN signal in the immunofluorescence assays was in fact due to degradation, there appeared to be a discrepancy in the timing of the degradation during Ad infection when comparing the results of biochemical methods (Fig. 1) with those of immunofluorescence assays (Fig. 3). One difference between these two methodologies is that the biochemical assay looks at a population of cells, while the immunofluorescence is a single-cell-based assay. During an infection, different cells may progress through the viral life cycle at different rates, and this may cause a difference in results when looking at time-sensitive data for a population rather than data for a single cell. To determine if this was the reason for the apparent discrepancy, a single-cell assay was implemented to visualize MRN degradation.
We utilized a dominant-negative CUL5 mutant (51) to determine if the disappearance of the MRN signal using immunofluorescence was due to proteasomal-dependent degradation. CUL5 is a component of the E3 ubiquitin ligase complex that E4-ORF6 and E1B-55K target. Woo and Berk showed that the N-terminal domain (NTD) of CUL5 can prevent the degradation of Mre11 during Ad infection (51). Cells were cotransfected with the CUL5-NTD expression plasmid and a DsRed-Mito expression plasmid, as a marker of transfected cells. Subsequently, the cells were infected with mutant inORF3 and immunostained at 8 hpi with antibodies against DBP and Mre11. In 91% of cells that had been transfected (DsRed-Mito positive) and were DBP positive, the Mre11 signal was restored (Fig. 4J to M and bottom), indicating that the loss of signal seen during mutant inORF3 infection is due to the degradation of Mre11 in these cells.
Terminal protein cleavage is not the basis for MRN inhibition of Ad replication.
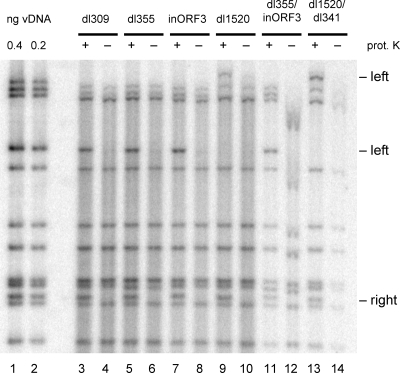
In order to visualize the nucleolytic processing that is thought to occur prior to Ad genome concatenation, an assay was utilized to determine if TP is cleaved off the viral genome by 10 hpi during an infection with viral mutants that are unable to inhibit the MRN complex. We sought to determine if endonucleolytic cleavage of the genome removed TP. Nuclear DNA was analyzed from virus-infected cells so as to exclude the viral genomes that entered the cell but failed to travel to the nucleus. Half of these extracts were treated with proteinase K, the other half were not treated, and all samples were digested with the restriction enzyme BglII. The samples were separated on an agarose gel and subjected to Southern blot analysis with a 32P-radiolabeled Ad5 whole-genome probe. The terminal fragments that were treated with proteinase K will not have TP attached and will run at 3,327 bp and 1,543 bp for the left and right termini, respectively, with the exception of the left-end fragments for the dl1520 mutants. The deletion made in this mutant removed the BglII site and created a left-end terminal fragment of ∼8 kb. For the samples that were not treated with proteinase K, TP may be covalently linked and will cause the terminal fragments to remain in the well of the gel. By comparing the quantity of terminal fragments in each sample, the percentage of genomes with TP attached can be determined. As expected, there was a significant decrease in the intensity of the terminal bands in the non-proteinase K-treated samples for the wild type (dl309) and the single mutants (dl355, inORF3, dl1520), indicating that TP was attached (Fig. 5). Similarly, there was a decrease in the non-proteinase K-treated samples with the double mutants (dl355/inORF3 and dl1520/dl341), confirming that TP is still covalently bound to the viral genome at 10 hpi, even in the presence of a functional MRN complex. Additionally, with all of the proteinase K-treated samples, the terminal DNA fragments displayed the same apparent mobility in the gel, even in the presence of functional MRN. We conclude that neither TP cleavage nor significant nucleolytic cleavage of the viral termini is the basis for MRN inhibition of Ad DNA replication.
FIG. 5.
TP cleavage is not the basis for MRN inhibition of Ad replication. A549 cells were infected with dl309 (lanes 3 and 4), dl355 (lanes 5 and 6), inORF3 (lanes 7 and 8), dl1520 (lanes 9 and 10), dl355/inORF3 (lanes 11 and 12), and dl1520/dl341 (lanes 13 and 14) and harvested at 10 hpi to prepare nuclear viral DNA (vDNA). Part of the samples were treated with proteinase K (prot. K; odd lanes, +), while the others were left untreated (even lanes, −). The samples were digested with BglII, run on a 1% agarose gel, and subjected to Southern blot analysis with a 32P-labeled whole-genome probe. Standards were run alongside the samples (lanes 1 and 2).
Sequence analysis of individual concatemer junctions reveals large deletions.
Previous work (48) showed that the concatemers produced during an infection of human KB cells with the E4 deletion mutant dl366 or dl808 were joined in any of the three possible orientations (left end to right end, left end to left end, and right end to right end). Furthermore, the junctions between monomers were characterized by a disperse set of terminal deletions, in that no specific junctional bands could be detected when intracellular viral DNA was analyzed with multiple-cutting restriction enzymes. The details of the mechanism by which concatemers arise are still not understood, although the presumption is that the absence of the E4-ORF3 and E4-ORF6 proteins allows the MRN complex to promote end joining of viral genomes. In systems where artificial breaks are induced in either plasmid DNA (6) or viral DNA (37), either of the E4 proteins can interfere with end joining.
Analysis of a set of individual concatemer junctions may give insight into the mechanism by which it arises. Accordingly, a cloning strategy was designed to investigate a subset of the junctions oriented in the right-end to left-end or left-end to left-end configuration. Table 1 shows the data for a set of 11 junctions, with the orientation and end points of the deletions presented, and one junction isolated and sequenced by Weiden and Ginsberg (48) using a different cloning strategy. Although this is a limited set, several conclusions can be drawn. As expected, the terminal deletions at the junction were heterogeneous in size, ranging from 528 bp to 5,220 bp. No specific sequence was favored at the junction site, and there was no evidence for the use of microhomologies at or near the site. It is notable that no junction contained any part of either ITR.
TABLE 1.
Sequence analysis of concatemeric junctions
| Clone no. | Orientationa | Leftward genome sequenceb | Common nucleotide(s)c | Rightward genome sequenced | No. of deletions (bp)e |
|---|---|---|---|---|---|
| 1 | R to L | ...CGAAAGCC31585 | AA | T317TTTGTGT... | 528 |
| 2 | R to L | ...ATAGGACT31507 | A | C500TCTTGAG... | 790 |
| 3 | R to L | ...GAAAACCT31420 | C666CATTTTG... | 1,044 | |
| 4 | R to L | ...GCACCAGC31452 | T | C728CCAACGA... | 1,074 |
| 5 | R to L | ...AAGGGCCA31481 | T797TACTCAC... | 1,114 | |
| 6 | R to L | ...ACCTATTA31424 | G814GCGCCCG... | 1,188 | |
| 7 | R to L | ...TAGGAGAG31294 | C718CCCGAAG... | 1,222 | |
| 8 | R to L | ...ATATTCTT31153 | A | C796TTACTCA... | 1,440 |
| 9 | L to L | ...GCCCACGG1683 | T247GGGCGTA... | 1,928 | |
| 10 | L to L | ...GCTCATTA2444 | A154CGGATGT... | 2,596 | |
| 11 | L to L | ...CCCGTCCA2710 | T | C758CCGACTC... | 3,465 |
| 12f | R to L | ...TGACACTA29029 | AC | T328CATAGCG... | 5,221 |
R to L, the data are presented so that the conventional right-hand end of the leftward genome is joined to the left-hand end of the rightward genome. L to L, the left-hand ends of both genomes joined. Note that these clones, because of the cloning strategy adopted, would have to be derived from junctions in which the ClaI site in the leftward genome had already been deleted.
Sequences are presented 5′ to 3′, ending with the last identifiable nucleotide from the leftward genome. The subscripted number at the end of the sequence represents the base pair position for the last nucleotide. Numbering is based on the last identifiable nucleotide for dl366 (total of 31,799 bp), with the exception of clone 12, which is based on dl808 (33,925 bp). The upstream BglII site used in cloning the right junctions (except clone 12) was at bp 30616. For the L to L junctions, the BglII site was at bp 3328.
These nucleotides are common to both the leftward and rightward genome sequences and, therefore, cannot be assigned to either genome.
Sequences are presented 5′ to 3′, beginning with the first identifiable nucleotide from the rightward genome. The subscripted number after the first nucleotide of the sequence represents the base pair position for the first nucleotide. All sequence numbers are based on dl366. The ClaI site used in cloning was at bp 917.
Calculated assuming that a hypothetical junction could form and retain all nucleotides from both genomes.
The flanking HindIII sites used in cloning were at bp 28957 and bp 2798.
DISCUSSION
The importance of the inhibition of the MRN complex during an Ad infection is illustrated by the redundant mechanisms utilized to inactivate the complex and is seen experimentally by a delay in the onset of virus replication and a significant decrease in viral DNA levels and virus yield with mutants that lack the ability to inhibit the MRN complex (7, 20, 24, 49). The E4-ORF3 protein is responsible for one mechanism, in which the MRN complex is sequestered into nuclear inclusions containing PML by 6 hpi (16, 43, 44). The E4-ORF6 and E1B-55K proteins form an E3 ubiquitin ligase complex that targets Mre11 for proteasome-dependent degradation (1, 21, 32, 39, 43). Since viral DNA replication is inhibited if MRN is not inactivated, one would anticipate that inhibition of the MRN complex would need to occur prior to the onset of viral DNA replication for both E4-induced mechanisms of MRN inactivation. This prediction holds true for E4-ORF3, but it was not apparent if this applied to MRN degradation induced by E4-ORF6/E1B-55K, since significant levels of MRN were evident at times of viral DNA replication (32, 43). If this did not take place, it either would suggest a new mechanism of MRN inhibition or may lead to insight into the initial recognition of the viral genome by the MRN complex.
The results of biochemical analyses of infected cell populations indicated that very little MRN degradation was evident at the time that viral DNA replication begins. In cells infected with a virus mutant that lacks E4-ORF3 (inORF3), MRN protein levels were reduced by only ∼twofold at the time when viral DNA replication was beginning (Fig. 1 and 2). Significant MRN degradation was not seen until 14 to 18 hpi, when the viral genome had been amplified by >100-fold. This suggested that there were significant levels of the MRN complex still present in the cell by the time viral DNA replication was occurring. As expected, viral DNA accumulation was significantly delayed for the double mutant that lacks both E4-ORF3 and E4-ORF6 (dl355/inORF3); however, at 15 hpi, the viral DNA began accumulating at a rate similar to that of the wild type (Fig. 2). This suggests that one of the following is true: the inhibition of viral DNA replication due to the activity of the MRN complex is reversible; it is not complete, and there is a threshold effect whereby some genomes escape MRN activity; or the E4 proteins have an additional, unknown function that accelerates viral DNA replication, and the inhibition seen is merely a delay due to the absence of stimulatory factors.
These results appear to be contradicted by single-cell assays, which showed that a checkpoint signaling response, the phosphorylation of ATM which is dependent on MRN activity, was inhibited by E4-ORF6/E1B-55K at the time when viral DNA accumulation was evident (Fig. 3). This applied to checkpoint signaling induced by Ad genomes delivered by the initial infection as well as checkpoint signaling induced by DNA damage. Further, single-cell immunofluorescence assays indicated the degradation of the MRN complex in infected cells staining positive for DBP prior to viral DNA accumulation (Fig. 4). The reduction in MRN levels was dependent on E4-ORF6 and E1B-55K and required a CUL5-containing E3 ubiquitin ligase complex (Fig. 4). It was previously found that active proteasomes are not necessary after 8 hpi for E1B-55K/E4-ORF6-dependent promotion of late gene expression (11). This is consistent with the findings presented here in that the E1B-55K/E4-ORF6 ubiquitin ligase activity functions early during infection to degrade the MRN complex.
The discrepancy observed between these analyses can be explained by noting the difference between the population-based and single-cell-based assays. At 8 hpi, only about half of cells express DBP to levels visible by immunofluorescence. When being stained at 18 hpi, after being infected with the same multiplicity of infection, nearly all the cells are positive for DBP (data not shown). This suggests that within a population of infected cells, the infections proceed at different rates. Two contributing factors in the rate of progression may depend on the various amounts of viral genomes that will enter the nucleus of each cell—the presence of more genomes may accelerate the viral life cycle. Also, in an unsynchronized population of cells, those cells already in S phase may proceed through the viral life cycle more quickly (18, 42). For these reasons, we believe that the twofold decrease in Mre11 levels detected by Western blot at 10 hpi (Fig. 1) is due to a significant loss of Mre11 in about half of the cells rather than a loss of half of the amount of Mre11 in all of the cells. Under these assumptions, it would follow that viral DNA replication is occurring earlier in some cells and that this would coordinate with those cells expressing higher levels of DBP. To this end, we can conclude that when cells are replicating viral DNA, they are expressing higher levels of DBP, the MRN complex has been degraded, and checkpoint signaling is inhibited, even when newly introduced DSBs are present. While we cannot show conclusively when the first rounds of replication are occurring, it seems likely that MRN degradation is complete, or at least nearly complete, prior to the onset of viral DNA replication. Finally, we conclude from the pATM assays (Fig. 3) that input Ad genomes from the initial viral infection are sufficient to be recognized by the MRN complex and induce a DSBR response. The latter conclusion is consistent with results indicating that Ad infection leads to the formation of Mdc1 foci early after infection (34).
Since Mre11 has nucleolytic activity and the ends of the Ad genome will need to be processed at least to remove the covalently attached TP in order to be ligated together, we examined the integrity of the termini of the Ad genome at 10 hpi under conditions where MRN was, or was not, inactivated. These results (Fig. 5) showed that there was no cleavage of TP from wild-type, single-mutant, or double-mutant genomes. This suggests that the MRN complex does not induce nucleolytic degradation of the Ad genome by 10 hpi, even though there are pATM foci forming, indicative of sites of DNA damage repair. We also did not see any evidence for concatemer junction formation at this time point, since the levels of the terminal restriction fragments in double-mutant virus infections corresponded equally to internal fragment; a reduction in the levels of terminal fragments should be observed relative to internal fragments if significant concatenation occurred. It is possible that large concatemeric Ad DNAs were excluded from isolation with the Hirt extraction procedure (22) that we used to isolate viral DNA. However, our results are entirely consistent with those of Weiden and Ginsberg, where Ad genome concatenation was found at late, but not early, times after infection (48). Also, our results are consistent with the recent report showing that concatenation does not cause the viral DNA replication inhibition (28).
The mechanism by which the concatemers arise in an infection with E4-deleted viruses is not known, although the working assumption is that they arise by the action of the cellular DNA end-joining mechanism(s). DNA-PKcs and DNA ligase IV have been implicated in the process due to studies that show that concatemers do not form in E4 mutant infections of M059J cells, which lack DNA-PKcs, and that E4-ORF6 contributes to the degradation of DNA ligase IV (2, 6, 16, 25, 43). Mammalian cell end joining is characterized by the conservation of sequences at both joined ends and the use of very limited or no sequence homology. This is borne out by the results of Table 1. Of the 12 clones analyzed, half showed no evidence for common sequence at the junction, and the others had either one (5 clones) or two (1 clone) common base pairs. Neighboring sequences, potentially lost in the joining reaction, also showed very limited and scattered homology (data not shown). These data strongly suggest that the joining reaction is not homology based and is most likely a consequence of NHEJ.
Two unresolved issues remain, however, namely the lack of simple terminal junctions with no deletions and the heterogeneity in the size of the deletions observed. These deletions could arise in one of at least three ways. First, deletions may occur prior to joining because genomes are incomplete, owing to some failure of completion of DNA synthesis, perhaps via MRN activity. Second, deletions could be introduced during the joining reaction itself because the end-joining proteins were directed to sites internal to the genome ends. TP and its associated proteins might block the ends. This idea, however, is not supported by the observation that full-length circles of Ad DNA can arise, at least under nonreplicative conditions (19). Finally, the deletions could have been made after the joining reaction because the large inverted palindrome created by the ITRs is unstable, and sequences are removed so that the palindromes are destroyed. This seems the least likely explanation. End joining after HO-induced cleavage of viral substrates creates large palindromic genomes without any evidence for “shedding” of junctional sequence (C. S. H. Young, unpublished data), and the full-length circles described by Graham et al. (19) are also stable.
The MRN complex will recognize the Ad genome and elicit a DSBR response if the viral proteins that inhibit the MRN complex (E4-ORF3, E4-ORF6, and E1B-55K) are not expressed. These viral proteins are not expressed to significant levels until several hours after the incoming genomes reach the nucleus, yet there are no detrimental effects of the MRN complex on the Ad genome until, or after, viral DNA replication is initiated, suggesting that the incoming genomes are protected from MRN recognition during the earliest stages of infection. It has been hypothesized that the Ad TP can protect the incoming genomes from recognition by the MRN complex and that, eventually, MRN will cleave the termini in a fashion similar to the way it cleaves Spo11 from meiotic-specific DSBs in S. cerevisiae (43, 50). Since this is not the case, it may be that there is some other unknown mechanism to mask the viral genome and protect it from MRN recognition or the detrimental effects that occur following MRN induction of a DSBR response. Further studies into the initial recognition of the viral genomes are under way.
Acknowledgments
We thank Arnold Levine for the antibody against DBP and Arnold Berk and Jennifer Woo for the CUL5-NTD expression vector. We thank Matthew Weitzman for advice concerning the TP cleavage assay and David Ornelles for the viral mutant dl1520/dl341. We acknowledge the excellent technical assistance of Mary Anderson and Ilana Shoshani. We thank the members of our laboratory for informed discussions.
This work was supported by NIH grants CA122677 and GM31452. K. A. K. was supported by NIH training grant AI007539.
Footnotes
Published ahead of print on 25 February 2009.
REFERENCES
- 1.Araujo, F. D., T. H. Stracker, C. T. Carson, D. V. Lee, and M. D. Weitzman. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 7911382-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, A., K. J. Rohleder, L. A. Hanakahi, and G. Ketner. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 817034-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421499-506. [DOI] [PubMed] [Google Scholar]
- 4.Barker, D. D., and A. J. Berk. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156107-121. [DOI] [PubMed] [Google Scholar]
- 5.Bhaskara, V., A. Dupre, B. Lengsfeld, B. B. Hopkins, A. Chan, J. H. Lee, X. Zhang, J. Gautier, V. Zakian, and T. T. Paull. 2007. Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes. Mol. Cell 25647-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263307-312. [DOI] [PubMed] [Google Scholar]
- 7.Bridge, E., and G. Ketner. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burma, S., B. P. Chen, M. Murphy, A. Kurimasa, and D. J. Chen. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 27642462-42467. [DOI] [PubMed] [Google Scholar]
- 9.Cerosaletti, K., and P. Concannon. 2004. Independent roles for nibrin and Mre11-Rad50 in the activation and function of Atm. J. Biol. Chem. 27938813-38819. [DOI] [PubMed] [Google Scholar]
- 10.Clatworthy, A. E., M. A. Valencia-Burton, J. E. Haber, and M. A. Oettinger. 2005. The MRE11-RAD50-XRS2 complex, in addition to other non-homologous end-joining factors, is required for V(D)J joining in yeast. J. Biol. Chem. 28020247-20252. [DOI] [PubMed] [Google Scholar]
- 11.Corbin-Lickfett, K. A., and E. Bridge. 2003. Adenovirus E4-34kDa requires active proteasomes to promote late gene expression. Virology 315234-244. [DOI] [PubMed] [Google Scholar]
- 12.D'Amours, D., and S. P. Jackson. 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signaling. Nat. Rev. Mol. Cell Biol. 3317-327. [DOI] [PubMed] [Google Scholar]
- 13.de Jager, M., C. Wyman, D. C. van Gent, and R. Kanaar. 2002. DNA end-binding specificity of human Rad50/Mre11 is influenced by ATP. Nucleic Acids Res. 304425-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai-Mehta, A., K. M. Cerosaletti, and P. Concannon. 2001. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol. Cell. Biol. 212184-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, J. D., and P. Hearing. 2003. Distinct roles of the adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J. Virol. 775295-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans, J. D., and P. Hearing. 2005. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J. Virol. 796207-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gatei, M., D. Young, K. M. Cerosaletti, A. Desai-Mehta, K. Spring, S. Kozlov, M. F. Lavin, R. A. Gatti, P. Concannon, and K. Khanna. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25115-119. [DOI] [PubMed] [Google Scholar]
- 18.Goodrum, F. D., and D. A. Ornelles. 1997. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J. Virol. 71548-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham, F. L., J. Rudy, and P. Brinkley. 1989. Infectious circular DNA of human adenovirus type 5: regeneration of viral DNA termini from molecules lacking terminal sequences. EMBO J. 82077-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harada, J. N., A. Shevchenko, A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 769194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26365-369. [DOI] [PubMed] [Google Scholar]
- 23.Hopfner, K. P., A. Karcher, D. S. Shin, L. Craig, L. M. Arthur, J. P. Carney, and J. A. Tainer. 2000. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101789-800. [DOI] [PubMed] [Google Scholar]
- 24.Huang, M. M., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 632605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jayaram, S., and E. Bridge. 2005. Genome concatenation contributes to the late gene expression defect of an adenovirus E4 mutant. Virology 342286-296. [DOI] [PubMed] [Google Scholar]
- 26.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17683-689. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi, J., A. Antoccia, H. Tauchi, S. Matsuura, and K. Komatsu. 2004. NBS1 and its functional role in the DNA damage response. DNA Repair (Amsterdam) 3855-861. [DOI] [PubMed] [Google Scholar]
- 28.Lakdawala, S. S., R. A. Schwartz, K. Ferenchak, C. T. Carson, B. P. McSharry, G. W. Wilkinson, and M. D. Weitzman. 2008. Differential requirements of the C terminus of Nbs1 in suppressing adenovirus DNA replication and promoting concatemer formation. J. Virol. 828362-8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, J.-H., and T. T. Paull. 2007. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 267741-7748. [DOI] [PubMed] [Google Scholar]
- 30.Lilley, C. E., R. A. Schwartz, and M. D. Weitzman. 2007. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 15119-126. [DOI] [PubMed] [Google Scholar]
- 31.Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Lin, J. H. Petrini, and M. B. Kastan. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404613-617. [DOI] [PubMed] [Google Scholar]
- 32.Liu, Y., A. Shevchenko, A. Shevchenko, and A. J. Berk. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 7914004-14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maser, R. S., K. J. Monsen, B. E. Nelms, and J. H. Petrini. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 176087-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathew, S. S., and E. Bridge. 2007. The cellular Mre11 protein interferes with adenovirus E4 mutant DNA replication. Virology 365346-355. [DOI] [PubMed] [Google Scholar]
- 35.Moreau, S., J. R. Ferguson, and L. S. Symington. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno-Herrero, F., M. de Jager, N. H. Dekker, R. Kanaar, C. Wyman, and C. Dekker. 2005. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature 437440-443. [DOI] [PubMed] [Google Scholar]
- 37.Nicolas, A. L., P. L. Munz, E. Falck-Pedersen, and C. S. Young. 2000. Creation and repair of specific DNA double-strand breaks in vivo following infection with adenovirus vectors expressing Saccharomyces cerevisiae HO endonuclease. Virology 266211-224. [DOI] [PubMed] [Google Scholar]
- 38.Paull, T. T., and M. Gellert. 2000. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc. Natl. Acad. Sci. USA 976409-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 153104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reich, N. C., P. Sarnow, E. Duprey, and A. J. Levine. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128480-484. [DOI] [PubMed] [Google Scholar]
- 41.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 2735858-5868. [DOI] [PubMed] [Google Scholar]
- 42.Shepard, R. N., and D. A. Ornelles. 2003. E4orf3 is necessary for enhanced S-phase replication of cell cycle-restricted subgroup C adenoviruses. J. Virol. 778593-8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418348-352. [DOI] [PubMed] [Google Scholar]
- 44.Stracker, T. H., D. V. Lee, C. T. Carson, F. D. Araujo, D. A. Ornelles, and M. D. Weitzman. 2005. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 796664-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stucki, M., and S. P. Jackson. 2006. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amsterdam) 5534-543. [DOI] [PubMed] [Google Scholar]
- 46.Uziel, T., Y. Lerenthal, L. Moyal, Y. Andegeko, L. Mittelman, and Y. Shiloh. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 225612-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volkert, F. C., and C. S. Young. 1983. The genetic analysis of recombination using adenovirus overlapping terminal DNA fragments. Virology 125175-193. [DOI] [PubMed] [Google Scholar]
- 48.Weiden, M. D., and H. S. Ginsberg. 1994. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. USA 91153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinberg, D. H., and G. Ketner. 1986. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J. Virol. 57833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weitzman, M. D., and D. A. Ornelles. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 247686-7696. [DOI] [PubMed] [Google Scholar]
- 51.Woo, J. L., and A. J. Berk. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]