SUMMARY
Hepatic metabolic derangements are key components in the development of fatty liver, insulin resistance, and atherosclerosis. SIRT1, a NAD+-dependent protein deacetylase, is an important regulator of energy homeostasis in response to nutrient availability. Here we demonstrate that hepatic SIRT1 regulates lipid homeostasis by positively regulating PPARα, a nuclear receptor that mediates the adaptive response to fasting and starvation. Hepatocyte-specific deletion of SIRT1 impairs PPARα signaling and decreases fatty acid β-oxidation, whereas overexpression of SIRT1 induces the expression of PPARα targets. SIRT1 interacts with PPARα and is required to activate PPARα co-activator PGC-1α. When challenged with a high-fat diet, liver-specific SIRT1 knockout mice develop hepatic steatosis, hepatic inflammation, and endoplasmic reticulum stress. Taken together, our data indicate that SIRT1 plays a vital role in the regulation of hepatic lipid homeostasis, and that pharmacological activation of SIRT1 may be important for the prevention of obesity-associated metabolic diseases.
Keywords: SIRT1, PPARα, hepatic fatty acid oxidation, PGC-1α, hepatic steatosis, inflammation
INTRODUCTION
Metabolic syndrome, a collection of abnormalities including obesity, type 2 diabetes, dyslipidemia, fatty liver, and a pro-inflammatory and prothrombotic state (Eckel et al., 2005; Grundy et al., 2004; Zimmet et al., 2001), affects more than 27% of adults in the United States (Ford et al., 2004; Hedley et al., 2004) and has become a major health concern worldwide. Central to the pandemic of this disease cluster is the dramatic increase in the incidence of obesity in most parts of the world. Obesity-induced ectopic accumulation of fat activates cellular stress signaling and inflammatory pathways (Hirosumi et al., 2002; Ozcan et al., 2004; Yuan et al., 2001), contributing to enhanced muscle insulin resistance, pancreatic β-cell failure, nonalcoholic steatohepatitis (NASH), and eventually to organ damage. Therefore, efforts to achieve a better understanding of the molecular mechanisms controlling lipid and energy metabolism are crucial to the development of new therapeutic strategies.
The liver is the central metabolic organ that regulates several key aspects of lipid metabolism including fatty acid β-oxidation, lipogenesis, and lipoprotein uptake and secretion, in response to nutritional and hormonal signals (van den Berghe, 1991). It is essential for the maintenance of systemic energy homeostasis in the body. For example, during the fed state, dietary glucose stimulates insulin secretion from the pancreas, which travels directly to the liver via the portal vein thus increasing hepatic lipogenesis and lipoprotein secretion. Conversely, upon fasting, hepatic fatty acid oxidation is enhanced in response to the release of pancreatic glucagon and adrenal cortisol. Dysregulation of lipid metabolic pathways results in the development of hepatic steatosis and contributes to the development of chronic hepatic inflammation, insulin resistance, and liver damage (Browning and Horton, 2004; Samuel et al., 2004; Sanyal, 2005).
The capacity of the liver to regulate metabolism is governed by a highly dynamic transcriptional regulatory network. The key regulatory factors include members of the adopted orphan nuclear receptors, particularly Peroxisome proliferators-activated receptor α (PPARα), Farnesoid X receptor (FXR), and Liver X receptor (LXR) (Chawla et al., 2001). This class of nuclear receptors forms heterodimers with the retinoid X receptor (RXR), and function as lipid sensors through the direct binding of a variety of lipids and their derivatives to their ligand-binding domain. Upon lipid binding, each of these receptors activates the transcription of a family of genes involved in lipid homeostasis, thereby modulating hepatic and systemic energy metabolism in response to nutrient availability (Shulman and Mangelsdorf, 2005). Recent studies have demonstrated that SIRT1, a nuclear NAD+-dependent protein deacetylase (Imai et al., 2000; Landry et al., 2000; Smith et al., 2000), plays an important role in the regulation of transcriptional networks in various critical metabolic processes. SIRT1 belongs to the class III family of histione deacetylases (HDACs), also known as sirtuins (Blander and Guarente, 2004). Members of the sirtuin family are longevity determinants in yeast, C. elegans, and Drosophila and are essential for lifespan extension provided by calorie restriction (Blander and Guarente, 2004). Because sirtuin activity is dependent on NAD+, an indicator of cellular nutrient status, they are believed to provide a functional link between metabolism, chromatin structure, and ultimately aging (Bishop and Guarente, 2007). The mammalian genome consists of seven sirtuins, collectively known as SIRT1 to SIRT7 (Frye, 2000). SIRT1 has been shown to deacetylate many nonhistone proteins, including p53 (Luo et al., 2001; Vaziri et al., 2001), NFκB (Yeung et al., 2004), FOXO transcriptional factors (Brunet et al., 2004; Motta et al., 2004), PGC-1α (Rodgers et al., 2005), LXR (Li et al., 2007), CLOCK and PER2 (Grimaldi et al., 2008; Nakahata et al., 2008), and TORC2 (Liu et al., 2008). Through its deacetylase activity, SIRT1 is able to either repress or activate the transcriptional activities of these targets, thereby regulating diverse metabolic and stress pathways (Guarente, 2006). In this report, we show that hepatic SIRT1 is an important regulator of lipid homeostasis, in particular fatty acid oxidation. Loss of SIRT1 in hepatocytes impairs the activity of PPARα, resulting in decreased fatty acid oxidation, leading to the development of hepatic steatosis and inflammation on a high-fat diet.
RESULTS
Hepatic deletion of SIRT1 alters PPARα signaling
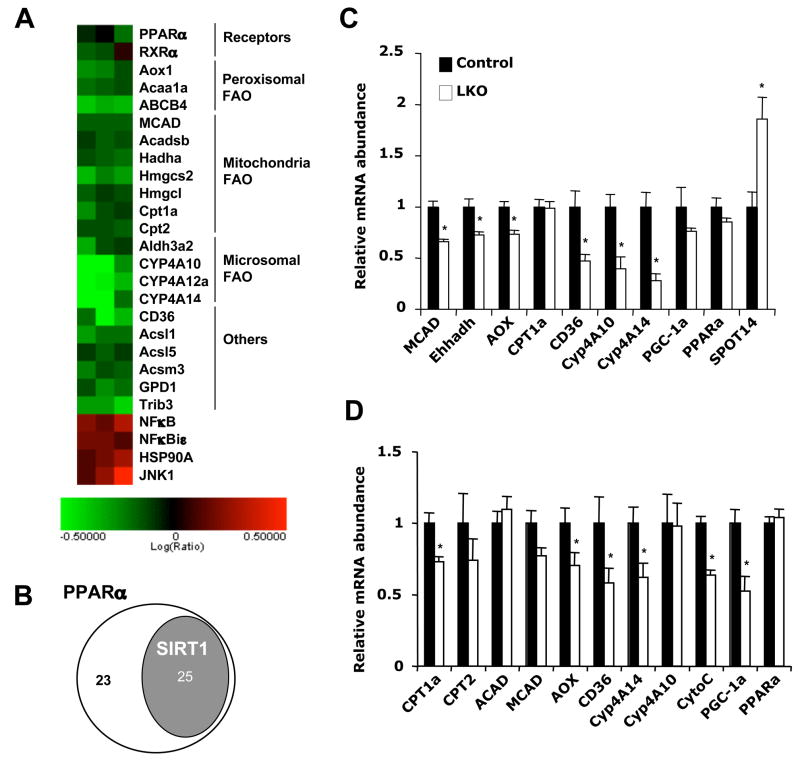
SIRT1 is an important regulator of nutrient homeostasis in several tissues involved in metabolism (Guarente, 2006). To examine the function of SIRT1 in the liver, we generated liver-specific SIRT1 knockout mice on the C57BL/6 background (SIRT1 LKO, see Experimental Procedures and Figure S1). Although SIRT1 LKO mice were phenotypically normal under a chow diet, microarray analysis and Ingenuity Pathway Analysis (IPA) of liver mRNA identified the PPARα signaling pathway as one of the main pathways affected by hepatic SIRT1 deficiency (Figure 1A, p=1.99×10−6). Previous studies have identified 48 PPARα response genes that are involved in hepatic fatty acid oxidation (FAO), ketone body synthesis, and fatty acid binding, transport, and activation (Rakhshandehroo et al., 2007). Subsequent analysis revealed that 25 of these genes were significantly decreased in SIRT1 LKO mice (Figure 1B and Table S1), indicating that loss of hepatic SIRT1 impairs PPARα mediated fatty acid metabolism. Quantitative real-time PCR (qPCR) confirmed decreased expression of several PPARα targets, including medium chain acyl-CoA dehydrogenase (MCAD), liver bifunctional enzyme (Ehhadh), palmitoyl acyl-CoA carboxylase (AOX), fatty acid transporter (CD36), and microsomal cytochrome P450 enzymes involved in the ω-oxidation of fatty acids (Cyp4A10 and Cyp4A14), in SIRT1 LKO mice compared to Lox controls (Control, Figure 1C). In contrast, the mRNA level of SPOT14, a gene known to be negatively regulated by PPARα (Ren et al., 1996), was significantly higher in the SIRT1 LKO mice.
Figure 1. Hepatic deletion of SIRT1 alters PPARα signaling.
(A) Microarray analysis of PPARα signaling pathway from control and SIRT1 LKO mice (n=6). The log ratios of SIRT1 LKO/control were presented by heat map.
(B) Venn-diagram representation of the subset of PPARα-regulated hepatic fatty acid metabolism genes that were significantly decreased in the liver of SIRT1 LKO mice (n=6, p<0.05).
(C and D) Quantitative real-time PCR (qPCR) analysis of PPARα target genes involved in fatty acid oxidation in the liver of control (black bars) and SIRT1 LKO mice (white bars) on the chow diet, sacrificed at 4pm (C) or after a 16 h fast (D). In this and other figures, error bars represent mean ± SEM. (n=6, *p<0.05).
PPARα is a lipid-sensing member of the nuclear receptor superfamily that regulates systemic fatty acid metabolism. Under normal physiological conditions in mice, PPARα levels in the liver peak in the late afternoon and decrease immediately after food intake ((Yang et al., 2006), www.nursa.org/10.1621/datasets.02002). Activation of PPARα by fatty acids and metabolites leads to transcriptional activation of various target genes involved in fatty acid oxidation, which are required for the normal adaptive response to starvation. Consequently, mice lacking PPARα accumulate excessive triglyceride in their liver and become hypoketonemic during fasting (Hashimoto et al., 2000; Kersten et al., 1999). To further confirm the involvement of SIRT1 in PPARα-mediated pathways, we examined expression levels of PPARα targets in SIRT1 LKO mice and Lox controls after overnight fasting. Consistent with our previous findings (Figure 1A–C), SIRT1 LKO mice displayed reduced expression of several PPARα target genes after overnight fasting (Figure 1D). Interestingly, the expression of PPARγ co-activator 1α (PGC-1α), a transcription co-factor known to activate PPARα (Vega et al., 2000), was also significantly lower in SIRT1 LKO mice. Taken together, these results suggest that SIRT1 may play a role in regulating PPARα signaling during physiological conditions of fasting.
SIRT1 regulates ligand-dependent PPARα activation
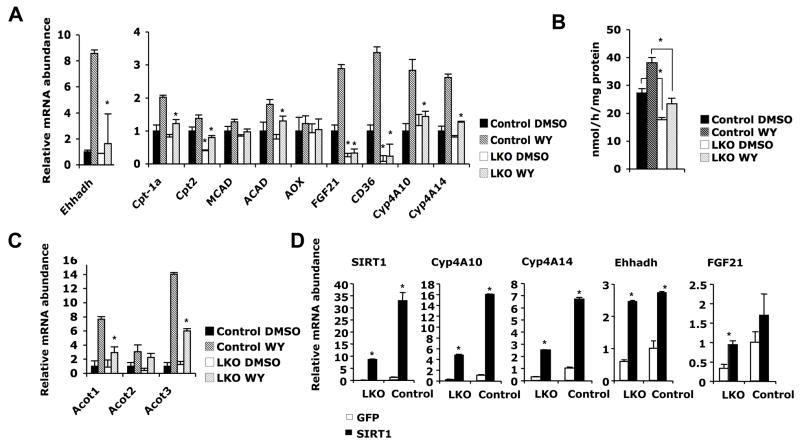
To elucidate the regulatory role of SIRT1 in PPARα signaling in liver, we treated primary hepatocytes from Lox control or SIRT1 LKO mice with a PPARα agonist WY14643. As shown in Figure 2A, WY treatment induced the expression of several genes involved in fatty acid oxidation in control hepatocytes. However, the induction of these genes was significantly lower in the SIRT1 deficient (LKO) hepatocytes (Figure 2A). Consistent with a reduction in fatty acid oxidation gene expression, β-oxidation of 3H-palmitate in SIRT1 deficient hepatocytes was significantly lower compared to that in control hepatocytes (Figure 2B). PPARα is also known to induce the expression of additional targets involved in fatty acid metabolism such as acyl-CoA thioesterases (Acots) (Hunt et al., 1999). As shown in Figure 2C, mRNA levels of Acot1 and Acot3 were also significantly lower in the SIRT1 deficient hepatocytes. Furthermore, lentivirus mediated overexpression of SIRT1 in LKO hepatocytes resulted in rescue of defective PPARα signaling, and further stimulated the transcriptional activity of PPARα in control hepatocytes (Figure 2D). Together, these data suggest that SIRT1 positively regulates PPARα in a cell autonomous fashion.
Figure 2. Loss of SIRT1 reduces the induction of PPARα targets and fatty acid oxidation in primary hepatocytes.
(A) SIRT1 deficiency in primary hepatocytes reduces the induction of fatty acid oxidation gene expression by PPARα agonist WY14643. Primary hepatocytes from control (black bars) or SIRT1 LKO mice (white bars) were treated as described in the Experimental procedures, and mRNA were analyzed by qPCR (n=3, *p<0.05).
(B) SIRT1 deficiency in primary hepatocytes reduces the rate of fatty acid oxidation. The oxidation rate of [3H]-palmitic acid in primary hepatocytes from control (black bars) and SIRT1 LKO (white bars) mice were measured as described in the Experimental Procedures (n=3, *p<0.05).
(C) SIRT1 deficient primary hepatocytes show reduced induction of other PPARα targets by WY14643.
(D) Overexpression of SIRT1 in primary hepatocytes induces the expression of PPARα targets. Primary hepatocytes from control and SIRT1 LKO mice were infected with lentiviruses expressing GFP (white bars) or SIRT1 (black bars) and mRNA were analyzed by qPCR (*p<0.05).
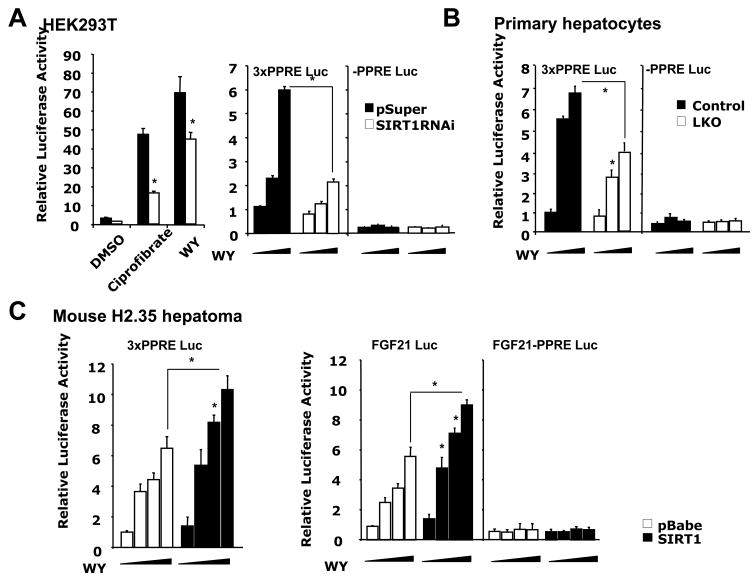
We then investigated the connection between SIRT1 and trans-activation of PPARα using luciferase reporter assays. HEK293T cells stably infected with an empty vector (pSuper) or a SIRT1 RNAi construct were transfected with a construct expressing a chimeric transcription factor with the Gal4 DNA binding domain (DBD) fused to the PPARα ligand-binding domain (LBD), followed by treatment with DMSO or PPARα agonists ciprofibrate (100μM) or WY (15μM). As expected, treatment with both PPARα agonists resulted in marked induction of luciferase activity in control HEK293T pSuper cells. In contrast, transcriptional response to these agonists was significantly reduced in HEK293T SIRT1 RNAi cells (Figure 3A, left panel). We then examined transactivation of PPARα using a luciferase reporter driven by three tandem-repeats of a consensus PPAR response element (3xPPRE). In control HEK293T pSuper cells, the PPARα agoinst WY stimulated PPRE driven luciferase in a dose dependent manner, while a mutant reporter (-PPRE) failed to respond to WY treatment (Figure 3A, right panel). The dose-dependent induction of the PPRE driven reporter was significantly blunted in HEK293T SIRT1 RNAi cells. Similar results were obtained in control and SIRT1 deficient primary hepatocytes (Figure 3B). We further examined the effect of elevated SIRT1 levels on the transactivation of the PPRE reporter (Figure 3C, left panel) and a luciferase reporter driven by the endogenous promoter of FGF21, a known PPARα target (Badman et al., 2007; Inagaki et al., 2007) (Figure 3C, right panel), in mouse H2.35 hepatoma cells. Overexpression of SIRT1 resulted in a dose-dependent increase in the luciferase expression from both reporters. Taken together, these results demonstrate that SIRT1 directly regulates PPARα transactivation through PPREs.
Figure 3. SIRT1 regulates ligand-dependent PPARα transactivation.
(A) Reduction of SIRT1 activity decreases ligand dependent PPARα transactivation. (Left panel) pSuper (black bars) or pSuper-SIRT1 RNAi (white bars) HEK293T cells were cotransfected with luciferase reporters and a construct encoding Gal4 DBD-PPARα LBD fusion protein, then treated with DMSO or PPARα agonists and analyzed for luciferase activity as described in Experimental Procedures. (Right panel) pSuper (black bars) and pSuper-SIRT1 RNAi (white bars) HEK293T cells were transfected with 3xPPRE-luciferase construct or a control construct lacking the PPRE, together with constructs expressing murine PPARα and RXRα. Cells were then treated with increasing doses of WY14643 and analyzed for luciferase activity as described in the Experimental Procedures. (*p<0.05)
(B) Loss of SIRT1 in primary hepatocytes reduces ligand dependent PPARα transactivation. (*p<0.05)
(C) Overexpression of SIRT1 stimulates ligand-dependent PPARα transactivation in mouse H2.35 hepatoma cells. (*p<0.05)
SIRT1 interacts with PPARα and is required for the activation of PGC-1α
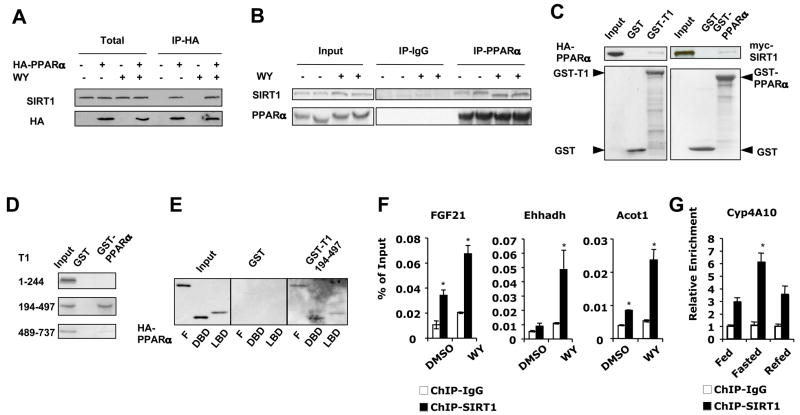
To further investigate the role of SIRT1 in regulating PPARα, we determined whether SIRT1 physically interacts with PPARα. As shown in Figure 4A, in HEK293T cells, endogenous SIRT1 was co-immunoprecipitated by anti-HA antibodies only when HA-PPARα was expressed, indicating a specific interaction between these two proteins. To confirm that SIRT1 and PPARα are present in common protein complexes in vivo, mouse liver nuclear extracts were precipitated with anti-PPARα antibodies. As shown in Figure 4B, endogenous SIRT1 was pulled-down specifically by anti-PPARα antibodies but not by IgG. Oral administration of WY for 2 h further increased the interaction between SIRT1 and PPARα, suggesting that SIRT1 preferentially associates with activated PPARα. Furthermore, the glutathione-S-transferase (GST)-SIRT1 fusion protein, but not GST control, pulled-down in vitro transcribed and translated HA-PPARα (Figure 4C, left panel). Reciprocally, the GST-PPARα fusion protein pulled down myc-SIRT1 (Figure 4C, right panel), suggesting that SIRT1 directly interacts with PPARα. We further found that PPARα strongly interacts with the aa 194–497 fragment of SIRT1 encoding the NAD+-dependent deacetylase catalytic core domain (Figure 4D). The SIRT1 core domain, on the other hand, associated with both the DBD and LBD of PPARα (Figure 4E), suggesting that SIRT1 interacts with PPARα at multiple sites.
Figure 4. SIRT1 interacts with PPARα.
(A) SIRT1 interacts with PPARα in HEK293T cells. Cell lysates from HEK293T cells transfected with empty vector or HA-PPARα were treated with DMSO or WY14643, were then immunoprecipitated (IP) with anti-HA antibodies.
(B) SIRT1 interacts with PPARα in the liver. Liver nuclear extracts from mice treated with vehicle or WY were immunoprecipitated with anti-PPARα antibodies.
(C) SIRT1 interacts with PPARα in vitro. GST-SIRT1 fusion protein was incubated with HA-PPARα (left), or GST-PPARα fusion protein was incubated with myc-SIRT1 (right) in the presence of 15 μM WY. Top panels, immunoblots of HA-PPARα or myc-SIRT1. Bottom panels, ponceau S staining of GST fusion proteins.
(D) GST-PPARα interacts with the catalytic core domain of SIRT1. GST-PPARα fusion protein was incubated with indicated myc-tagged SIRT1 fragments in the presence of 15 μM WY.
(E) The catalytic core domain of SIRT1 interacts with the both DNA-binding domain (DBD) and ligand-binding domain (LBD) of PPARα. GST-SIRT1 (aa 194–497) fusion protein was incubated with HA-tagged full-length (F), DBD, or LBD of PPARα.
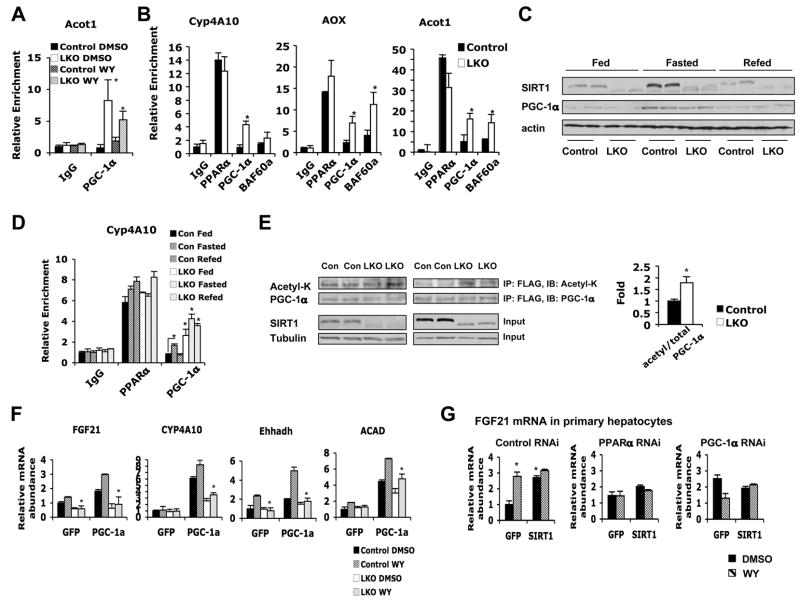
(F) SIRT1 localizes to the PPRE of PPARα targets. Primary hepatocytes treated with DMSO or WY for 4 h were subjected to ChIP with IgG control (white bars) or anti-SIRT1 (black bars) antibodies and analyzed by qPCR with primers flanking PPRE in the promoters of FGF21, Acot1 and Ehhadh.
(G) SIRT1 is recruited to the PPRE of Cyp4A10 upon fasting. ChIP assays were performed with chromatin extracts from livers of control mice that were fed, fasted for 24 h, or fasted for 24 h then refed for 24 h.
The interaction of SIRT1 with PPARα suggested that SIRT1 may associate with PPREs on promoters of PPARα target genes. To test this possibility, we analyzed the association of SIRT1 with PPREs using a chromatin immunoprecipitation (ChIP) assay. As shown in Figure 4F, treatment with WY significantly increased the association of SIRT1 to the PPREs of a number of PPARα targets compared to IgG controls, indicating a ligand-dependent recruitment of SIRT1 to the promoter of PPARα target genes. Furthermore, SIRT1 was recruited to the PPRE of the Cyp4A10 gene in liver by a 24 h fasting, and dissociated from this PPRE upon refeeding (Figure 4G), implying that the association of SIRT1 to PPRE is regulated by the nutritional status.
Previous studies have demonstrated that PGC-1α, an inducible co-activator for a number of nuclear receptors and additional transcriptional factors, physically interacts with PPARα thus stimulating its transcriptional activity (Vega et al., 2000). The association of PGC-1α with PPARα results in further recruitment of BAF60a and other SWI/SNF chromatin-remodeling complexes to activate transcription of PPARα targets (Li et al., 2008). Since mRNA levels of PGC-1α were consistently lower in livers of SIRT1 LKO mice after fasting (Figure 1D), we postulated that the observed reduction of PPARα signaling was due to insufficient recruitment of PGC-1α to target promoters. However, to our surprise, PPRE-associated PGC-1α levels were dramatically increased in SIRT1 LKO primary hepatocytes (Figure 5A), and PPRE-associated PGC-1α as well as BAF60a levels were also significantly elevated in SIRT1 LKO mouse livers after fasting (Figure 5B). Further analyses indicated that total PGC-1α protein was still induced by fasting and repressed by refeeding in the SIRT1 LKO liver (Figure 5C), but PPRE-associated PGC-1α was accumulated regardless of feeding status (Figure 5D). These results suggest that hepatic SIRT1 is required for maintenance of the recruitment/dissociation cycle of PGC-1α in response to the metabolic status. PGC-1α is a direct substrate of SIRT1, and deacetylation of this co-activator by SIRT1 stimulates its activity (Rodgers et al., 2005). Consistent with this notion, acetylated PGC-1α levels were significantly increased in SIRT1 LKO primary hepatocytes after treatment with WY (Figure 5E), and the co-activation activity of PGC-1α on a number of PPARα targets was significantly blunted in SIRT1 deficient primary hepatocytes (Figure 5F, Figure S2B).
Figure 5. SIRT1 is required to activate PGC-1α.
(A) PGC-1α accumulates on the PPRE of Acot1 in SIRT1 LKO primary hepatocytes. Control and SIRT1 LKO primary hepatocytes treated with DMSO or WY for 4 h were subjected to ChIP with IgG control or anti-PGC-1α antibodies and analyzed by qPCR with primers flanking PPRE in the promoter of Acot1.
(B) Accumulation of PGC-1α and BAF60a on the PPRE of PPARα targets after fasting in the SIRT1 LKO liver. ChIP assays were performed with chromatin extracts from livers of control (black bars) and SIRT1 LKO (white bars) mice fasted for 16 h.
(C) PGC-1α protein levels are induced by fasting and repressed by refeeding in both control and SIRT1 LKO mice. Liver extracts from control and SIRT1 LKO mice that were fed, fasted for 24 h, or fasted for 24 h then refed for 24 h, were immunoblotted with indicated antibodies.
(D) Accumulation of PGC-1α on the PPRE regardless of feeding status. ChIP assays were performed with chromatin extracts from livers of control and SIRT1 LKO mice that were fed, fasted for 24 h, or fasted for 24 h then refed for 24 h.
(E) SIRT1 deficiency in hepatocytes increases the acetylation of PGC-1α. FLAG-PGC-1α from control (Con) and SIRT1 LKO (LKO) primary hepatocytes were treated and immuno-purified as described in the Experimental Procedures, and analyzed with anti-acetyl-lysine (acetyl-K) antibodies. (*p<0.05).
(F) Decreased PGC-1α co-activation activity on the expression of PPARα target genes in SIRT1 deficient primary hepatocytes. Primary hepatocytes from control and SIRT1 LKO mice were infected with adenoviruses expressing GFP or PGC-1α, and treated with DMSO or WY as described in the Experimental Procedures (*p<0.05).
(G) SIRT1-mediated induction of FGF21 requires PPARα and PGC-1α. Primary hepatocytes from control mice were electroporated with a negative control siRNA or siRNAs against PPARα or PGC-1α. Cells were then infected with lentiviruses expressing GFP or SIRT1, and treated as described in the Experimental Procedures (*p<0.05).
To determine whether PPARα or PGC-1α is required for the SIRT1-mediated induction of PPARα target genes, we electroporated siRNA against either PPARα or PGC-1α into primary hepatocytes (Figure S2C), followed by infection with GFP or SIRT1 lentiviruses. As shown in Figure 5G, in control hepatocytes, overexpression of SIRT1 significantly induced the expression of FGF21, whereas this induction was abolished in PPARα RNAi cells, indicating that SIRT1 induces the expression of PPARα targets through PPARα. The requirement of PPARα was supported by the luciferase reporter assays in Figure 3 (particularly Figure 3C) which indicate that SIRT1 regulates the expression of PPARα targets through the PPRE. RNAi of PGC-1α caused a compensative increase of PPARα levels in hepatocytes (Figure S2F), resulting in higher basal levels of FGF21 (Figure 5G, right panel, DMSO). However, overexpression of SIRT1 failed to further increase expression of FGF21, indicating that SIRT1 requires PGC-1α to activate PPARα. Interestingly, our data show that PGC-1α is under the transcriptional control of PPARα and SIRT1 in primary hepatocytes (Figure S2E).
Hepatic deletion of SIRT1 impairs lipid homeostasis upon high-fat feeding
Activation of PPARα by fasting promotes lipolysis in white adipose tissues and stimulates fatty acid oxidation and ketogenesis in the liver. The regulation of PPARα by SIRT1 suggests that loss of function of SIRT1 could impair PPARα-mediated lipid homeostasis in vivo. Indeed, SIRT1 LKO mice fed a chow diet accumulated about two folds of triglycerides in the liver after 16 h fasting (Figure S3A), although their serum free fatty acid levels remained normal (Figure S3E).
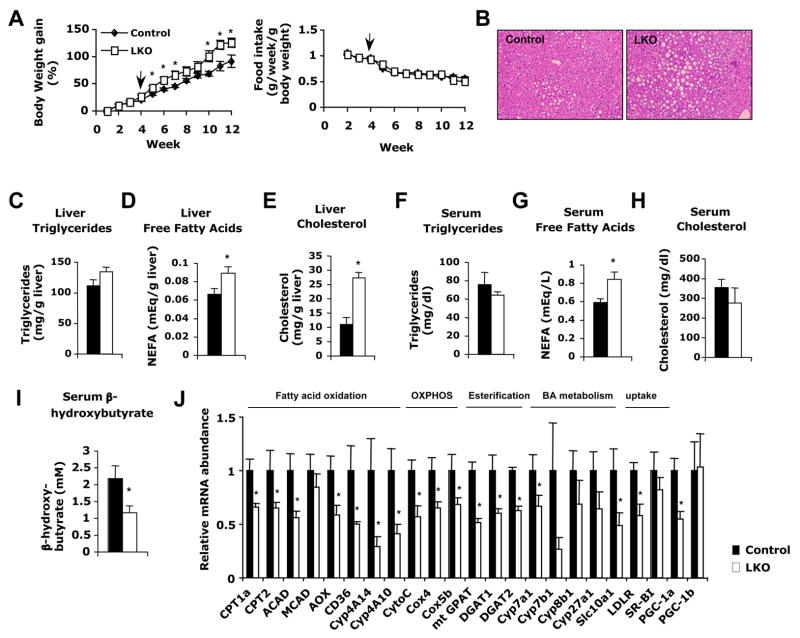
To further examine the pathophysiological effects of blunted PPARα signaling in vivo, Lox control and SIRT1 LKO mice were fed ad libitum a western diet providing 40% Kcal fat and 0.21% cholesterol for 11 weeks, then sacrificed after 16 h fasting. Although SIRT1 LKO mice showed no body weight abnormality on a chow diet (data not shown), they gained significantly more weight than control mice on the western diet (Figure 6A). Consistent with diet-induced obesity, SIRT1 LKO mice showed greater lipid accumulation in the liver compared to control mice, as revealed by H&E staining of liver sections (Figure 6B) and enzymatic colorimetric quantification of extracted liver triglycerides, NEFA, and cholesterol (Figure 6C–E). Their serum free fatty acid levels were also significantly increased as compared to control mice (Figure 6G). Additionally, serum β-hydroxybutyrate, a marker for fatty acid oxidation and ketogenesis in the liver, was significantly lower in the SIRT1 LKO mice (Figure 6I).
Figure 6. Hepatic loss of SIRT1 function impairs lipid homeostasis upon high-fat diet feeding.
(A) Body weight gain and food intake curves of control and SIRT1 LKO mice (n=10–11) under western diet. Arrow indicated the beginning of western diet feeding.
(B–E) SIRT1 LKO mice accumulate more lipids in the liver after fasting as indicated by (B) Hematoxylin and eosin staining of liver sections from control and SIRT1 LKO mice; and total liver triglycerides (C), free fatty acids (D), and cholesterol (E) levels.
(F–I) SIRT1 deficiency in the liver increases serum free fatty acids but reduces serum β-hydroxybutyrate after fasting. Serum triglycerides (F), free fatty acids (G), cholesterol (H), and β-hydroxybutyrate (I) were analyzed as described.
(J) Relative expression of genes encoding key factors in hepatic fatty acid oxidation, oxidative phosphorylation (OXPHOS), fatty acid esterification, bile acid (BA) metabolism, and cholesterol uptake. (n=4–5, *p<0.05).
Increases in hepatic lipids, particularly NEFA, along with a decrease in serum β-hydroxybutyrate after fasting, indicate a defect in the fatty acid oxidation pathway. In line with these observations, levels of PPARα targets known to regulate fatty acid oxidation were significantly lower in SIRT1 LKO mouse livers (Figure 6J). A similar reduction in the expression of mitochondrial oxidative phosphorylation genes (Cytochrome C, Cox4 ad Cox5b) was observed in these mice. Again, the decreased expression of the above-mentioned PPARα target genes was accompanied by a 40% decrease in the expression of PGC-1α mRNA (Figure 6J). In addition, SIRT1 LKO mice displayed significantly lower mRNA levels of mitochondrial glycerol-3-phosphate acyltransferase (mtGPAT), acyl coenzyme A: diacylglycerol acyltransferase 1 (DGAT1), and acyl coenzyme A: diacylglycerol acyltransferase 2 (DGAT2), genes involved in the esterification of fatty acids to glycerol for the synthesis of triglycerides, suggesting that the increase in liver free fatty acids (Figure 6D) in SIRT1 LKO mice may also be a consequence of impaired fatty acid esterification.
The considerably higher liver cholesterol levels in SIRT1 LKO mice prompted us to analyze expression of genes involved in cholesterol synthesis, transport, uptake, efflux and degradation. While the major cholesterol uptake genes LDLR and SR-BI were decreased or unchanged (Figure 6J), there were no observable differences in levels of cholesterol synthesis (SREBP2, HMGCoA reductase) or efflux genes (ABCA1, ABCG1, ABCG8) (data not shown), indicating that the elevated hepatic cholesterol is not due to increased uptake, synthesis, or decreased efflux. Instead, Cyp7A1, the rate-limiting enzyme involved in the conversion of cholesterol to bile acids (Kalaany and Mangelsdorf, 2006), as well as other enzymes in the bile acid synthesis pathways, were decreased in SIRT1 LKO mice (Figure 6J), suggesting that the accumulation of hepatic cholesterol may be the result of diminished bile synthesis.
Hepatic deletion of SIRT1 leads to hepatic inflammation and ER stress on the high-fat diet
It is being increasingly recognized that obesity is associated with inflammation and cellular stress signaling (Reviewed by (Wellen and Hotamisligil, 2005)). Excess lipid accumulation in peripheral tissues promotes macrophage infiltration, thus stimulating local inflammation and eventually insulin resistance (Kanda et al., 2006; Weisberg et al., 2003). Excess lipid accumulation in tissues has also been linked to the development of endoplasmic reticulum (ER) stress, which has also been implicated in inhibition of the insulin signaling pathway (Ozcan et al., 2004). The accumulation of lipids in the livers of SIRT1 LKO mice led us to examine whether SIRT1 LKO mice fed a high-fat diet suffer from increased ER stress and inflammation in the liver.
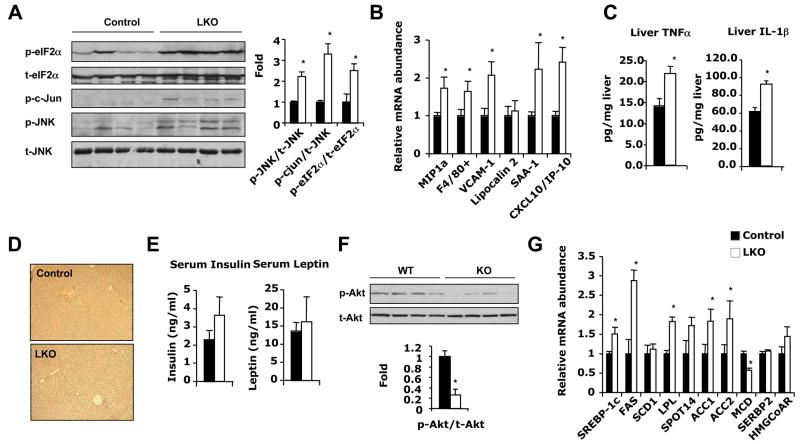
ER stress provokes an ER transmembrane protein kinase, pancreatic ER kinase (PERK), to phosphorylate the α subunit of the translation initiation factor (eIF2α) thus blocking cellular protein translation (Harding et al., 1999; Shi et al., 1998). Comparison of levels of phosphorylated eIF2α in liver extracts of control and SIRT1 LKO mice revealed that SIRT1 LKO mice had a 2.2-fold increase in p-eIF2α protein compared to Lox controls (Figure 7A). Hyper-activation of c-Jun N-terminal kinase (JNK) through phosphorylation is another marker of ER stress (Urano et al., 2000). Phosphorylation of JNK (p-JNK) and its downstream target c-Jun (p-c-Jun) were significantly increased by 2.1-fold and 3.2-fold respectively in SIRT1 LKO mice compared to control mice (Figure 7A). Together, these findings indicate that accumulation of lipids in SIRT1 LKO results in elevated ER stress in the liver.
Figure 7. Hepatic deletion of SIRT1 causes hepatic inflammation and ER stress upon high-fat diet feeding.
(A) SIRT1 deficiency in liver increases ER stress. ER stress markers were analyzed by immunoblotting as described.
(B) SIRT1 LKO mice display increased hepatic inflammation. Expression of macrophage markers and pro-inflammatory genes in the livers of control (black bars) and SIRT1 LKO mice (white bars) was analyzed by qPCR.
(C) SIRT1 LKO mice display increased hepatic levels of pro-inflammatory cytokines. Liver extracts from control and SIRT1 LKO mice were analyzed for TNFα and IL-1β by ELISA.
(D) Normal deposition of collagen in the SIRT1 LKO livers. The liver sections from control and SIRT1 LKO mice were stained with Sirius red for collagen. Mice in (A to D) were fed a western diet and scarified after 16 h fasting. (n=4–5, *p<0.05).
(E) Serum insulin and leptin from control (black bars) and SIRT1 LKO (white bars) mice.
(F) SIRT1 LKO mice display signs of hepatic insulin resistance.
(G) SIRT1 deficiency in the liver increases hepatic lipogenesis.
Mice in (E to G) were fed a western diet and scarified after 4 h fasting. (n=4–5, *p<0.05).
Infiltration of macrophages into tissues is a hallmark of local inflammation. Analysis of mRNA from livers of SIRT1 LKO mice revealed a 70–80% increase in macrophage markers, including macrophage inflammation protein (MIP1a) and F4/80+ (Figure 7B). Notably, several pro-inflammatory molecules that are dramatically induced in PPARα deficient livers (Stienstra et al., 2007), such as vascular cell adhesion molecule (VCAM-1), serum amyloid A-1 (SAA-1), and interferon (IFN)-γ inducible protein (CXCL10), were significantly higher in livers of SIRT1 LKO mice compared to Lox controls (Figure 7B). TNFα and IL-1β, two major pro-inflammatory cytokines induced by fat accumulation, were also significantly increased in the liver SIRT1 LKO mice (Figure 7C). These observations indicate that SIRT1 LKO mice are prone to development of hepatic inflammation. However, livers of high-fat diet fed SIRT1LKO mice showed very low levels of collagen deposition (Figure 7D), a hallmark of liver fibrosis, suggesting a full-blown steatohepatitis has not yet been developed under the current high-fat diet feeding conditions.
Since both ER stress and inflammation directly contribute to the development of insulin resistance (Wellen and Hotamisligil, 2005), our observations imply that SIRT1 LKO mice would show signs of altered insulin signaling. Indeed, after feeding with a high-fat diet, Ser473 phosphorylation of Akt, a key molecule in the insulin signaling pathway, was ~80% lower in livers of SIRT1 LKO mice (Figure 7F), despite a trend of higher blood insulin levels (Figure 7E). Interestingly, high insulin levels in these mice were still able to stimulate the expression of key synthesis/uptake genes, such as SREBP-1c, fatty acid synthase (FAS), acetyl CoA carboxylase (ACC1, ACC2), and lipoprotein lipase (LPL), on both chow (Figure S3I) and high-fat diets (Figure 7G), indicating that insulin signaling is selectively impaired in the SIRT1 LKO mice.
DISCUSSION
Hepatic nuclear receptors play an important role in the regulation of lipid metabolism, storage, transport and elimination in response to nutrient and hormonal cues. Dysfunction of these receptors is linked to a number of age-associated metabolic diseases, including diabetes, obesity and atherosclerosis (Chawla et al., 2001). In the present study, we identify PPARα signaling as a central pathway affected by hepatic deletion of SIRT1. We observed that blunted PPARα signaling in SIRT1 LKO mice resulted in decreased fatty acid oxidation and ketogenesis (Figure 2), contributing to the development of hepatic steatosis, inflammation, and ER stress on a high-fat diet (Figure 6 and 7).
Several lines of evidence suggest a link between SIRT1 and PPARα. For instance, both SIRT1 and PPARα are activated by fasting and food restriction (Cohen et al., 2004; Hashimoto et al., 2000; Kersten et al., 1999; Rodgers et al., 2005). PGC-1α, a key coactivator for PPARα signaling (Li et al., 2008; Vega et al., 2000), is a direct substrate of SIRT1 (Rodgers et al., 2005). Here we show that PPARα signaling is significantly impaired in SIRT1 LKO mice, while increased SIRT1 levels stimulate PPARα activity (Figure 2), providing a direct link between SIRT1 and PPARα. Furthermore, our data suggest that SIRT1 regulates PPARα signaling primarily through the activation of PGC-1α (Figure 5). In SIRT1 deficient hepatocytes, PGC-1α is still recruited to the PPREs on the promoter of fatty acid oxidation genes (Figure 5A and 5B). However, it remains in a hyper-acetylated state (Figure 5E) that inhibits its ability to promote transcription (Figure 5F). How acetylation affects the activity of PGC-1α is still not well understood. Since PGC-1α is highly accumulated on the promoter regions of PPARα target genes in SIRT1 deficient cells (Figure 5A and 5B), it is possible that SIRT1 is required to efficiently remove less-activated PGC-1α from the promoters to allow recruitment of active PGC-1α for additional rounds of transcription. This mechanism has been well described for other SIRT1-mediated transcriptional activations (Kitamura et al., 2005; Li et al., 2007; Pagans et al., 2005).
In addition to fatty acid oxidation, PGC-1α also stimulates hepatic gluconeogenesis in response to fasting (Herzig et al., 2001; Yoon et al., 2001). The decreased coactivation activity of PGC-1α in SIRT1 LKO mice suggests that gluconeogenesis may be impaired. However, we failed to observe notable defects in the hepatic gluconeogenesis in the SIRT1 LKO mice (Figure S5). Fasting glucose levels in SIRT1 LKO mice were normal under a chow diet, and were slightly increased under the western diet or a high-fat diet (Figure S5A). mRNA levels of two rate-limiting enzymes in the hepatic gluconeogenesis pathway, phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase), were also normal (Figure S5B). These observations are not surprising, given the fact that SIRT1 also deacetylates and represses TORC2 (Liu et al., 2008) and FOXO1 (Motta et al., 2004), two additional key factors involved in promoting gluconeogenesis in the early and late fasting phases, respectively. Therefore, the net effect of loss of SIRT1 on gluconeogenesis is determined by the complex compensatory patterns of multiple factors.
An intriguing observation in the present study is that the effect of SIRT1 on fatty acid metabolism is coupled with altered cholesterol metabolism. Not only do SIRT1 LKO mice accumulate massive amounts of cholesterol when fed a western diet (Figure 5E), they also accumulate significantly higher hepatic cholesterol when fed a high-fat diet without cholesterol (Figure S4). These observations suggest increased de novo synthesis is the major source of hepatic cholesterol. However, the expression levels of two key mediators of cholesterol synthesis, SREBP2 and HMGCoA reductase, were normal in SIRT1 LKO mice on the western diet. Hence, the increased synthesis may be due to an increase in the availability of substrates through accumulated hepatic free fatty acids (Figure 6D).
The metabolic phenotypes observed in SIRT1 LKO mice are in line with several previous reports. For example, activation of SIRT1 by the polyphenol resveratrol and several synthetic pharmacologic activators has been shown to protect against high-fat induced obesity and metabolic derangements (Baur et al., 2006; Lagouge et al., 2006; Milne et al., 2007). Manipulation of SIRT1 levels in the liver has been reported to affect the expression of a number of genes involved in glucose and lipid metabolism (Rodgers and Puigserver, 2007). Additionally, recent studies demonstrated that modest overexpression of SIRT1 resulted in a protective effect against high-fat induced hepatic steatosis and glucose intolerance (Banks et al., 2008; Pfluger et al., 2008). Our observations from SIRT1 LKO mice suggest that hepatic SIRT1 mediates a fine balance between energy influx and energy expenditure in the liver. However, it is important to note that although insulin signaling is impaired in the livers of SIRT1 LKO mice (Figure 7F), they show normal insulin sensitivity and fuel metabolism in white adipose tissue and muscle, and thus do not develop systemic glucose intolerance (Figure S5C).
Several of the observed metabolic alterations in the SIRT1 LKO mice are in contrast from those observed in some SIRT1 knockout animal studies (unpublished observations; (Chen et al., 2008)). For example, SIRT1 LKO mice gain significantly more weight than wild type mice and develop hepatic steatosis when fed with high-fat diets (Figure 6A and Figure S4). However, whole body SIRT1 knockout mice are protected from high-fat diet induced obesity and fatty liver (Li and Guarente, unpublished observations). A distinguishing difference between these two knockout models is their overall growth condition. SIRT1 LKO mice have no obvious phenotypic abnormalities under normal dietary conditions, whereas whole body SIRT1 knockouts suffer severe growth retardation, which can likely be attributed to a number of developmental defects (Cheng et al., 2003; McBurney et al., 2003). In addition, defective SIRT1 function in other tissues is likely to systemically affect energy metabolism in the whole body SIRT1 knockout mice, making it difficult to dissect hepatic specific function of SIRT1.
In summary, we have shown that hepatic SIRT1 plays an important role in the regulation of lipid metabolism in response to nutrient and hormonal signals. While it has been reported that SIRT1 systemically regulates energy homeostasis in many metabolic tissues by modulating a variety of signaling pathways, we show that in the liver, a major target of this sirtuin is the PPARα/PGC-1α signaling pathway and fatty acid oxidation. Our findings provide a direct link between SIRT1 and hepatic fatty acid metabolism, and suggest that therapeutic strategies designed to modulate SIRT1 activity may be beneficial for the treatment of hepatic diseases as well as obesity-associated metabolic syndrome.
EXPERIMENTAL PROCEDURES
Animal experiments
SIRT1 allele with floxed exon 4 (Cheng et al, 2003) was backcrossed 5 times into the C57BL/6 background. It was then bred with mice expressing the Cre recombinase driven by the albumin promoter (Jackson laboratory) to generate liver specific SIRT1 knockout mice (SIRT1 LKO) in over 98% C57BL/6 background. SIRT1 LKO mice and their age-matched littermate Lox controls (Cre−/−, SIRT1 flox/flox) older than 6-week of age were fed ad libitum either a standard laboratory chow diet or a high-fat western diet (D12079B, Research Diets) for 11 weeks. All animal experiments were conducted in accordance with guidelines of NIEHS/NIH Animal Care and Use Committee.
Histological and biochemical analysis
Paraffin-embedded liver sections were stained with hematoxylin and eosin for morphology or sirius red for collagen. Serum NEFA, cholesterol, and triglycerides were measured using commercially available kits (WAKO and Sigma). Serum insulin and leptin levels were measured by ELISA (Meso scale discovery). Liver lipids were extracted as described (Danno et al., 1992), and liver triglycerides, NEFA, and cholesterol were quantified by commercially available kits (Sigma and Wako).
Cell culture
HEK293T cells stably infected with pSuper or pSuper-SIRT1 RNAi have been described (Li et al., 2007). Mouse primary hepatocytes were isolated from control or SIRT1 LKO mice using collagenase perfusion, seeded on collagen-coated plates in seeding medium (high glucose DMEM, 10% FBS, 100nM insulin, 1uM dexamethasone), and maintained in maintenance medium (low glucose DMEM, 0.1% bovine serum albumin). To induce the expression of fatty acid oxidation genes, primary hepatocytes were pre-incubated with 125μM palmitic acid/BSA for overnight in the presence or absence of 30μM WY14643 in low glucose medium, followed by incubation with 125μM palmitic acid/BSA and 1 mM Carnitine in glucose free DMEM medium for 4 h.
Fatty acid β-oxidation
Primary hepatocytes isolated from SIRT1 LKO mice or Lox controls were pre-incubated with 500μM palmitic acid/BSA in maintenance medium for 24 h in the presence or absence of 30μM WY14643 followed by incubation with 125μM 3H palmitic acid (Sigma)/BSA and 1 mM Carnitine in PBS for 2 h. 3H2O was measured as described previously (Finck et al., 2006).
Luciferase assay
For trans-activation experiments, cells were transfected with indicated fire-fly luciferase reporter and control pRL-TK (Renilla Luciferase, Promega). Cells were incubated with or without PPARα agonists as indicated for 24 h and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). The final firefly luciferase activity was normalized to the co-expressed renilla luciferase activity.
Western blot analysis, Co-immunoprecipitation and chromatin immunoprecipitation (ChIP) analysis
Liver whole-cell homogenates were prepared with NP40 buffer (50mM Tris-HCL, pH 8.0, 150mM NaCl, 0.5% NP40) containing Complete ™ protease and phosphatase inhibitors (Roche), and then immunoblotted using antibodies against p-JNK, JNK, p-Elf2α, Elf2α, p-cJun, p-Akt and Akt (Cell Signaling Technology Inc.).
For endogenous immuno-precipitation between SIRT1 and PPARα, control mice were oral gavaged with vehicle or 30 mg/kg WY for 2 h, liver nuclear extracts were then immunoprecipitated with anti-PPARα antibodies (H-98, Santa Cruz biotechnology Inc.).
ChIP analysis was performed as described by Upstate Biotechnology with antibodies against SIRT1, PGC-1α (Santa Cruz biotech and Chemicon), PPARα (Affinity BioReagents), BAF60a (BD Biosciences), or normal rabbit IgG. DNA fragments were subjected to qPCR using primers flanking PPRE on various PPARα targets.
Protein acetylation analysis
Primary hepatocytes from Lox control (Control) and SIRT1 LKO (LKO) mice were infected with lentiviruses expressing FLAG-PGC-1α. 72 h after infection, hepatocytes were treated with WY14643 for 3 h, and incubated with 25 μM MG132, 1 μM TSA, and WY for an additional hour. FLAG-PGC-1α was immuno-purified with anti-FLAG m2 beads (Sigma) and analyzed with anti-acetyl-lysine polyclonal antibodies (Cell Signaling Technology Inc.).
Statistical analysis
Values are expressed as mean ± standard error of mean (SEM). Significant differences between means was analyzed by two-tailed, unpaired Student’s t test and differences were considered significant at p< 0.05.
Acknowledgments
We thank Drs. Anton Jetten, Paul Wade, and John Cidlowski for critical reading of the manuscript; Dr. Frederic Alt at Harvard Medical School for providing SIRT1 exon 4 floxed allele. We also thank the NIEHS microarray facility for performing the microarray experiments and Jennifer Collins for analyzing the microarray data; NIEHS Laboratory of Experimental Pathology for histological staining and serum hormone ELISA; NIEHS viral core facility for lentiviruses. This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to X.L. (Z01 ES102205).
Footnotes
ACCESSION NUMBERS
The NCBI GEO accession number for the microarray data reported in this paper is.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E. Hepatic Fibroblast Growth Factor 21 Is Regulated by PPARalpha and Is a Key Mediator of Hepatic Lipid Metabolism in Ketotic States. Cell Metab. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, Accili D. SirT1 Gain of Function Increases Energy Efficiency and Prevents Diabetes in Mice. Cell metabolism. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nature reviews. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. The Journal of clinical investigation. 2004;114:147–152. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science (New York, NY. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science (New York, NY. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes & development. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003 doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science (New York, NY. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Danno H, Jincho Y, Budiyanto S, Furukawa Y, Kimura S. A simple enzymatic quantitative analysis of triglycerides in tissues. J Nutr Sci Vitaminol (Tokyo) 1992;38:517–521. doi: 10.3177/jnsv.38.517. [DOI] [PubMed] [Google Scholar]
- Eckel RH, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet. 2005;365:1415–1428. doi: 10.1016/S0140-6736(05)66378-7. [DOI] [PubMed] [Google Scholar]
- Ford ES, Giles WH, Mokdad AH. Increasing prevalence of the metabolic syndrome among u.s. Adults. Diabetes Care. 2004;27:2444–2449. doi: 10.2337/diacare.27.10.2444. [DOI] [PubMed] [Google Scholar]
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochemical and biophysical research communications. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- Grimaldi B, Nakahata Y, Kaluzova M, Masubuchi S, Sassone-Corsi P. Chromatin remodeling, metabolism and circadian clocks: The interplay of CLOCK and SIRT1. Int J Biochem Cell Biol. 2008 doi: 10.1016/j.biocel.2008.08.035. [DOI] [PubMed] [Google Scholar]
- Grundy SM, Brewer HB, Jr, Cleeman JI, Smith SC, Jr, Lenfant C. Definition of metabolic syndrome: report of the National Heart, Lung, and Blood Institute/American Heart Association conference on scientific issues related to definition. Arterioscler Thromb Vasc Biol. 2004;24:e13–18. doi: 10.1161/01.ATV.0000111245.75752.C6. [DOI] [PubMed] [Google Scholar]
- Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Cook WS, Qi C, Yeldandi AV, Reddy JK, Rao MS. Defect in peroxisome proliferator-activated receptor alpha-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J Biol Chem. 2000;275:28918–28928. doi: 10.1074/jbc.M910350199. [DOI] [PubMed] [Google Scholar]
- Hedley AA, Ogden CL, Johnson CL, Carroll MD, Curtin LR, Flegal KM. Prevalence of overweight and obesity among US children, adolescents, and adults, 1999–2002. Jama. 2004;291:2847–2850. doi: 10.1001/jama.291.23.2847. [DOI] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M, Hotamisligil GS. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- Hunt MC, Nousiainen SE, Huttunen MK, Orii KE, Svensson LT, Alexson SE. Peroxisome proliferator-induced long chain acyl-CoA thioesterases comprise a highly conserved novel multi-gene family involved in lipid metabolism. J Biol Chem. 1999;274:34317–34326. doi: 10.1074/jbc.274.48.34317. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V, Elmquist JK, Gerard RD, Burgess SC, Hammer RE, Mangelsdorf DJ, Kliewer SA. Endocrine Regulation of the Fasting Response by PPARalpha-Mediated Induction of Fibroblast Growth Factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Kalaany NY, Mangelsdorf DJ. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol. 2006;68:159–191. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- Kanda H, Tateya S, Tamori Y, Kotani K, Hiasa K, Kitazawa R, Kitazawa S, Miyachi H, Maeda S, Egashira K, Kasuga M. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. The Journal of clinical investigation. 2006;116:1494–1505. doi: 10.1172/JCI26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. The Journal of clinical investigation. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura YI, Kitamura T, Kruse JP, Raum JC, Stein R, Gu W, Accili D. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005;2:153–163. doi: 10.1016/j.cmet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu C, Li N, Hao T, Han T, Hill DE, Vidal M, Lin JD. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 2008;8:105–117. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Molecular cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Iii JY, Olefsky J, Guarente L, Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008 doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Molecular and cellular biology. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science (New York, NY. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Pagans S, Pedal A, North BJ, Kaehlcke K, Marshall BL, Dorr A, Hetzer-Egger C, Henklein P, Frye R, McBurney MW, Hruby H, Jung M, Verdin E, Ott M. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol. 2005;3:e41. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhshandehroo M, Sanderson LM, Matilainen M, Stienstra R, Carlberg C, de Groot PJ, Muller M, Kersten S. Comprehensive Analysis of PPARalpha-Dependent Regulation of Hepatic Lipid Metabolism by Expression Profiling. PPAR research. 2007;2007:26839. doi: 10.1155/2007/26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Thelen A, Jump DB. Peroxisome proliferator-activated receptor alpha inhibits hepatic S14 gene transcription. Evidence against the peroxisome proliferator-activated receptor alpha as the mediator of polyunsaturated fatty acid regulation of s14 gene transcription. J Biol Chem. 1996;271:17167–17173. doi: 10.1074/jbc.271.29.17167. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel VT, Liu ZX, Qu X, Elder BD, Bilz S, Befroy D, Romanelli AJ, Shulman GI. Mechanism of hepatic insulin resistance in non-alcoholic fatty liver disease. J Biol Chem. 2004;279:32345–32353. doi: 10.1074/jbc.M313478200. [DOI] [PubMed] [Google Scholar]
- Sanyal AJ. Mechanisms of Disease: pathogenesis of nonalcoholic fatty liver disease. Nat Clin Pract Gastroenterol Hepatol. 2005;2:46–53. doi: 10.1038/ncpgasthep0084. [DOI] [PubMed] [Google Scholar]
- Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Molecular and cellular biology. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman AI, Mangelsdorf DJ. Retinoid x receptor heterodimers in the metabolic syndrome. N Engl J Med. 2005;353:604–615. doi: 10.1056/NEJMra043590. [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci U S A. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stienstra R, Mandard S, Patsouris D, Maass C, Kersten S, Muller M. Peroxisome proliferator-activated receptor alpha protects against obesity-induced hepatic inflammation. Endocrinology. 2007;148:2753–2763. doi: 10.1210/en.2007-0014. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science (New York, NY. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- van den Berghe G. The role of the liver in metabolic homeostasis: implications for inborn errors of metabolism. Journal of inherited metabolic disease. 1991;14:407–420. doi: 10.1007/BF01797914. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Molecular and cellular biology. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. The Journal of clinical investigation. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. The Journal of clinical investigation. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. The EMBO journal. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- Yuan M, Konstantopoulos N, Lee J, Hansen L, Li ZW, Karin M, Shoelson SE. Reversal of obesity- and diet-induced insulin resistance with salicylates or targeted disruption of Ikkbeta. Science (New York, NY. 2001;293:1673–1677. doi: 10.1126/science.1061620. [DOI] [PubMed] [Google Scholar]
- Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]