Abstract
A close association must be maintained between the male pronucleus and the centrosomes during pronuclear migration. In C. elegans, simultaneous depletion of inner nuclear membrane LEM proteins EMR-1 and LEM-2, depletion of the nuclear lamina proteins LMN-1 or BAF-1, or the depletion of nuclear import components leads to embryonic lethality with small pronuclei. Here, a novel centrosome detachment phenotype in C. elegans zygotes is described. Zygotes with defects in the nuclear envelope had small pronuclei with a single centrosome detached from the male pronucleus. ZYG-12, SUN-1, and LIS-1, which function at the nuclear envelope with dynein to attach centrosomes, were observed at normal concentrations on the nuclear envelope of pronuclei with detached centrosomes. Analysis of time-lapse images showed that as mutant pronuclei grew in surface area, they captured detached centrosomes. Larger tetraploid or smaller histone::mCherry pronuclei suppressed or enhanced the centrosome detachment phenotype respectively. In embryos fertilized with anucleated sperm, only one centrosome was captured by small female pronuclei, suggesting the mechanism of capture is dependent on the surface area of the outer nuclear membrane available to interact with aster microtubules. We propose that the limiting factor for centrosome attachment to the surface of abnormally small pronuclei is dynein.
Keywords: Nuclear envelope, centrosome, male pronucleus, pronuclear migration, lamin, LEM, BAF, microtubule aster, outer nuclear membrane, dynein
Introduction
After fertilization, male and female pronuclei migrate towards one another before the first mitotic division of the zygote. This essential step in early animal development is usually mediated by microtubules and microtubule motors; the process is dependent on a tight association between the male pronucleus and the centrosome (reviewed in Reinsch and Gonczy, 1998). In most animals, including humans, Drosophila, C. elegans, and sea urchins, the sperm contributes the only functional centrioles, which nucleate the centrosomes of the 1-cell zygote. Parthonogenic animals and rodents are the only known exceptions to this rule where centrioles are derived de novo from the oocyte (Schatten, 1994; Sutovsky et al., 1999). Shortly after fertilization, the male pronucleus decondenses and the centrosome duplicates. Both centrosomes maintain a tight association with the growing male pronucleus. As the microtubule aster grows from centrosomes at the male pronucleus, the complex is pushed towards the center of the one-celled embryo. Meanwhile, the female pronucleus completes meiosis, forms a nuclear envelope and migrates towards the male pronucleus along microtubules emanating from the male aster (reviewed in Reinsch and Gonczy, 1998; Schatten, 1994). The mechanisms of how centrosomes remain closely attached to male pronuclei during pronuclear migration remain mostly unknown.
The centrosome also remains tightly associated to the nuclear envelope in most interphase cells (Bornens, 1977; Malone et al., 2003) and the close association of centrosomes with the nucleus is required for microtubule dependent nuclear positioning (Morris, 2000; Starr and Han, 2003). Among other cell-types, nuclear to centrosome interactions are essential for the formation of polarity in tissue culture fibroblasts and the migration of neurons (Gomes et al., 2005; Tsai et al., 2005). Therefore it is important to characterize the mechanisms of how centrosomes attach to the nuclear envelope to better understand both early animal development and nuclear positioning in more general terms.
A two-step model for centrosome attachment in the early C. elegans embryo has been proposed (Malone et al., 2003). In the first step, dynein plays a central role. The centrosome is brought into close proximity of the nuclear envelope by the microtubule motor dynein, which is connected to the outer nuclear membrane and moves along microtubules toward the centrosome. Genetic evidence suggests that dynein functions in part to initialize the attachment of centrosomes to nuclei. In Drosophila or C. elegans embryos depleted of dynein components, centrosome separation fails and about 15% of centrosomes fail to attach to the nuclear envelope (Gonczy et al., 1999; Robinson et al., 1999). Dynein also functions as the major centrosome repulsive force. In wild-type zygotes, the two centrosomes localize to opposite sides of the male pronucleus. However, in dhc-1(RNAi) embryos, centrosomes fail to separate, even when attached to the nucleus (Gonczy et al., 1999). It was surprising to implicate a minus end directed motor in centrosome separation. Gonczy et al. (1999) propose that dynein complexes anchored to the cytoplasmic surface of the nuclear envelope pull on the leading edge of centrosomes where the longer astral microtubules are. When astral microtubules along the entire surface of the nucleus are of equal size, dynein forces are balanced and centrosome movement stops with the centrosomes at opposite sides of the nucleus (Gonczy et al., 1999). In support of a role for dynein in centrosome attachment to the nucleus and separation, knockdown of either dli-1 or lis-1, which encode dynein accessory proteins, cause similar phenotypes (Cockell et al., 2004; Yoder and Han, 2001). In the above model, dynein and accessory proteins must be recruited to the cytoplasmic surface of the nuclear envelope. The C. elegans outer nuclear membrane protein ZYG-12 binds to DLI-1 and is necessary for recruitment of DHC-1, ARP-1 and LIS-1 to the nuclear envelope (Malone et al., 2003).
zyg-12 also plays a central role in the second centrosome attachment step, when the close association of the nucleus and the centrosome is maintained by a dynein-independent interaction between the centrosome and the outer nuclear membrane. Two genes, zyg-12 and sun-1 (also called mtf-1), are essential for centrosome attachment and embryonic development in the C. elegans zygote (Fridkin et al., 2004; Malone et al., 2003). zyg-12(ct350) and sun-1(RNAi) embryos are defective in the attachment of both centrosomes to the male pronucleus in 100% of 1-cell embryos (Malone et al., 2003). SUN-1 and ZYG-12 have conserved SUN and KASH domains respectively that likely interact in the intermembrane space of the nuclear envelope and together function to bridge the nuclear envelope (Starr, 2007; Starr and Fischer, 2005; Tzur et al., 2006; Wilhelmsen et al., 2006; Worman and Gundersen, 2006). ZYG-12 at the outer nuclear membrane is thought to interact with centrosomal pools of ZYG-12, attaching the centrosome to the surface of the nucleus (Malone et al., 2003). In the developing eye disc of Drosophila, the KASH protein Klarsicht and the SUN protein Klaroid play analogous roles (Kracklauer et al., 2007; Patterson et al., 2004). Like ZYG-12, Klarsicht is also thought to function through dynein (Mosley-Bishop et al., 1999; Patterson et al., 2004; Welte, 2004).
Dynein, dynein regulators, and SUN and KASH proteins, which in part function to recruit dynein to the nuclear surface, play a central role in centrosome attachment and pronuclear migration. Additionally, inner nuclear membrane and nuclear lamina proteins play poorly defined roles in centrosome attachment. Disruption of the nuclear envelope induced by dsRNA treatment of nuclear import components in the Ran pathway or mel-28 lead to a centrosome detachment phenotype and severe defects in mitosis of C. elegans embryos (Askjaer et al., 2002; Galy et al., 2006). Furthermore, lamin and the integral inner nuclear membrane protein emerin have recently been shown to function during centrosome attachment; fibroblasts from emerin-defective human patients or lamin A/C mutant mice have a centrosome detachment phenotype (Lee et al., 2007; Salpingidou et al., 2007). Although the exact mechanisms are not clear, it is likely that emerin is functioning at the outer nuclear membrane for centrosome attachment (Salpingidou et al., 2007). Thus, components of the nuclear envelope and the molecular machines that function to target them there are required for proper centrosome attachment to the nuclear envelope. Here we probe the role of the nuclear envelope and lamina for attachment of centrosomes to the male pronucleus in the early C. elegans embryo.
The nuclear envelope is a specialized extension of the endoplasmic reticulum. The inner and outer nuclear membranes are continuous with one another at nuclear pores. In metazoans, the inner nuclear membrane is closely associated with the nuclear lamina, which provides the structural strength and shape to the nuclear envelope (Gruenbaum et al., 2005). Major components of the nuclear lamina include lamin intermediate filaments, the small chromatin-binding barrier-to-autointigration factor (BAF), and LEM domain integral membrane proteins (LAP2β, emerin, and MAN1) of the inner nuclear membrane. These three groups of proteins form a structural triad, interacting with one another in a mutually dependent manner to build the nuclear lamina (Gruenbaum et al., 2005). In humans, B-type lamins are ubiquitously expressed and essential, while A/C-type lamins are expressed at certain times and are not essential for cell viability. However mutations in the lamin A/C gene lead to a wide variety of crippling developmental diseases termed laminopathies (Mounkes et al., 2003; Worman and Bonne, 2007). Individually, LEM proteins are not essential at a cellular level, but mutations in emerin or MAN1 also lead to a variety of tissue-specific diseases (Somech et al., 2005). LEM proteins likely play essential redundant roles in the nuclear envelope (Liu et al., 2003). We describe a novel role for nuclear lamina proteins in centrosome attachment to the nuclear envelope.
C. elegans is an excellent model system to study the functions of nuclear lamina proteins in a developmental context for a variety of reasons. First, one can rapidly study loss-of-function phenotypes by treating with dsRNA. Second, since early C. elegans development is so thoroughly described and easily observed, mutant developmental phenotypes can be studied by time-lapse imaging. Finally, C. elegans might be the simplest organism with a nuclear lamina clearly conserved with higher animals (Gorjanacz et al., 2007; Oegema and Hyman, 2006). C. elegans has a single lamin gene (lmn-1), a BAF homologue (baf-1), and only three LEM domain genes, an emerin homologue (emr-1), a single MAN1 and Lem2 homologue (lem-2) and lem-3, which lacks a transmembrane domain (Gruenbaum et al., 2002; Liu et al., 2000; Liu et al., 2003; Margalit et al., 2005; Zheng et al., 2000). The loss-of-function mutant phenotypes of many nuclear lamina proteins have been previously described in C. elegans. lmn-1 and baf-1 are essential for early cell divisions (Liu et al., 2000; Margalit et al., 2005; Zheng et al., 2000). Single dsRNA depletion of either emr-1 or lem-2 have no major phenotypes, but simultaneous depletion of both LEM-coding genes causes a synthetic embryonic lethal phenotype similar to the lmn-1 or baf-1 mutations (Liu et al., 2003).
We demonstrate an essential role for the nuclear lamina in attaching the centrosome to the nuclear envelope in newly fertilized C. elegans embryos. The centrosome detachment defect does not affect the localization of ZYG-12, SUN-1, or the dynein-associated LIS-1. Instead, our data implies that the size of the male pronucleus corresponds to the number of centrosomes attached. We present data in support of a new model where abnormally small nuclei only have the surface area to attach one centrosome and as the nucleus expands, it reaches a size where it is able to attach both centrosomes. Data from embryos fertilized with anucleated sperm suggest the surface area of the nucleus available to interact with microtubules is likely the limiting factor for the association of centrosomes to small mutant pronuclei.
Materials and Methods
C. elegans strains and RNA interference
C. elegans strains were grown and maintained by standard protocols and the N2 strain was wildtype (Brenner, 1974). The integrated GFP::tubulin, histone::mCherry strain OD57 (McNally et al., 2006) and the integrated GFP::tubulin strain AZ244 (Praitis et al., 2001) were used for live filming of the centrosome separation phenotype. The ZYG-12::GFP line WH223 (Malone et al., 2003), the β-tubulin::GFP; histone::GFP line TY3558 (Strome et al., 2001), and the LMN-1::YFP line XA3502 (Galy et al., 2003) were previously described. The γ-tubulin::GFP line OD44 was provided by Arshad Desai (Univ. California, San Diego; (McNally et al., 2006). The SUN-1::GFP line was WH340 (from C. J. M. and J. White, University of Wisconsin). In a few experiments as an alternative to emr-1(RNAi), the emr-1(gk119) knockout allele, strain VC237, was used (gift of the C. elegans Reverse Genetics Core Facility at UBC and the International C. elegans Gene Knockout Consortium); VC237 was crossed to OD57 to make UD116. For the LIS-1::GFP experiments, the line WH239 was used (Malone et al., 2003). The tetraploid strain used was SP346 and contains no markers (Madl and Herman, 1979). For the anucleate sperm experiments, fem-1(hc17) strain BA17 feminized animals (Nelson et al., 1978) were crossed to ZYG-12::GFP or tubulin::GFP; histone::GFP males. These were mated to emb-27(g48) strain GG48 (Sadler and Shakes, 2000). Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
Double-stranded RNA (dsRNA) was synthesized in vitro using T7 polymerase (Promega, Madison, WI) off of PCR templates with overhanging T7 polymerase promoters according to standard protocols (Ahringer, 2006; Fire et al., 1998). Templates for emr-1 and lem-2 dsRNA were PCR amplified from approximately 1 kb genomic regions cloned into the vector L4440 and obtained from the RNAi feeding library (Geneservice Ltd, Cambridge, UK; (Kamath et al., 2003). Templates for lmn-1 and baf-1 dsRNA were amplified from full-length cDNAs yk561f10 and yk333d11 respectively (gifts of Yuji Kohara, National Institute of Genetics, Japan(Kohara, 1996). The dsRNA was injected into both gonads of young adult hermaphrodites (Ahringer, 2006) and embryos were observed for phenotypes as described below 24–48 hours later. In some experiments, embryos from hermaphrodites fed bacteria expressing dsRNA were observed (Ahringer, 2006).
Microscopy
For fixed studies, embryos were released from adult hermaphrodites by cutting animals on poly-L-lysine-coated slides. They were freeze-cracked by immersion in liquid N2, fixed in methanol at room temperature for 5 min, washed in PBST (1× PBS containing 0.05% Tween 20), and blocked in 5% dry milk in PBS for 30 min (Miller and Shakes, 1995). Fixed embryos were stained with an anti–tubulin monoclonal antibody at 1/400 dilution in PBS (clone DM 1A; Sigma-Aldrich, St. Louis, MO), anti-nuclear pore monoclonal antibody at a 1/1000 dilution in PBS (mAb414; Covance, Richmond, CA), rabbit anti-GFP antibody at a 1/500 dilution (Invitrogen, Carlsbad, CA) for the LMN-1::GFP experiments, rabbit anti-GFP antibody at a 1/400 dillution (Novus Biologicals, Littleton, CO) for the LIS-1::GFP experiments, or a rat anti-ZYG-12 polyclonal antibody at a 1/100 dilution (Malone et al., 2003) at room temperature for 1 hour or 4°C overnight. The slides were then washed with PBST and incubated with Cy3-conjugated goat anti-mouse IgG or Cy3-conjugated goat anti-rat IgG (Jackson Laboratories, West Grove, PA) diluted in PBS at room temperature for 1 h. After a final wash with PBST and DAPI (4′, 6′-diamidino-2-phenylindole) staining, the slides were washed with PBS and mounted in mounting medium (1.8% n-propyl galate, 80% glycerol) for observation.
For live filming of pronuclear migration, early embryos were collected from untreated or emr-1(RNAi); lem-2(RNAi) treated hermaphrodites from the GFP::tubulin, histone::mCherry strain OD57 or the GFP::tubulin strain AZ244 and placed on a thin pad of 5% agar under a coverslip for filming (Malone et al., 2003; McNally et al., 2006). Images were collected in the DIC, red, and green channels at 15–30 second intervals from the point of identification of a newly fertilized embryo through pronuclear migration, until nuclear envelope breakdown, or until development apparently stopped. Nuclei were measured under DIC optics by determining the diameter at two orthogonal axes and taking the average. Centrosome-to-nuclear envelope distance was measured from the center of the tubulin::GFP signal to the edge of the male pronucleus using by DIC optics. For the anucleate sperm experiments, feminized animals expressing ZYG-12::GFP or tubulin::GFP and histone::GFP were crossed to emb-27(g48) males (Sadler and Shakes, 2000); both mutations are temperature sensitive, so the cross was done at 25°C. The crosses were set up as controls or with feminized animals treated with dsRNA by either injection or feeding. Newly fertilized zygotes were filmed at 10-second intervals in the GFP channel as above.
Most images were acquired with a Leica fluorescence microscope using an UplanApo 63× 1.4 objective. Images and time-lapse movies were analyzed and merged with Leica FW4000 software. Images were adjusted for contrast and prepared for publication using Adobe Photoshop or Illustrator. Movies were cropped, rotated and compressed for viewing using Adobe After Effects CS3. The ZYG-12 images were taken at the MCB imaging facility at UC Davis using an Olympus FV1000 Laser Scanning Confocal microscope. Relative intensities of nuclear envelope and centrosome staining of ZYG-12 were measured using ipLab software.
Results
Defects in the nuclear lamina disrupt centrosome attachment to the nuclear envelope
We examined the role of the nuclear lamina and inner nuclear membrane proteins in early C. elegans embryos. The C. elegans homologues of emerin, MAN1/Lem2, BAF, and lamin interact in a complex in vitro and have a mutually dependent structural relationship in vivo, suggesting that they work in a common pathway (Liu et al., 2000; Liu et al., 2003; Margalit et al., 2005). It has been shown that simultaneous RNAi against both genes encoding inner nuclear membrane LEM proteins in C. elegans, emr-1 and lem-2, or single dsRNA depletion of either lmn-1 or baf-1 leads to embryonic lethality with a high level of chromosomal segregation defects (Liu et al., 2003). To examine the early mitotic defects more closely, we employed a similar RNAi approach. We observed the same chromosomal segregation defects as previously reported where 60–90% of two or four cell embryos showed chromatin bridges or other defects in anaphase, demonstrating that we can effectively knock-down emr-1, lem-2, baf-1, and lmn-1 (data not shown). In addition to the expected chromosomal segregation defects, we observed an earlier centrosome detachment defect that has not been previously described.
We were interested in observing the behavior of the male pronucleus and the centrosomes that are normally attached to it. However, sperm are resistant to RNAi (Fraser et al., 2000). The sperm nucleus is very condensed and lacks a nuclear envelope (Kimble and Ward, 1988; Ward et al., 1981; Wolf et al., 1978). Neither LMN-1 nor nuclear pore complexes localize to the nucleus of mature sperm (Browning and Strome, 1996; Liu et al., 2000) and instead it is surrounded by a halo of RNA and the novel protein SPE-11 (Browning and Strome, 1996; Ward et al., 1981). Thus, the nuclear lamina of the sperm pronucleus is derived from proteins pre-loaded in the oocyte and our RNAi approach, which knocked-down lamina components in the oocyte, was appropriate to examine the role of the developing lamina in male pronuclei.
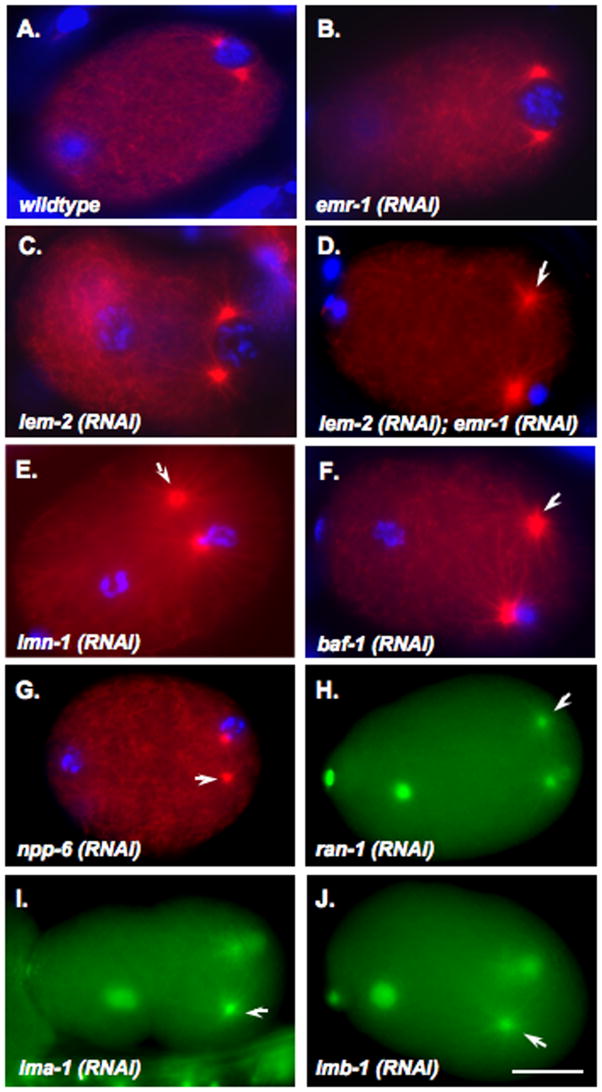
Upon fertilization, the sperm contributes the sole centrosome of the zygote. The centrosome duplicates and daughter centrosomes move to opposite sides of the male pronucleus before pronuclear migration, centration and rotation of the centrosome/pronuclear complex, and formation of the first mitotic spindle (Oegema and Hyman, 2006). We examined the behavior of centrosomes in newly fertilized zygotes prior to pronuclear migration treated with dsRNA against components of the nuclear lamina, fixed, and stained for tubulin. In the control zygotes or zygotes treated with dsRNA targeted against either emr-1 or lem-2, there were always two centrosomes tightly associated with the male pronucleus (Fig. 1A–C). In zygotes treated with dsRNA against both emr-1 and lem-2, one centrosome failed to associate with the male pronucleus while the other was closely associated (Fig. 1D). 13 of 22 fixed emr-1(RNAi); lem-2(RNAi) embryos observed prior to pronuclear migration had one detached centrosome. Likewise, a large portion of embryos from lmn-1(RNAi) (5 of 11) and baf-1(RNAi) zygotes (16 of 25) exhibited the same phenotype (Fig. 1E–F). The centrosome detachment phenotype observed here is only partially penetrant because it is difficult to stage fixed embryos. Live imaging (see below) suggests that detached centrosomes are often captured before pronuclear migration. Thus, the penetrance of the phenotype is actually much higher. To ensure that we observed detached centrosomes and not microtubule asters without centrosomes, lmn-1(RNAi) embryos expressing γ-tubulin::GFP were examined and foci of γ-tubulin localization detached from the male pronucleus were observed (Sup. Fig. 1). We therefore conclude that the nuclear lamina is required for normal attachment of the second centrosome to the nuclear envelope of the male pronucleus.
Fig. 1.
Embryos treated with dsRNA against the nuclear lamina or nuclear import components show centrosome detachment defects. One-cell embryos prior to pronuclear migration are shown. (A–G) Embryos were fixed and stained for tubulin in red and chromatin in blue. (H–J) Live embryos expressing β-tubulin::GFP and histone::GFP are shown. Embryos are from hermaphrodites injected with dsRNA against the following: (A) no RNA wild-type control, (B) emr-1, (C) lem-2, (D) emr-1 and lem-2, (E) lmn-1, (F) baf-1, (G) npp-1, (H) ran-1, (I) ima-1, and (J) imb-1. In (D–J), arrows mark centrosomes detached from male pronuclei. Posterior is right and the scale bar is 10 μm.
Nuclear pores, ZYG-12, and dynein localize in nuclear lamina mutants with detached centrosomes
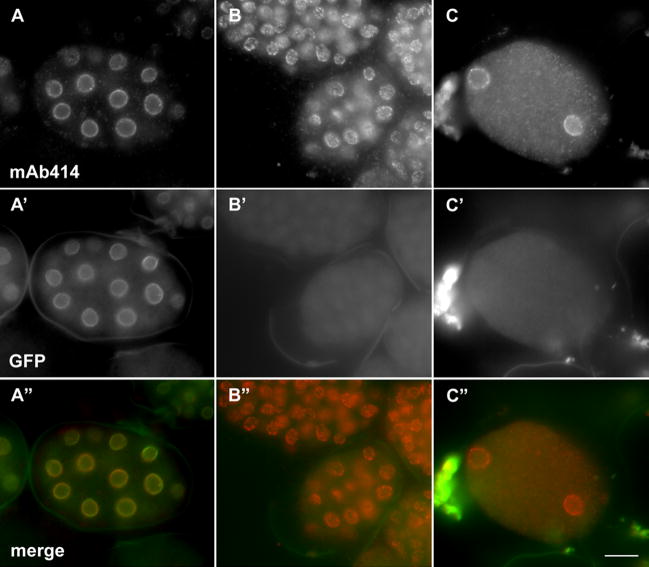
One potential model for the centrosome detachment phenotype is that by disrupting components of the nuclear lamina we actually disrupted the integrity of the entire nuclear envelope. Therefore, gross nuclear envelope morphology was examined by staining for nuclear pore complexes (Fig. 2 and Sup. Fig. 2). The monoclonal antibody 414 (mAb414) recognizes a conserved family of nuclear pore components and has previously been established as a marker to monitor the morphology of the nuclear envelope in C. elegans (Browning and Strome, 1996; Davis and Blobel, 1986; Lee et al., 2000). It was previously shown that in lmn-1(RNAi) or baf-1(RNAi) embryos, nuclear pore complexes failed to localize properly (Liu et al., 2000; Margalit et al., 2005). Although we observed some nuclear pore complexes to be abnormally clustered in lmn-1(RNAi) embryos, in many nuclei the nuclear pore complexes appeared fairly normal as compared to wildtype (Fig. 2). There may be a slight increase in the cytosolic levels of mAb414 antigen in lmn-1(RNAi) embryos (Fig. 2), which would be consistent with a slight defect in nuclear pore formation. However, this nearly normal nuclear pore complex localization was observed in embryos despite strong lmn-1 disruption as assayed by loss of LMN-1::YFP (Fig 2). The nuclear pore complex phenotype was more severe in older embryos, agreeing with previous reports, although those reports also acknowledge that nuclear pores in many dsRNA-treated embryos appear normal (Liu et al., 2000; Margalit et al., 2005). To observe nuclear pore complex localization in nuclei with detached centrosomes, newly fertilized zygotes from dsRNA-treated animals were fixed and simultaneously stained with mouse monoclonal antibodies against tubulin and nuclear pores. lem-2(RNAi); emr-1(RNAi) zygotes, as well as lmn-1(RNAi) or baf-1(RNAi) zygotes, had disrupted centrosome attachment defects as above (arrows in Sup. Fig. 2). In all three cases, the nuclear pores were distributed uniformly around the rim of the nucleus, indistinguishable from the controls (Sup. Fig. 2). Together, these data suggest that despite disrupting the nuclear lamina, the gross overall structure and morphology of the nuclear envelope was not completely disrupted in our dsRNA-treated embryos.
Fig. 2.
Nuclear pore localization in lmn-1(RNAi) embryos. LMN-1::YFP (line XA3502) hermaphrodites were (A) untreated or (B–C) injected with dsRNA against lmn-1. Embryos were fixed and stained for nuclear pore complexes with the antibody mAb414 in the red channel (shown in top panels) and YFP was observed in the green (A′-C′). (A″-C″) A merge of LMN-1::YFP and nuclear pore complexes is shown. Posterior is right and the scale bar is 10 μm.
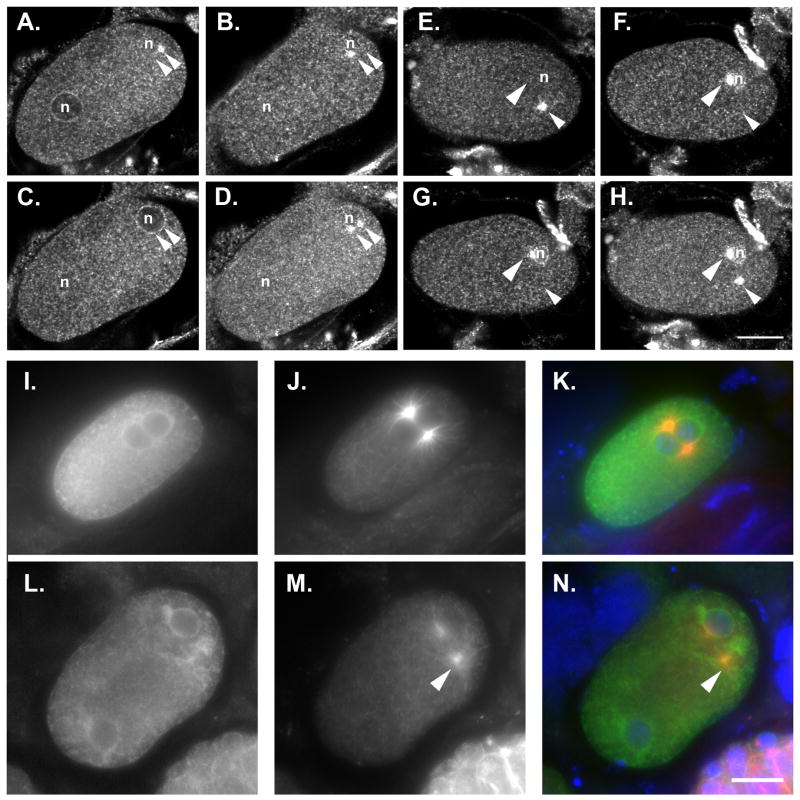
The nuclear membrane protein SUN-1 recruits ZYG-12 to the outer nuclear membrane where it functions to attach both centrosomes to the nuclear envelope (Malone et al., 2003). It is not yet known how SUN-1 is targeted to the nuclear envelope, although lmn-1 is not required for SUN-1 localization (Fridkin et al., 2004). The zyg-12(null) phenotype (both centrosomes detached) is different from the one we report here (only one centrosome detached), but one potential mechanism for the nuclear lamina to control centrosome attachment is to recruit SUN-1 and ZYG-12 to the nuclear envelope. We therefore examined the levels of ZYG-12 on the nuclear membrane in lem-2(RNAi); emr-1(RNAi) zygotes. dsRNA-treated zygotes with one detached centrosomes were examined for localization of ZYG-12 using both a ZYG-12::GFP line (not shown) and an antibody against ZYG-12. In both cases ZYG-12 localized normally to both centrosomes and the nuclear envelope (Fig. 3A–H). Anti-ZYG-12 fluorescence intensity ratios of the nuclear envelope to the centrosomes in the same embryo were determined by confocal microscopy. In seven control zygotes, the average ratio was 0.41 with a standard error of ± 0.05. In six lem-2(RNAi); emr-1(RNAi) zygotes the average ratio was 0.38 ± 0.04. We conclude that disruptions of the nuclear lamina do not affect levels of ZYG-12 at the nuclear envelope. We also observed the effect of dsRNA against nuclear lamina components on the localization of SUN-1::GFP. SUN-1::GFP localized normally to the nuclear envelope in lmn-1(RNAi) embryos (Sup. Fig. 3). There may have been an increase of intranuclear SUN-1::GFP in the dsRNA-treated embryos that could be due to nuclear envelope defects.
Fig. 3.
ZYG-12 and LIS-1 localization to the nuclear envelope is not disrupted in dsRNA treated embryos. (A–D) Three confocal Z-sections through a single wild-type embryo stained with anti-ZYG-12 antibodies. (A) The female pronucleus and one centrosome are shown. (B) The second centrosome is shown. (C) The male pronucleus is shown. (D) A merged image is shown. (E–H) three confocal Z-sections through a single embryo treated with dsRNA against emr-1 and lem-2. (E) One centrosome is shown. (F) The second centrosome is shown. (G) The second centrosome and the male pronucleus are shown. (H) A merged image is shown; the female pronucleus was in a different frame. Arrowheads mark centrosomes and pronuclei are marked by an n. (I–K) An untreated LIS-1::GFP embryo. (L–N) A lmn-1(RNAi) LIS-1::GFP embryo. Embryos were fixed and stained for tubulin in the red channel (J and M) and for GFP in the green channel (I and L). (K and N) A merge of LIS-I::GFP, tubulin and DNA stained with DAPI is shown. Arrowheads mark centrosomes detached from the male pronucleus. Posterior is right and the scale bar is 10 μm.
Dynein on the surface of the outer nuclear membrane plays a central role in centrosome attachment (Gonczy et al., 1999; Malone et al., 2003). Recruitment of dynein to the nuclear periphery requires ZYG-12 at the outer nuclear membrane (Malone et al., 2003), but it is not known whether ZYG-12 is sufficient for dynein localization. We therefore wanted to test whether dynein localized to the surface of small pronuclei. To approximate DHC-1 at the nuclear envelope, we examined LIS-1::GFP localization to the nuclear periphery, which requires DHC-1 at the nuclear periphery (Cockell et al., 2004; Malone et al., 2003). Fixed zygotes were examined and stained for GFP and tubulin to identify defective pronuclei with a detached centrosome. At a qualitative level with samples prepared in parallel and images taken at the same exposures, LIS-1::GFP localized at normal concentrations in small lmn-1(RNAi) male pronuclei with centrosomes detached (Fig. 3I–N) suggesting that dynein was also present at the nuclear periphery. A new phenotype, where both centrosomes were detached from the male pronucleus, was often observed in LIS-1::GFP; lmn-1(RNAi) embryos. This is likely a neomorphic effect from overexpression of LIS-1::GFP. Because of the variability of the overexpression, it was difficult to quantify the relative amounts of LIS-1::GFP in dsRNA-treated versus untreated embryos. Since both ZYG-12 and LIS-1::GFP localize in dsRNA-treated embryos, we conclude that dynein is likely localized at normal concentrations in the mutant small pronuclei.
The surface area of pronuclei dictates the number of centrosomes that are able to attach to the nuclear envelope
It has been previously shown that emr-1(RNAi);lem-2(RNAi), lmn-1(RNAi), or baf-1(RNAi) zygotes have small interphase nuclei with abnormally condensed chromatin (Liu et al., 2000; Liu et al., 2003; Margalit et al., 2005). We also observed that the size of male pronuclei from all three dsRNA treatments was significantly smaller than untreated controls (Figs. 1–3 and data not shown). We therefore hypothesized that the detached centrosome phenotype is due to a reduced surface area of the nucleus. Furthermore, we proposed that mutations in other genes that also cause a small nucleus phenotype would have a similar centrosome detachment phenotype. Previous C. elegans whole-genome RNAi screens and studies on nuclear import have shown that defects in many nuclear pore components and members of the nuclear import pathway lead to abnormally small pronuclei (Askjaer et al., 2002; Franz et al., 2005; Galy et al., 2003; Sonnichsen et al., 2005). In fact, there are currently 27 unique hits in Phenobank (http://worm.mpi-cbg.de/phenobank2/cgi-bin/MenuPage.py) for pronuclear appearance or size (Sonnichsen et al., 2005). lmn-1 is on the list, as well as thirteen genes encoding components of nuclear pores, six involved in nuclear import, and T24F1.2, a likely nuclear envelope component and the homologue of mammalian NET5 (Gunsalus et al., 2005; Schirmer et al., 2003). The effects of the other six on nuclear size are not as easily explained. This group consists of a component of signal peptidase (weak effect), a fatty-acid co-A ligase, alpha actinin, two novel genes, Y54GA.2 and H02I12.5, with other more severe phenotypes, and ooc-5, which has other defects in oogenesis (Basham and Rose, 2001). We tested the roles in centrosome attachment for four additional members of this group. dsRNA treatment against the nuclear pore component npp-6, the Ran GTPase ran-1, α-importin ima-1, or β-importin imb-1 all caused small pronuclei and the detached centrosome phenotype similar to direct disruptions of the nuclear lamina (Fig. 1 G-J). It is likely that most of the class of 27 cause the same defects in nuclear envelope formation leading to small pronuclei and the centrosome detachment phenotype.
Analysis of fixed images of dsRNA-treated embryos suggested that male pronuclei with one centrosome detached were smaller than male pronuclei treated with the same dsRNA with both centrosomes attached. In the case of baf-1(RNAi) embryos, pronuclei with both centrosomes attached had an average surface area of 96 μm2, which was significantly smaller than wildtype (152 μm2), but significantly larger than the average diameter of male pronuclei with one detached centrosome (47 μm2; p<0.00001). The fact that the small size of male pronuclei correlated with detached centrosomes could explain why only about half of our fixed images of dsRNA-depleted embryos showed the detached centrosome phenotype. Perhaps the fixed images with both centrosomes attached were from later developmental time points where the male pronucleus had time to grow and capture the second centrosome.
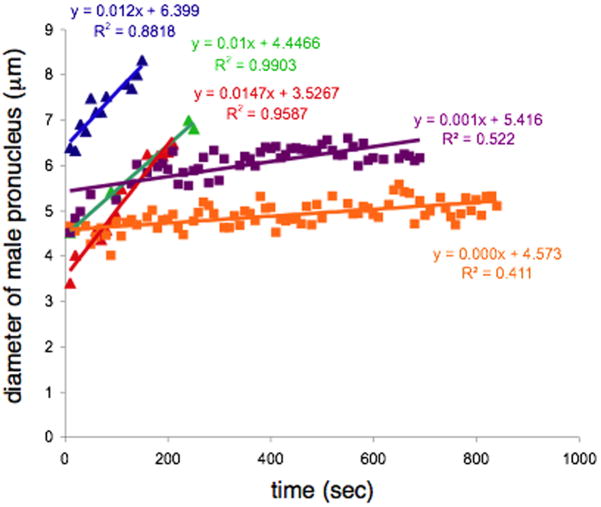
Based on the above data, we proposed that the centrosome detachment phenotype might be transient. Soon after fertilization the pronucleus might be too small to attach both centrosomes and at later time points the male pronucleus could grow in surface area, allowing the second centrosome to be reeled in and attach. To test this hypothesis, time-lapse imaging of pronuclear size and behavior of centrosomes was employed. We filmed early one-cell embryos from both control and emr-1(RNAi); lem-2(RNAi) or lmn-1(RNAi) hermaphrodites expressing α-tubulin::GFP to follow centrosomes (Figs. 4–7). The size of the male pronucleus was determined using DIC optics. A detached centrosome was defined as the center of the tubulin::GFP signal being greater than 3 μm from the edge of the male pronucleus. In three time-lapse movies in the untreated tubulin::GFP background, the diameter of male pro-nuclei grew at a consistent rate of about 0.012 μm/sec (Fig. 4), a rate similar to previous reports (Cowan and Hyman, 2006). Male pronuclei from dsRNA-treated embryos grew significantly slower (Fig. 4), and sometimes appear to arrest prior to pronuclear migration. This made it difficult to precisely define time zero to a uniform developmental event, such as pronuclear meeting or nuclear envelope breakdown prior to mitosis. We therefore defined time zero as the arbitrary time we started filming a fertilized embryo. We also examined the timing of nuclear envelope formation in wildtype. LMN-1::YFP was observed before we could detect the male pronucleus using DIC optics, significantly earlier than α-tubulin::GFP can be detected on centrosomes (data not shown). Therefore, in wildtype, the nuclear envelope forms before the centrosome begins to nucleate significant numbers of microtubules.
Fig. 4.
Growth rate of male pronuclei. Plots show the rate of growth of the diameter of the male pronucleus over time. Data points from three wildtype embryos are shown with triangles (blue, green, red). Data points from two lmn-1(RNAi)-treated embryos are shown with squares (purple, orange). The fits to a linear curve are shown in the corresponding colors.
Fig. 7.
Time-lapse imaging of centrosome attachment in emr-1; lem-2 defective embryos. (A) An emr-1(gk119); lem-2(RNAi) embryo in OD57 (tubulin::GFP; histone::mCherry). The free centrosome is captured by the female pronucleus (middle panel) and the male pronucleus is not in the spindle (bottom). (B) An emr-1(RNAi); lem-2(RNAi) embryo in AZ244 (tubulin::GFP). Very early both centrosomes are attached (top), then one detached (middle), and finally both are attached (bottom). Images from a time series are shown; fluorescent images from green and red channels merged (A–B) or the green (C–D) are shown on the left and corresponding DIC images are on the right. Time in minutes:seconds from the start of filming and surface area of male pronuclei in μm2 are shown in the corners of panels. Arrowheads mark centrosomes detached form the nucleus. Posterior is right and the scale bar is 10 μm.
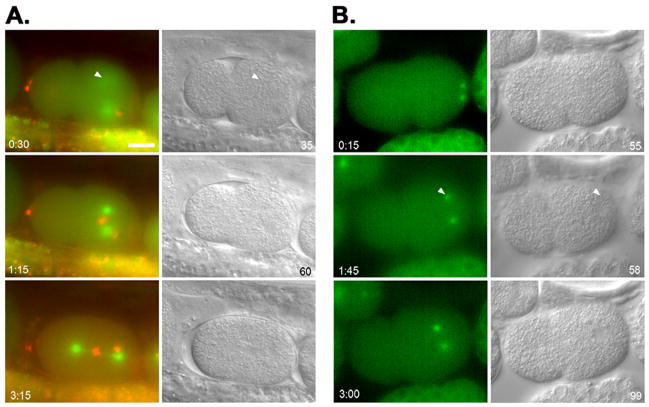
Seventeen live dsRNA-treated embryos with a detached centrosome were filmed and categorized into three major classes. Nine were captured by the male pronucleus, four were captured by the female pronucleus or the male and female pronuclei after pronuclear meeting, and 4 failed to be captured. Of the nine where the detached centrosome was captured by the male pronucleus prior to pronuclear migration, seven were filmed early enough so that we could observe the size of the male pronucleus growing. In two embryos the centrosome was captured by the male pronucleus within a minute of the start of filming, too late to observe an increase in the size of the male pronucleus. In the seven embryos filmed long enough to observe capture of the centrosome by the growing male pronucleus, the average surface area of male pronuclei was 67 μm2 at the start of filming (a representative embryo with a detached centrosome is shown in Fig. 5B and Supplemental Movie 2, an untreated embryo is shown in Fig. 5A and Supplemental Movie 1, the statistical data is in Fig. 6A, and the complete data set is in Sup. Fig. 4A–B). The male pronuclei grew to an average of 114 μm2 at the time the second centrosome was captured. This difference was significant with p<0.0001. However, even at the time of centrosome capture, the size of the male pronucleus was still significantly smaller than wild-type control male pronuclei with both centrosomes attached prior to pronuclear migration (136 μm2; n=5; p=0.025; Fig. 6A). These data support our hypothesis that disruption of the nuclear lamina delays the decondensation of the male pronucleus, and that the ability of the male pronucleus to attach to both centrosomes is size dependent.
Fig. 5.
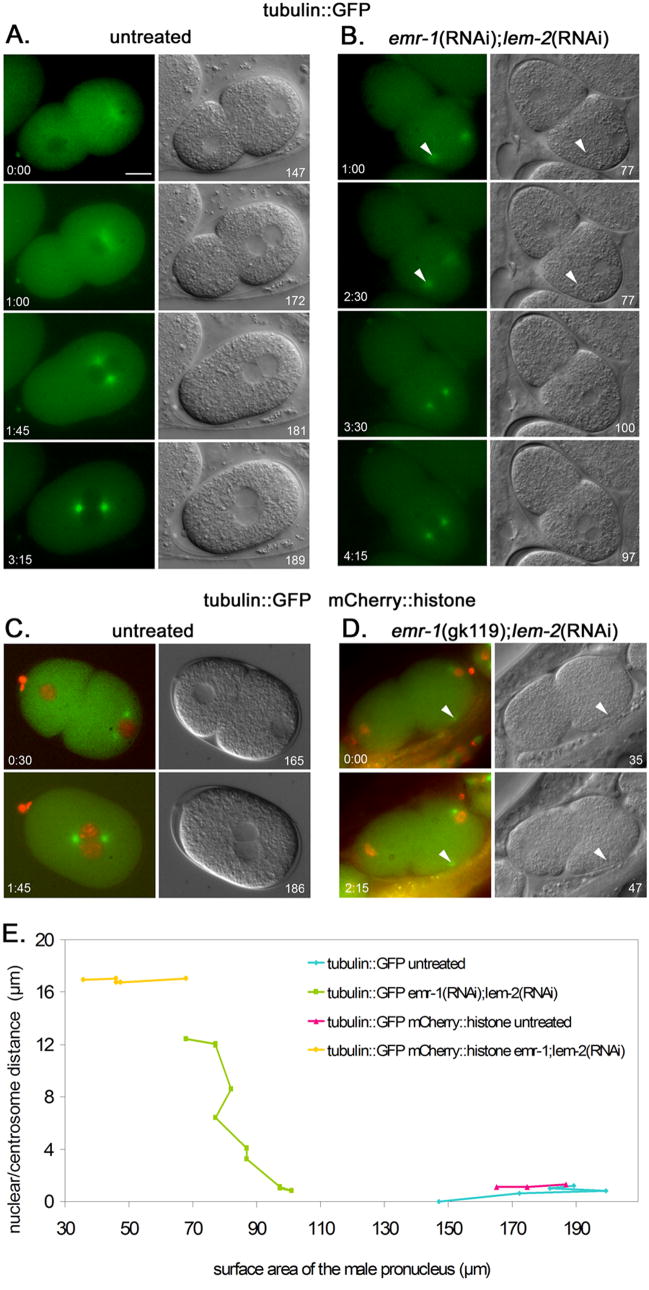
Time-lapse imaging of centrosome attachment in emr-1; lem-2 defective embryos. (A) An untreated AZ244 (tubulin::GFP) embryo. (B) An emr-1(RNAi); lem-2(RNAi) embryo in AZ244. (C) An untreated OD57 (tubulin::GFP; histone::mCherry) embryo. (D) An emr-1(gk119); lem-2(RNAi) embryo in OD57. Images from a time series are shown; fluorescent images from the green channel (A–B) or the green and red merged (C–D) are shown on the left and corresponding DIC images are on the right. Time in minutes:seconds from the start of filming and surface area of male pronuclei in μm2 are shown in the corners of panels. Arrowheads mark centrosomes detached form the nucleus. Posterior is right and the scale bar is 10 μm. (E) A plot of the surface area of male pronuclei versus the distance between the edge of the nucleus and the center of the centrosome. Points from different time points are connected. The same four embryos used in A–D are plotted.
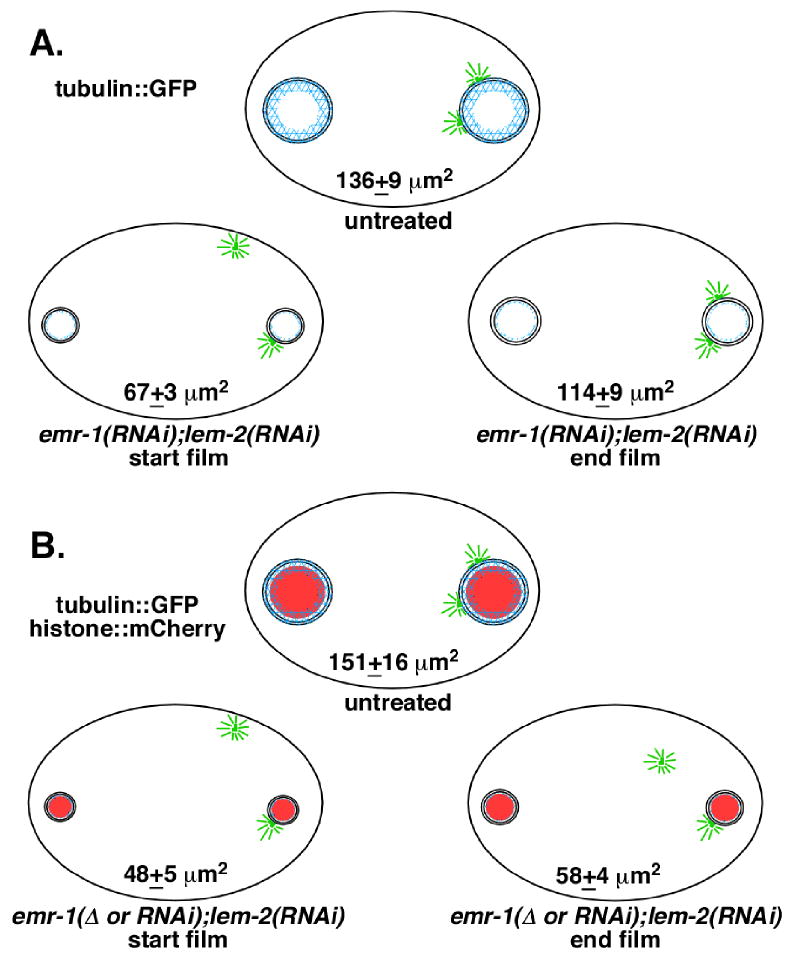
Fig. 6.
The timing of centrosome attachment is correlated with the surface area of the nucleus. Newly fertilized embryos are drawn with male pronuclei on the right (posterior). Average surface areas of male pronuclei with associated standard errors are as indicated. Centrosomes are drawn in green, the nuclear lamina in blue, and the mCherry chromatin in red. (A) Filmed embryos in the tubulin::GFP strain AZ244. Five untreated and seven emr-1(RNAi); lem-2(RNAi) mutant embryos were used for this data set. The sizes of male pronuclei were measured at the start of filming when only one centrosome was attached and then later when both centrosomes became attached, prior to pronuclear migration. The nuclei grew significantly between the two time points (p<0.0001). (B) Filmed embryos in the tubulin::GFP; histone::mCherry strain OD57. Six untreated and six emr-1(gk119); lem-2(RNAi) or emr-1(RNAi); lem-2(RNAi) mutant embryos were used in this data set. At the start of filming the mutant embryos were significantly smaller than the tubulin::GFP dsRNA-treated embryos (p=0.002) and the centrosomes were never captured by the male pronuclei.
In the other classes where the male pronucleus failed to capture the centrosome, chromosomal segregation defects would be predicted to follow. In one case, the female pronucleus captured the detached centrosome, then made contact with the second centrosome without fully associating with the male pronucleus. A spindle was formed around only the female pronucleus, resulting in chromosomes outside of the mitotic spindle (Fig. 7A).
We were able to observe that centrosome duplication occurs very close to the male pronucleus in the lamina defective embryos. Two of the films began early enough so that we initially saw both centrosomes attached next to each other in close vicinity to the forming male pronucleus, but one centrosome then quickly drifted away from the male pronucleus (Fig. 7B). In wildtype, male pronuclei start out just as compact as lamina defective pronuclei, although they expand much more quickly. It was previously reported that immediately after centrosome duplication, in a low percentage of embryos, one centrosome is detached (Malone et al., 2003). We did not see any untreated embryos with a detached centrosome, but we were using different markers, which might not have been incorporated into the centrosome from the zygote cytoplasm as early, and we may not have been filming early enough.
It is unlikely that the delay or failure of centrosomes to attach to the male pronucleus was due to phototoxicity. Untreated tubulin::GFP embryos were filmed under the same blue light conditions as above and emr-1(RNAi); lem-2(RNAi) embryos in N2 wildtype with no exposure to ultraviolet light. In both cases we were able to film long enough for pronuclear migration and at least the start of the first mitosis. In wildtype we were easily able to film to the four-cell stage of development, much longer than when we observed development in the tubulin::GFP background (data not shown). Furthermore, fixed embryos that were never filmed had similar centrosome detachment phenotypes (Fig. 1). After a few minutes our filmed tubulin::GFP embryos may have shown some effects of phototoxicity. However, any effects were limited, since even in wildtype depletion of emr-1 and lem-2 severely delayed pronuclear migration.
Further support for the hypothesis that over-condensed male pronuclei can only attach to one centrosome came from data generated by filming zygotes from emr-1(gk119 or RNAi); lem-2(RNAi) treated hermaphrodites expressing tubulin::GFP and histone::mCherry. Untreated, the surface area of nuclei in the tubulin::GFP; histone::mCherry line are no different than the tubulin::GFP line (Fig. 6). Fortuitously, the combination of the nuclear lamina disruption and the addition of the mCherry tag to a histone (or something else in the transgenic background) led to a more condensed male pronucleus than dsRNA treatment in the tubulin::GFP strain. We filmed six emr-1(RNAi); lem-2(RNAi) or emr-1(gk119); lem-2(RNAi) treated embryos in the tubulin::GFP; histone::mCherry background with a centrosome detached from the male pronucleus (a representative embryo with a detached centrosome is shown in Fig. 5D and Supplemental Movie 4, an untreated embryo is shown in Fig. 5C and Supplemental Movie 3, the statistical data is in Fig. 6B, and the complete data set is in Sup. Fig. 4C–D). While male pronuclei grew a little in size, they did not approach the average size that the pronuclei grew to at the time of centrosome attachment in the tubulin::GFP background with unlabeled histones (Fig. 6). Subsequently, none were able to capture the second centrosome in a similar time frame to the dsRNA-depleted embryos in the unmodified histone strain. These data lend further support to the nuclear size model.
The mechanism of how the histone::mCherry strain OD57 acted to enhance the small pronuclear defect in the emr-1(RNAi); lem-2(RNAi) embryos is unknown. To test whether the enhancement was due to an overexpression of tagged histone, emr-1(RNAi); lem-2(RNAi) zygotes in the β-tubulin::GFP; histone::GFP (TY3558) background were examined. Five embryos with a single detached centrosome from the male pronucleus were filmed. In three of the five embryos, the male pronucleus grew and the second centrosome became attached before pronuclear migration (Supplemental Movie 5). These male pronuclei started at an average surface area of 77 ± 8.5 μm2 (average ± standard error) and grew to 126 ± 15 μm2 at the time the second centrosome became attached. In the other two embryos, the female pronucleus met the male pronucleus before capturing the second centrosome. The size of the male pronuclei and the centrosome detachment phenotype in the histone::GFP background were statistically the same as the data in the wild-type histone background and statistically larger than in the histone::mCherry background (compare to numbers in Fig. 6). Similar results were seen in baf-1(RNAi) embryos in the histone::GFP background (data not shown). We therefore conclude that either the histone::mCherry fusion or something else in the OD57 strain enhanced the small pronuclear phenotype of the nuclear envelope component knockdowns.
To test whether a large pronucleus with a defective lamina was able to attach to both centrosomes, we treated tetraploid animals with lmn-1(RNAi) by feeding. We reasoned that sperm from tetraploid lines would contain twice the normal amount of chromatin and therefore have a bigger surface area (Madl and Herman, 1979). A potential caveat to this experiment is that the volume of the oocyte is also scaled up in tetraploid lines. Fixed embryos prior to pronuclear migration were stained for tubulin and chromatin and were analyzed as above. The RNAi feeding approach was validated by observing severe chromosomal segregation defects in fixed embryos on the same slides, high embryonic lethality, and loss of LMN-1::YFP expression in parallel experiments (data not shown). Male pronuclei from untreated tetraploid hermaphrodites had an average surface area of 235 ± 22 μm2 (n=15; ± std. error), while male pronuclei from tetraploid hermaphrodites fed lmn-1(RNAi) had an average surface area of 106 ± 10 μm2 (n=18), smaller than untreated (p<0.0001), but larger than lamina defective fixed pronuclei from diploids (65 ± 4.5 μm2; n=58; p<0.0001) (Fig. 8A). 15/18 fixed embryos from lmn-1(RNAi) tetraploid hermaphrodites observed prior to pronuclear migration had both centrosomes attached normally to the male pronucleus (Fig. 8B) as compared to only 24/58 of the fixed lamin defective embryos from diploid hermaphrodites. 1/18 of the embryos from the tetraploid background had a single detached centrosome and 2/18 had both centrosomes detached (Fig 8C), a phenotype we never saw in the diploid background. The three pronuclei with centrosome detachment phenotypes were not significantly smaller than the others. We also noticed that the pronuclei from tetraploid animals often appeared to be in a prophase-like condensed state. Furthermore, pronuclei from tetraploids were often misshapen as compared to wildtype, suggesting that there might be other defects in the nuclear envelope other than size.
Fig. 8.
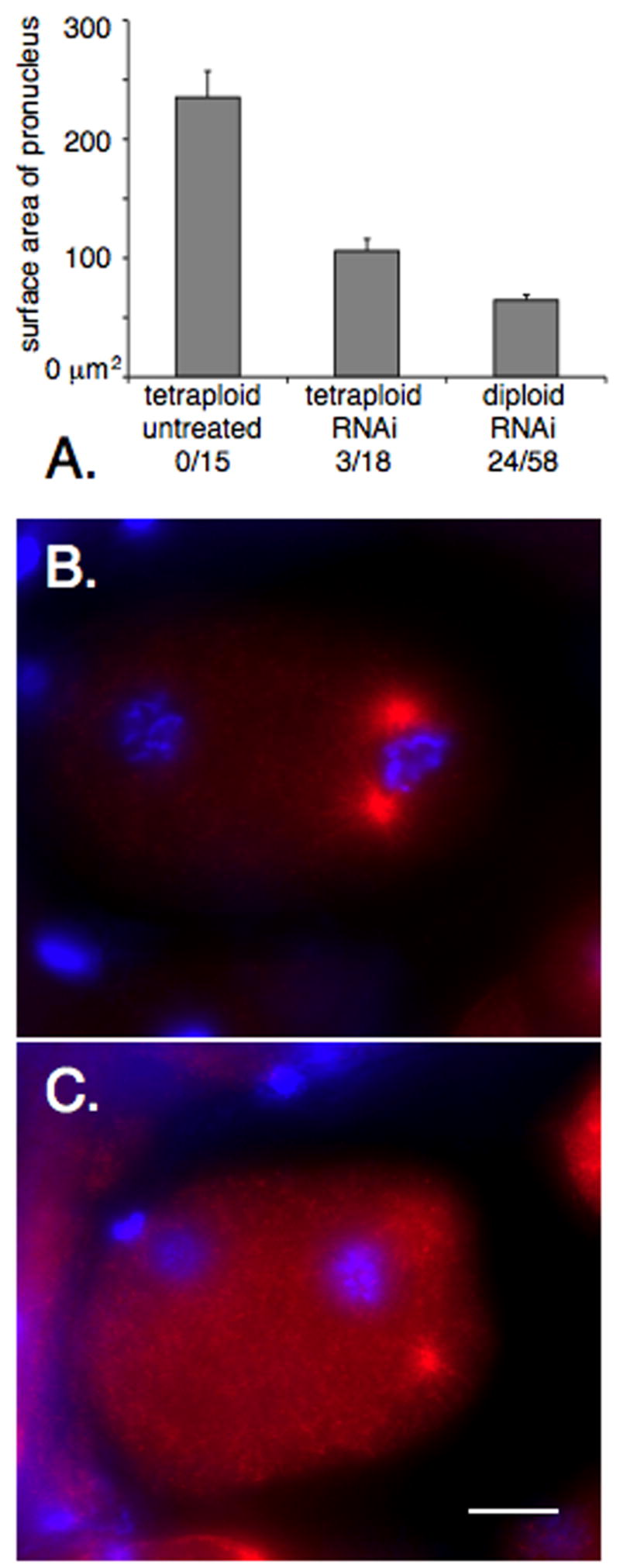
Both centrosomes usually attach to male pronuclei in lmn-1(RNAi) embryos from a tetraploid background. (A) Size of male pronuclei from fixed images prior to pronuclear migration in untreated embryos from tetraploid hermaphrodites, embryos from tetraploid lmn-1(RNAi) hermaphrodites, or embryos treated with dsRNA against lmn-1, baf-1, or emr-1 and lem-2. Error bars show standard error. Each of the three samples are statistically different (p,0.0001). The number of embryos with detached centrosomes out of the total number observed are shown below each bar. (B–C) Fixed embryos from a lmn-1(RNAi) treated tetraploid hermaphrodite stained for tubulin in red and chromatin in blue. (B) An embryo with both centrosomes attached to the male pronucleus; 15 of 18 observed embryos were in this class. (C) An embryo with both centrosomes detached (one is slightly out of frame); 2 of 18 embryos. Posterior is right and the scale bar is 10 μm.
Small nuclei can only interact with single microtubule asters leading to the centrosome detachment defect
To explain the centrosome detachment phenotype in embryos with reduced nuclear size, we proposed two possible models. In the centrosome model, the attachment of one centrosome itself inhibits the second one from attaching to the nucleus. In the aster microtubule model, the association of microtubules from one centrosome with the nucleus inhibits the microtubules from the second aster from interacting with the nucleus. To test these models, we used dsRNA-treated fem-1 worms to mate with emb-27 males, which have sperm with centrosomes but no pronuclei (Sadler and Shakes, 2000). The centrosome model predicts that the two centrosomes should move to the nucleus with similar speed until one attaches and inhibits the attachment of the other. The microtubule model predicts that only one centrosome should undergo significant movement towards the female pronucleus.
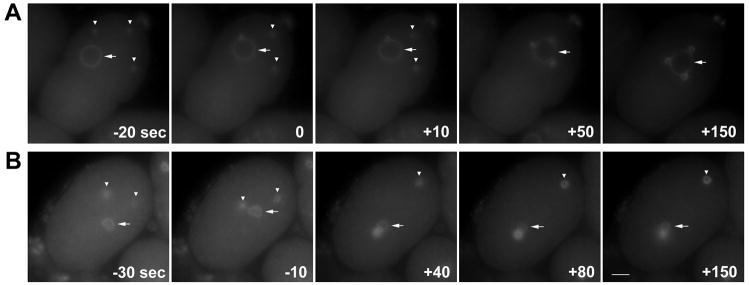
Mutations in the APC component emb-27 produce male sperm without nuclei that are still able to fertilize embryos, establish polarity, and contribute a centrosome to the embryo (Sadler and Shakes, 2000). In normal oocytes from feminized fem-1(hc17) mothers fertilized with anucleate emb-27(g48) sperm, the centrosome from the sperm duplicates after female meiosis and nuclear envelope formation is complete. We filmed the behavior of centrosomes over time following either a ZYG-12::GFP marker or tubulin::GFP; histone::GFP markers (data not shown). Often, emb-27(g48) embryos contribute more than one centrosome (Diane Shakes, personal communication). We frequently observed more than two centrosomes in our assay. Regardless of the number of centrosomes, in normal embryos fertilized with anucleate sperm (n=8), all the centrosomes migrated towards and connected with the female pronucleus at with the same dynamics (Fig. 9A). All the centrosomes became closely associated with the female pronucleus within a minute of one another. However, in female pronuclei that were small due to dsRNA against lmn-1 (n=6), npp-6 (n=2), or emr-1; lem-2 (n=3), only one centrosome migrated towards the female pronucleus (Fig. 9B) while the other remained detached for the duration of the film (at least two minutes, and sometimes much longer). These data support the aster microtubule model that the surface area of the nucleus available to interact with the microtubule aster is limiting the number of centrosomes that can attach to the nucleus. A normal sized female pronucleus is of sufficient size to interact with at least three centrosomes (Fig. 9A). However, when the surface area is small, only one aster is able to interact and thus only one centrosome can attach.
Fig. 9.
Centrosome attachment to female pronuclei in embryos fertilized with anucleate sperm. Time-lapse images of embryos from fem-1(hc17) mothers fertilized by anucleate emb-27(g48) sperm are shown. An untreated embryo (A) and an embryo from a mother treated with dsRNA against lmn-1 (B) are shown. Centrosomes and the nuclear envelope were followed by ZYG-12::GFP. Time in seconds is shown; t=0 when the first centrosome is attached in close proximity to the nuclear envelope. Arrows mark the nucleus and arrowheads mark detached centrosomes.
Discussion
The size of the male pronucleus predicts how many centrosomes will attach
Sperm contribute the sole centrosome to the newly fertilized zygote. Close attachment between the male pronucleus and the centrosome is required for normal pronuclear migration towards one another and synchronous entry of both the male and female pronucleus into the first mitotic event (Hachet et al., 2007). We show that disruption of either nuclear lamina components or nuclear import machinery leads to a unique centrosome detachment phenotype where exactly one centrosome is detached. In other published examples of centrosome detachment defects, both centrosomes detach at an equal frequency (Askjaer et al., 2002; Galy et al., 2006; Gonczy et al., 1999; Malone et al., 2003; Patterson et al., 2004; Robinson et al., 1999). With the exception of rare pronuclei from tetraploid animals or in embryos that overexpress LIS-1::GFP, in which many other things are likely disrupted, we only observe a single centrosome detached in our dsRNA treatments while the other centrosome remained attached normally.
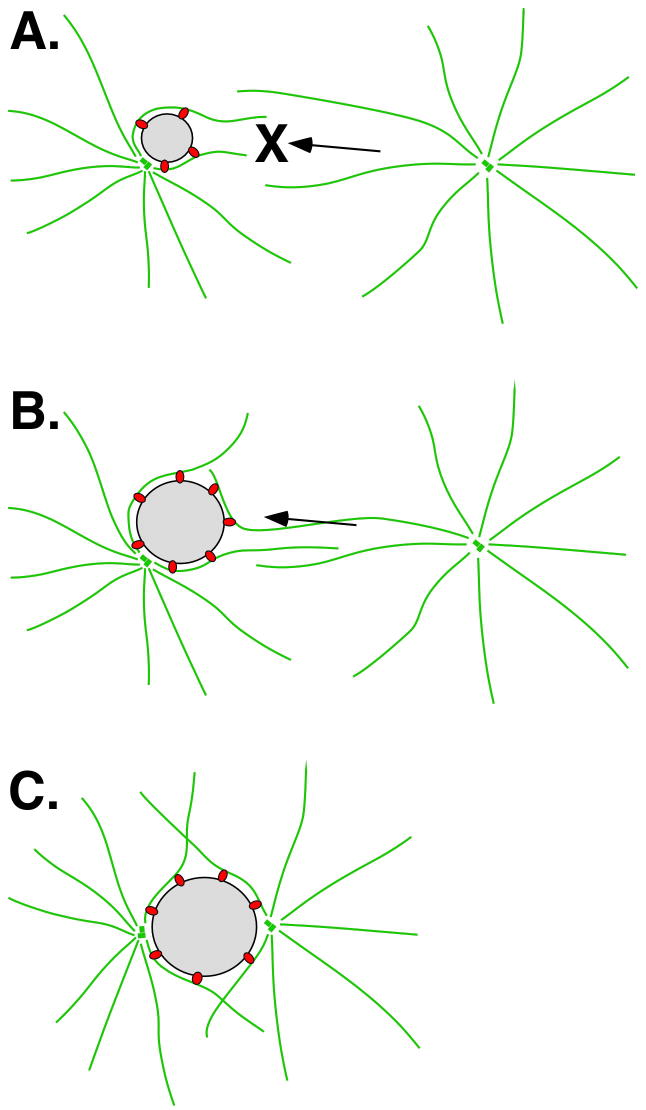
Our data suggest a model where centrosome attachment to the nuclear membrane is dependent on the surface area of the pronucleus (Fig. 10). In our model, a small, over-condensed nucleus is able to attach only a single centrosome. Time-lapse imaging of the centrosome detachment phenotype revealed that in dsRNA-treated zygotes, the male pronucleus slowly grew and reached a certain surface area threshold, at which time the detached centrosome was captured (Figs. 5–6). To test the size-dependent aspects of our model, emr-1(RNAi); lem-2(RNAi) zygotes were observed in various backgrounds; ones that approximated wild-type and ones that increased or decreased the size of the mutant pronuclei. The time-lapse data captured in tubulin::GFP or tubulin::GFP; histone::GFP backgrounds closely approximated wildtype. However, lamina disruption in transgenic animals expressing histone::mCherry led to synthetically smaller pronuclei. Alternatively, we examined centrosome attachment in male pro-nuclei from lmn-1(RNAi) tetraploid animals, which were significantly larger. As the model predicts, centrosomes remained detached at a much higher frequency from the smaller histone::mCherry pronuclei and both centrosomes were more likely attached to larger pronuclei from a tetraploid line. This suggests that the disruptions employed here to generate the single detached centrosome phenotype act through the same mechanism—by reducing the growth rate and therefore the surface area of the male pronucleus. We conclude that an intact nuclear lamina is required for normal decondensation of pronuclei and subsequent attachment of the centrosome to the outer nuclear membrane of male pronuclei.
Fig. 10.
The nuclear size model for centrosome attachment. (A) A lamina-defective, small male pronucleus (grey circle) has a limited amount of dynein (red) and is therefore only able to interact with only a single centrosome and its microtubule aster (green) while the other is blocked (X). (B) As the pronucleus grows past a certain threshold, additional dynein is recruited and some is now able to interact with aster microtubules from the second aster. (C) Once the nucleus has sufficient surface area and dynein to interact with both asters, both centrosomes become attached to the nucleus. Dynein on the outer nuclear surface pushes the centrosomes to opposite sides of the nucleus. See text for details.
What is rate limiting for centrosome attachment to small pronuclei?
We hypothesized that a reduced surface area of a mutant pronucleus limits the amount of a factor or factors on the outer nuclear membrane that interact with microtubules and/or centrosomes. These factors could be interacting directly with the centrosome or through the microtubule asters associated with centrosomes. The behavior of centrosomes from the anucleate sperm experiment (Fig. 9) suggested that the limiting factors on the pronucleus interact with aster microtubules and not the centrosome itself. In wild-type zygotes fertilized with anucleate sperm, all centrosomes migrated simultaneously towards female pronuclei and attached soon after one another. However, only one centrosome migrated towards and attached to a small, lamina defective female pronucleus. We therefore propose that a small pronucleus is limited in some factor or factors (red circles in Fig. 10) that interact with the microtubule aster at some distance away from the centrosome. Once a small nucleus is interacting with one aster, the second aster is blocked (Fig 10A). As the nucleus grows in surface area, more factors are proportionally recruited to the nuclear envelope and room for the second aster to interact becomes available. Once the second aster interacts with the growing nucleus, the centrosome rapidly moves towards and attaches to the nucleus (Fig. 10B–C).
The limiting factor on the outer nuclear membrane that interacts with microtubule asters is most likely dynein. Dynein at the nuclear envelope has been previously proposed to be involved in centrosome attachment and separation (Gonczy et al., 1999; Malone et al., 2003). ZYG-12 is required for dynein localization to the nuclear periphery, and dynein is required for LIS-1 localization (Cockell et al., 2004; Malone et al., 2003). Both ZYG-12 and LIS-1 localized to normal concentrations to the nuclear periphery of lamina-defective, small pronuclei, suggesting that dynein does too (Fig. 3). We propose that the overall amounts of dynein on the surface of a small nuclear envelope would fail to capture enough aster microtubules to keep both centrosome attached. One aster would “win,” blocking the other from interacting with the limited pool of dynein on the nuclear surface. As the nuclear envelope grows, so too would the number of dynein complexes, although the concentration of dynein would remain constant. At a certain nuclear surface area threshold, enough dynein would be free of the first aster to interact with the second (Fig. 10).
Our surface-area-dependent centrosome-attachment model could explain mechanisms of pronuclear migration in the ctenophore Beroe ovata. Beroe embryos are very large (about 1 mm) and are often fertilized by multiple sperm; embryos develop normally even when fertilized with more than ten sperm. Each male pronucleus remains very condensed and forms a large (50–70 μm in diameter) microtubule aster. The female pronucleus migrates long distances to interact and fuse with a single male pronucleus prior to the first mitotic division (Carre et al., 1991; Carre and Sardet, 1984; Rouviere et al., 1994). During pronuclear migration, the male pronucleus is about three μm in diameter compared to the female pronucleus that is 15–20 μm in diameter, which translates into to a nuclear surface area in the female pronucleus that is 25–45 times larger than that of the male pronucleus (Carre et al., 1991). We propose that one reason Beroe male pronuclei remain small is so that they cannot interact with an aster other than its own. Meanwhile, the female pronucleus has a large surface area so that it can interact with microtubules at a low concentration far from the center of the male asters. Dynein on the female pronucleus would then interact with microtubules and move it towards a male pronucleus. At some point, dynein on the surface of the female pronucleus interacts predominantly with microtubules from a single male pronuclear-associated aster, and the female moves rapidly towards and fuses with that male pronucleus.
Multiple mechanisms for centrosome attachment throughout development
The centrosome detachment phenotype presented in this paper is a result of a defective nuclear envelope. However, normal C. elegans sperm, and therefore male pronuclei immediately after fertilization, lack a nuclear envelope, but nonetheless keep the centrioles attached to the nucleus (Ward et al., 1981; Wolf et al., 1978). Thus, the nuclear envelope-based mechanism described here cannot be the only mechanism for attaching centrosomes to the male pronucleus. Spermatid nuclei and centrioles are surrounded by an electron-dense halo likely made of ribonucleoproteins (Ward et al., 1981). Mutations in fer-2, fer-3, and fer-4 all disrupt the perinuclear halo, but the molecular identity of these proteins is unknown (Ward et al., 1981). Mutations in spe-11 are paternal effect embryonic lethal and have a disrupted sperm perinuclear halo (Hill et al., 1989). The novel SPE-11 protein localizes to the sperm perinuclear halo and diffuses into the cytoplasm of the embryo shortly after fertilization (Browning and Strome, 1996). Perhaps the perinuclear halo of sperm is made of an unknown RNA and the fer-2, -3, -4 and spe-11 gene products. Such a ribonucleoprotein complex could function to keep the centrioles closely attached to the tightly condensed male pronucleus immediately after fertilization. Then, the nuclear envelope begins to form, the nucleus begins to decondense, the centrosomes begin to mature by nucleating microtubules, and the perinuclear halo dissolves into the cytoplasm. It is at this time that components of the nuclear envelope take over in the mechanism of centrosome attachment that we propose. Nematodes appear unique in that their sperm lack a nuclear envelope (Wolf et al., 1978). Thus, the relative contributions of these two mechanisms of attachment, the RNA halo versus the new nuclear envelope, vary among organisms and the nuclear envelope-based mechanism we uncover here is likely to play an even more important role in other phyla.
A third mechanism for centrosome attachment to the nucleus must exist for later in embryogenesis. As the embryo divides, cell and nuclear volumes drastically decrease. In fact there is correlation between the nuclear and cytoplasmic volumes (Neumann and Nurse, 2007). Furthermore, ZYG-12 is not required for nuclear/centrosomal attachment in the later stages of embryogenesis and development. The mechanisms that take over are not known, but because the cytoplasm is so much smaller and centrosomes have an inherent property of self-centering (Holy et al., 1997), one would imagine that it becomes much simpler to keep the centrosome close to the nucleus in the center of most cells. We therefore conclude that the mechanism of centrosome attachment described here only operates during the first few cell divisions of embryogenesis, a critical time to precisely control nuclear and spindle positioning within a giant cytoplasm during rapid and precise cell divisions.
The role of the nuclear lamina in centrosome attachment
We have demonstrated a role for the nuclear lamina in centrosome attachment during C. elegans development. Disruption of the nuclear lamina by dsRNA against two LEM domain proteins (emr-1 and lem-2), the BAF homologue (baf-1), or the single C. elegans lamin (lmn-1) leads to a specific and reproducible phenotype in the newly fertilized C. elegans zygote. The male pronucleus is over-condensed and the attachment between one centrosome and the nuclear envelope fails. This likely leads to chromosomal segregation defects in the first mitotic division and embryonic lethality. We also showed that RNAi against components of the nuclear import machinery leads to the same small-nuclei, centrosome-detachment phenotype. It is likely that defects in nuclear import disrupt the nuclear lamina, as shown in yeast (King et al., 2006). Therefore, all the treatments employed here to make pronuclei smaller likely disrupt centrosome positioning through the same mechanism. We propose that defects in nuclear import disrupt the formation of the nuclear lamina and male pronucleus decondensation. This leads to a small pronucleus at the time of centrosome duplication and as the microtubule aster is beginning to form. The mechanism of how a defective nuclear lamina leads to an over condensed and small nucleus is unknown. One possibility is that the nuclear lamina recruits a histone acetyltransferase to decrease compaction of chromatin. The inner nuclear membrane protein human SUN1 has recently been shown to function in chromatin decondensation by such a mechanism (Chi et al., 2007).
Our data showed that the overall structure of the nuclear envelope in our nuclear lamina-disrupted nuclei was surprisingly normal. Nuclear pores localized normally in the small nuclei. This is in contrast to what has been previously reported for lmn-1(RNAi) or baf-1(RNAi), where nuclear pores cluster (Liu et al., 2000; Margalit et al., 2005). Liu et al. (2000) state that “many (but not all)” of lmn-1(RNAi) cells have displaced nuclear pores. At the early stage we observed, we conclude that lmn-1 is not required to evenly space nuclear pores. Perhaps as development progresses, the role of the nuclear lamina in maintaining the spacing of nuclear pores becomes more important. An alternative explanation to the differences between the results reported here and those of Liu et al. (2000) and Margalit et al. (2005) is that our dsRNA treatment was less severe than theirs. However, this is unlikely, as dsRNA was injected, the observed terminal lethal phenotype was the same, and lamin was depleted below detectable levels of LMN-1::YFP transgenic animals treated with lmn-1(RNAi). The outer nuclear membrane protein ZYG-12 and the inner nuclear membrane protein SUN-1 also localized normally in nuclear lamina defective embryos. This was previously reported for lmn-1(RNAi) (Fridkin et al., 2004), but we show it to be also true in emr-1(RNAi); lem-2(RNAi) embryos. The mechanism of SUN-1 and ZYG-12 localization to and maintenance at the nuclear envelope remains to be determined.
Mutations in components of the human nuclear lamina, most notably emerin and lamin, lead to a wide variety of crippling diseases termed laminopathies. These diseases disrupt a number of different tissues, but the pathologies of how mutations in the nuclear lamina lead to disease are poorly understood (Mounkes et al., 2003; Somech et al., 2005; Worman and Bonne, 2007). A strong centrosome detachment phenotype has been reported for interphase emerin deficient fibroblasts from Emery Dreifuss muscular dystrophy patients (Salpingidou et al., 2007). Salpingidou et al. (2007) show that emerin is present in both the inner and outer nuclear membrane and show that emerin interacts with tubulin. They propose a model where emerin on the outer nuclear membrane directly interacts with microtubules to attach the centrosome to the nuclear envelope (Salpingidou et al., 2007). A similar phenotype has recently been described for mouse embryonic fibroblasts deficient in Lamin A/C (Lee et al., 2007). It would be interesting to determine whether the surface area of the nuclei in these cells is related to the centrosome detachment phenotype. Although rough examinations of the limited data presented in the figures does not support a drastic reduction in nuclear size, these complete data were not reported (Salpingidou et al., 2007). One major difference in this finding and the ones reported here, is that in C. elegans, emerin functioned redundantly with the MAN1 and Lem2 homologue and no centrosome detachment phenotypes were observed in emr-1 mutant animals (also see Lee et al., 2000). We propose that emerin functions in at least two pathways of differing importance to different cell types to attach centrosomes to the outer nuclear membrane; as a member of the outer nuclear membrane where it could attach directly to microtubules (Salpingidou et al., 2007) and as described here as a component of the nuclear lamina where it contributes to the growth of the nuclear envelope and surface area of the nucleus.
Supplementary Material
Time lapse images of an AZ244 (tubulin::GFP) untreated embryo. Images were taken at 15-second intervals for 7 minutes. DIC and green fluorescent images were merged. Posterior is right.
Time lapse images of an AZ244 emr-1(RNAi); lem-2(RNAi) embryo. Images were taken at 15-second intervals for 10.5 minutes. DIC and green fluorescent images were merged. Posterior is right.
Time lapse images of an OD57 (tubulin::GFP; histone::mCherry) untreated embryo. Images were taken at 15-second intervals for 5.75 minutes. Green and red fluorescent images were merged. Posterior is right.
Time lapse images of an OD57 emr-1(gk119); lem-2(RNAi) embryo. Images were taken at 15-second intervals for 13.5 minutes. Green and red fluorescent images were merged. Posterior is right.
Time lapse images of an TY3558 emr-1(RNAi); lem-2(RNAi) embryo. Images were taken at 30-second intervals for 11 minutes. Green fluorescent images show tubulin::GFP and histone::GFP. Posterior is left.
The centrosome detachment phenotype disrupts γ-tubulin localization. A live lmn-1(RNAi) embryo expressing γ-tubulin::GFP is shown. DIC and green fluorescent images are overlaid. The arrow shows a centrosome detached from the nucleus. Posterior is right and the scale bar is 10 μm.
In embryos with detached centrosomes, nuclear pores localize to the nuclear envelope normally. One-cell embryos prior to pronuclear migration are shown. In all panels, embryos are simultaneously stained for tubulin and nuclear pores in red and chromatin in blue. Embryos are from hermaphrodites injected with dsRNA against the following: (A) no RNA wild-type control, (B) emr-1 (C) lem-2, (D) emr-1 and lem-2, (E) lmn-1, (F) baf-1. In (E) the second centrosome is out of frame. In (D–F) arrows mark centrosomes detached from male pronuclei. Posterior is right and the scale bar is 10 μm.
SUN-1::GFP localizes normally in embryos with a detached centrosome phenotype. (A) an untreated newly fertilized embryo expressing SUN-1::GFP. (B) a newly fertilized embryo from a lmn-1(RNAi) mother with a detached centrosome expressing SUN-1::GFP at normal levels. GFP is green (A–B), tubulin is red (A′–B′) and DNA is blue in the merge (A″–B″).
Time-lapse imaging of centrosome attachment in emr-1; lem-2 defective embryos. The data for the representative data shown in Fig. 5 and the statistical data shown in Fig. 6 are shown here. (A) Untreated AZ244, tubulin::GFP, embryos. (B) emr-1(RNAi); lem-2(RNAi) embryos in the AZ244 background. (C) Untreated OD57, tubulin::GFP histone::mCherry, embryos. (D) emr-1(RNAi or gk119); lem-2(RNAi) embryos in the OD57 background. Plots of the surface area of male pronuclei versus the distance between the edge of the nucleus and the center of the centrosome are shown on the left. Each colored line represents one series of time-lapse images. The beginning and end of the green arrow (B, D) and the associated dashed and solid lines represent the average size of emr-1(RNAi); lem-2(RNAi) male pronuclei in the AZ244 background at the start of filming and at the time of capture of the second centrosome respectively. The beginning and end of the red arrow (B, D) and the associated dashed and solid lines represent the average size of emr-1(RNAi or gk119); lem-2(RNAi) male pronuclei in the OD57 background at the start and end of filming respectively. On the right, the surface area of each male pronucleus at the beginning of filming, when both the nucleus and detached centrosome was observed (B, D), or in early pronuclear migration (A, C), is shown. The surface area of male pronuclei is also shown at the time the female pronucleus meets the male (A, C), at the time the second centrosome is attached by the male pronucleus (B) or at the end of filming (D). In the cases when the detached centrosome failed to attach, the embryo was filmed until there was no further change in the embryo’s appearance (the embryo was likely dead). In two embryos (marked with a †), the detached centrosome was captured by the female pronucleus. The embryos shown in Fig. 5 and Supplemental Movies 1–4 are marked with an *.
Acknowledgments
We thank Lesilee Rose and Frank McNally for reagents, advice on filming live embryos, and comments on the manuscript, Michael Paddy for assistance with the confocal microscope, and Diane Shakes, Wendy Hanna-Rose, and Evelyn Houliston for helpful discussions. We thank Matt McGee, Kang Zhou, and the other members of the Starr and Malone labs for helpful comments and support. We thank the anonymous reviewers for many suggestions. This work was supported in part by a Basil O’Conner Starter Scholar Research Award grant number 5-FY05-45 from the March of Dimes Foundation to D.A.S. and grant 1 R01 GM073874-1 from the National Institutes of Health to D.A.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahringer J. In: Reverse Genetics. community TCer., editor. WormBook; 2006. [Google Scholar]
- Askjaer P, et al. Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol Biol Cell. 2002;13:4355–70. doi: 10.1091/mbc.E02-06-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basham SE, Rose LS. The Caenorhabditis elegans polarity gene ooc-5 encodes a Torsin-related protein of the AAA ATPase superfamily. Development. 2001;128:4645–56. doi: 10.1242/dev.128.22.4645. [DOI] [PubMed] [Google Scholar]
- Bornens M. Is the centriole bound to the nuclear membrane? Nature. 1977;270:80–2. doi: 10.1038/270080a0. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning H, Strome S. A sperm-supplied factor required for embryogenesis in C. elegans. Development. 1996;122:391–404. doi: 10.1242/dev.122.1.391. [DOI] [PubMed] [Google Scholar]
- Carre D, et al. In vitro fertilization in ctenophores: sperm entry, mitosis, and the establishment of bilateral symmetry in Beroe ovata. Dev Biol. 1991;147:381–91. doi: 10.1016/0012-1606(91)90296-f. [DOI] [PubMed] [Google Scholar]
- Carre D, Sardet C. Fertilization and early development in Beroe ovata. Dev Biol. 1984;105:188–95. doi: 10.1016/0012-1606(84)90274-4. [DOI] [PubMed] [Google Scholar]
- Chi YH, et al. Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in de-condensation of mitotic chromosomes. J Biol Chem. 2007 doi: 10.1074/jbc.M703098200. [DOI] [PubMed] [Google Scholar]
- Cockell MM, et al. lis-1 is required for dynein-dependent cell division processes in C. elegans embryos. J Cell Sci. 2004;117:4571–82. doi: 10.1242/jcs.01344. [DOI] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Cyclin E-Cdk2 temporally regulates centrosome assembly and establishment of polarity in Caenorhabditis elegans embryos. Nat Cell Biol. 2006;8:1441–7. doi: 10.1038/ncb1511. [DOI] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Franz C, et al. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J. 2005;24:3519–31. doi: 10.1038/sj.emboj.7600825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–30. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Fridkin A, et al. Matefin, a Caenorhabditis elegans germ line-specific SUN-domain nuclear membrane protein, is essential for early embryonic and germ cell development. Proc Natl Acad Sci U S A. 2004;101:6987–92. doi: 10.1073/pnas.0307880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, et al. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr Biol. 2006;16:1748–56. doi: 10.1016/j.cub.2006.06.067. [DOI] [PubMed] [Google Scholar]
- Galy V, et al. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell. 2003;14:5104–15. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes ER, et al. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–63. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Gonczy P, et al. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;147:135–50. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjanacz M, et al. What can Caenorhabditis elegans tell us about the nuclear envelope? FEBS Lett. 2007 doi: 10.1016/j.febslet.2007.03.052. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, et al. The expression, lamin-dependent localization and RNAi depletion phenotype for emerin in C. elegans. Cell Sci. 2002;115:923–9. doi: 10.1242/jcs.115.5.923. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, et al. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Gunsalus KC, et al. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature. 2005;436:861–5. doi: 10.1038/nature03876. [DOI] [PubMed] [Google Scholar]
- Hachet V, et al. Centrosomes promote timely mitotic entry in C. elegans embryos. Dev Cell. 2007;12:531–41. doi: 10.1016/j.devcel.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Hill DP, et al. A sperm-supplied product essential for initiation of normal embryogenesis in Caenorhabditis elegans is encoded by the paternal-effect embryonic-lethal gene, spe-11. Dev Biol. 1989;136:154–66. doi: 10.1016/0012-1606(89)90138-3. [DOI] [PubMed] [Google Scholar]
- Holy TE, et al. Assembly and positioning of microtubule asters in microfabricated chambers. Proc Natl Acad Sci U S A. 1997;94:6228–31. doi: 10.1073/pnas.94.12.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–7. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kimble J, Ward S. Germ-line Development and Fertilization. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1988. pp. 191–214. [Google Scholar]
- King MC, et al. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–7. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- Kohara Y. Large scale analysis of C. elegans cDNA. Tanpakushitsu Kakusan Koso. 1996;41:715–20. [PubMed] [Google Scholar]
- Kracklauer MP, et al. Drosophila klaroid Encodes a SUN Domain Protein Required for Klarsicht Localization to the Nuclear Envelope and Nuclear Migration in the Eye. Fly. 2007;1:75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- Lee JS, et al. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J. 2007;93:2542–52. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, et al. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol Biol Cell. 2000;11:3089–99. doi: 10.1091/mbc.11.9.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, et al. Essential roles for caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell. 2000;11:3937–47. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, et al. MAN1 and emerin have overlapping function(s) essential for chromosome segregation and cell division in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2003;100:4598–603. doi: 10.1073/pnas.0730821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl JE, Herman RK. Polyploids and sex determination in Caenorhabditis elegans. Genetics. 1979;93:393–402. doi: 10.1093/genetics/93.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, et al. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–36. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- Margalit A, et al. Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc Natl Acad Sci U S A. 2005;102:3290–5. doi: 10.1073/pnas.0408364102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K, et al. Katanin controls mitotic and meiotic spindle length. J Cell Biol. 2006;175:881–91. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Shakes DC. Immunofluorescence microscopy. Methods Cell Biol. 1995;48:365–94. [PubMed] [Google Scholar]
- Morris NR. Nuclear migration. From fungi to the mammalian brain. J Cell Biol. 2000;148:1097–101. doi: 10.1083/jcb.148.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley-Bishop KL, et al. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr Biol. 1999;9:1211–20. doi: 10.1016/s0960-9822(99)80501-6. [DOI] [PubMed] [Google Scholar]
- Mounkes L, et al. The laminopathies: nuclear structure meets disease. Curr Opin Genet Dev. 2003;13:223–30. doi: 10.1016/s0959-437x(03)00058-3. [DOI] [PubMed] [Google Scholar]
- Nelson GA, et al. Intersex, a temperature-sensitive mutant of the nematode Caenorhabditis elegans. Dev Biol. 1978;66:386–409. doi: 10.1016/0012-1606(78)90247-6. [DOI] [PubMed] [Google Scholar]
- Neumann FR, Nurse P. Nuclear size control in fission yeast. J Cell Biol. 2007;179:593–600. doi: 10.1083/jcb.200708054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Hyman AA. In: Cell Division. Community TCeR., editor. WormBook; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson K, et al. The functions of klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol Biol Cell. 2004;15:600–10. doi: 10.1091/mbc.E03-06-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, et al. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–26. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinsch S, Gonczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;111:2283–95. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- Robinson JT, et al. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J Cell Biol. 1999;146:597–608. doi: 10.1083/jcb.146.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouviere C, et al. Characteristics of pronuclear migration in Beroe ovata. Cell Motil Cytoskeleton. 1994;29:301–11. doi: 10.1002/cm.970290403. [DOI] [PubMed] [Google Scholar]
- Sadler PL, Shakes DC. Anucleate Caenorhabditis elegans sperm can crawl, fertilize oocytes and direct anterior-posterior polarization of the 1-cell embryo. Development. 2000;127:355–66. doi: 10.1242/dev.127.2.355. [DOI] [PubMed] [Google Scholar]
- Salpingidou G, et al. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J Cell Biol. 2007;178:897–904. doi: 10.1083/jcb.200702026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten G. The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev Biol. 1994;165:299–335. doi: 10.1006/dbio.1994.1256. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, et al. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–2. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- Somech R, et al. Nuclear envelopathies--raising the nuclear veil. Pediatr Res. 2005;57:8R–15R. doi: 10.1203/01.PDR.0000159566.54287.6C. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–9. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Starr DA. Communication between the cytoskeleton and the nuclear envelope to position the nucleus. Mol Biosyst. 2007;3:583–9. doi: 10.1039/b703878j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fischer JA. KASH ‘n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays. 2005;27:1136–46. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- Starr DA, Han M. ANChors away: an actin based mechanism of nuclear positioning. J Cell Sci. 2003;116:211–6. doi: 10.1242/jcs.00248. [DOI] [PubMed] [Google Scholar]
- Strome S, et al. Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol Biol Cell. 2001;12:1751–64. doi: 10.1091/mbc.12.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutovsky P, et al. Biogenesis of the centrosome during mammalian gametogenesis and fertilization. Protoplasma. 1999;206:249–262. [Google Scholar]
- Tsai JW, et al. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170:935–45. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur YB, et al. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat Rev Mol Cell Biol. 2006;7:782–8. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- Ward S, et al. Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. J Cell Biol. 1981;91:26–44. doi: 10.1083/jcb.91.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte MA. Bidirectional transport along microtubules. Curr Biol. 2004;14:R525–37. doi: 10.1016/j.cub.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K, et al. KASH-domain proteins in nuclear migration, anchorage and other processes. J Cell Sci. 2006;119:5021–9. doi: 10.1242/jcs.03295. [DOI] [PubMed] [Google Scholar]
- Wolf N, et al. Spermatogenesis in males of the free-living nematode, Caenorhabditis elegans. J Ultrastruct Res. 1978;63:155–69. doi: 10.1016/s0022-5320(78)80071-9. [DOI] [PubMed] [Google Scholar]
- Worman HJ, Bonne G. “Laminopathies”: A wide spectrum of human diseases. Exp Cell Res. 2007 doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Gundersen GG. Here come the SUNs: a nucleocytoskeletal missing link. Trends Cell Biol. 2006;16:67–9. doi: 10.1016/j.tcb.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Yoder JH, Han M. Cytoplasmic dynein light intermediate chain is required for discrete aspects of mitosis in Caenorhabditis elegans. Mol Biol Cell. 2001;12:2921–33. doi: 10.1091/mbc.12.10.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, et al. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc Natl Acad Sci U S A. 2000;97:8997–9002. doi: 10.1073/pnas.150240197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time lapse images of an AZ244 (tubulin::GFP) untreated embryo. Images were taken at 15-second intervals for 7 minutes. DIC and green fluorescent images were merged. Posterior is right.
Time lapse images of an AZ244 emr-1(RNAi); lem-2(RNAi) embryo. Images were taken at 15-second intervals for 10.5 minutes. DIC and green fluorescent images were merged. Posterior is right.
Time lapse images of an OD57 (tubulin::GFP; histone::mCherry) untreated embryo. Images were taken at 15-second intervals for 5.75 minutes. Green and red fluorescent images were merged. Posterior is right.
Time lapse images of an OD57 emr-1(gk119); lem-2(RNAi) embryo. Images were taken at 15-second intervals for 13.5 minutes. Green and red fluorescent images were merged. Posterior is right.
Time lapse images of an TY3558 emr-1(RNAi); lem-2(RNAi) embryo. Images were taken at 30-second intervals for 11 minutes. Green fluorescent images show tubulin::GFP and histone::GFP. Posterior is left.
The centrosome detachment phenotype disrupts γ-tubulin localization. A live lmn-1(RNAi) embryo expressing γ-tubulin::GFP is shown. DIC and green fluorescent images are overlaid. The arrow shows a centrosome detached from the nucleus. Posterior is right and the scale bar is 10 μm.
In embryos with detached centrosomes, nuclear pores localize to the nuclear envelope normally. One-cell embryos prior to pronuclear migration are shown. In all panels, embryos are simultaneously stained for tubulin and nuclear pores in red and chromatin in blue. Embryos are from hermaphrodites injected with dsRNA against the following: (A) no RNA wild-type control, (B) emr-1 (C) lem-2, (D) emr-1 and lem-2, (E) lmn-1, (F) baf-1. In (E) the second centrosome is out of frame. In (D–F) arrows mark centrosomes detached from male pronuclei. Posterior is right and the scale bar is 10 μm.
SUN-1::GFP localizes normally in embryos with a detached centrosome phenotype. (A) an untreated newly fertilized embryo expressing SUN-1::GFP. (B) a newly fertilized embryo from a lmn-1(RNAi) mother with a detached centrosome expressing SUN-1::GFP at normal levels. GFP is green (A–B), tubulin is red (A′–B′) and DNA is blue in the merge (A″–B″).
Time-lapse imaging of centrosome attachment in emr-1; lem-2 defective embryos. The data for the representative data shown in Fig. 5 and the statistical data shown in Fig. 6 are shown here. (A) Untreated AZ244, tubulin::GFP, embryos. (B) emr-1(RNAi); lem-2(RNAi) embryos in the AZ244 background. (C) Untreated OD57, tubulin::GFP histone::mCherry, embryos. (D) emr-1(RNAi or gk119); lem-2(RNAi) embryos in the OD57 background. Plots of the surface area of male pronuclei versus the distance between the edge of the nucleus and the center of the centrosome are shown on the left. Each colored line represents one series of time-lapse images. The beginning and end of the green arrow (B, D) and the associated dashed and solid lines represent the average size of emr-1(RNAi); lem-2(RNAi) male pronuclei in the AZ244 background at the start of filming and at the time of capture of the second centrosome respectively. The beginning and end of the red arrow (B, D) and the associated dashed and solid lines represent the average size of emr-1(RNAi or gk119); lem-2(RNAi) male pronuclei in the OD57 background at the start and end of filming respectively. On the right, the surface area of each male pronucleus at the beginning of filming, when both the nucleus and detached centrosome was observed (B, D), or in early pronuclear migration (A, C), is shown. The surface area of male pronuclei is also shown at the time the female pronucleus meets the male (A, C), at the time the second centrosome is attached by the male pronucleus (B) or at the end of filming (D). In the cases when the detached centrosome failed to attach, the embryo was filmed until there was no further change in the embryo’s appearance (the embryo was likely dead). In two embryos (marked with a †), the detached centrosome was captured by the female pronucleus. The embryos shown in Fig. 5 and Supplemental Movies 1–4 are marked with an *.