Abstract
Two types of acquired loss of heterozygosity are possible in cancer: deletions and copy-neutral uniparental disomy (UPD). Conventionally, copy number losses are identified using metaphase cytogenetics while detection of UPD is accomplished by microsatellite and copy number analysis and as such, is not often used clinically. Recently, introduction of single nucleotide polymorphism (SNP) microarrays have allowed for the systematic and sensitive detection of UPD in hematological malignancies and other cancers. In this study, we have applied 250K SNP array technology to detect previously cryptic chromosomal changes, particularly UPD, in a cohort of 301 patients with myelodysplastic syndromes (MDS), overlap MDS/myeloproliferative disorders (MPD), MPD, and acute myeloid leukemia (AML). We show that UPD is a common chromosomal defect in myeloid malignancies, particularly in chronic myelomonocytic leukemia (CMML; 48%) and MDS/MPD-unclassifiable (38%). Furthermore, we demonstrate that mapping minimally overlapping segmental UPD regions can help target the search for both known and unknown pathogenic mutations, including newly identified missense mutations in the proto-oncogene c-Cbl in 7/12 patients with UPD11q. Acquired mutations of c-Cbl E3 ubiquitin ligase may explain the pathogenesis of a clonal process in a subset of MDS/MPD, including CMML.
INTRODUCTION
Among chromosomal aberrations involved in the pathogenesis of hematological malignancies, somatic uniparental disomy (UPD) is increasingly recognized as a common molecular defect that results in copy-neutral loss of heterozygosity (LOH). It is likely that this defect is random and occurs either as a result of mitotic recombination or as an attempt to correct loss of chromosomal material.(1) Important as a clonal marker, UPD may participate in the malignant pathological process, particularly if UPD results in duplication of either an activating or loss of function mutation, or even perhaps an aberrant germ-line genetic variant. UPD can also lead to increased or decreased gene expression through alteration of an encoded epigenetic pattern.(2) Perhaps the most well-known example of UPD involved in hematologic malignancies is UPD9p,(3) which led to the identification of the JAK2 V617F mutation in myeloproliferative disorders (MPD).(4-6)
Routine detection of UPD was not easily possible in the past and required systematic, labor-intensive microsatellite and copy-number analysis limited in resolution. Recently, the advent of single nucleotide polymorphism array (SNP-A) technology has allowed for the efficient and effective detection of segmental UPD in addition to other, previously undetectable micro-deletions and duplications. Previously, we and others have shown that clonal UPD occurs frequently in myelodysplastic syndromes (MDS), secondary acute myeloid leukemia (AML), MPD, and MDS/MPD overlap disease entities.(7-9) Other studies have shown that in patients with AML, regions of UPD can correlate with homozygous somatic mutations affecting proteins including FLT3 and CEBPA.(10-12) However, systematic analysis of commonly affected areas of UPD using SNP-A technology in a broader cohort of patients with myeloid malignancies has not been performed. In this study, we have applied high-density 250K SNP-A to patients with malignant myeloid disorders to identify segmental UPD, map shared/overlapping lesions, suggest candidate genes which may be involved in disease pathogenesis, and examine relationships between UPD and corresponding clinical phenotypes.
MATERIALS AND METHODS
Patients
Bone marrow aspirates and/or blood was collected from 301 patients with myeloid malignancies (mean age 64 years; range 17-87) seen between 2002-2008 at participating institutions. Informed consent for sample collection was obtained according to protocols approved by the Cleveland Clinic and Johns Hopkins University IRBs. Samples obtained from 116 healthy individuals at the Cleveland Clinic (CCF) were used as controls. In addition, a cohort of 61 CEPH (Utah residents with ancestry from northern and western Europe; CEU) HapMap individuals was used for comparison;(13) however, it should be noted that the criteria used to assign membership in the CEPH population have not been specified, except that all donors were residents of Utah.(14)
DNA extraction
DNA was extracted from patient specimens using the ArchivePure DNA Blood Kit (5Prime, Gaithersburg, MD, USA) as per the manufacturer’s instructions. The concentration of the DNA was determined using a ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA) and the quality determined by gel electrophoresis. CD3+ lymphocytes were isolated by magnetic bead separation using the RoboSep instrument (StemCell Technologies, Vancouver, Canada).
SNP-A analysis
The Gene Chip Mapping 250K Assay Kit (Affymetrix, Santa Clara, CA, USA) was used for SNP-A analysis and utilized per the manufacturer’s instructions as previously described.(15) Lesions identified by SNP-A were compared with the Cancer Genome Anatomy Project database (http://cgap.nci.nih.gov) and our own internal control series to exclude known copy number variants. To confirm regions of LOH detected by 250K SNP-A, we repeated samples when possible (N=95) on ultra-high density Affymetrix 6.0 arrays and analyzed using Genotyping Console v2.0 (Affymetrix). Signal intensity was analyzed and SNP calls determined using Gene Chip Genotyping Analysis Software Version 4.0 (GTYPE). Copy number (CN) and areas of UPD were investigated using a Hidden Markov Model and CN Analyzer for Affymetrix GeneChip Mapping 250K arrays (CNAG v3.0) as previously described.(15, 16)
Mutational screening
Screening for the JAK2 V617F and c-MPL W515L mutations was performed using a DNA tetra-primer ARMS assay as previously described.(8, 17, 18) For FLT3-ITD detection, capillary electrophoresis was used: PCR amplification primers were prepared as follows: ITD-F: 5′-FAMGCAATTTAGGTATGATGAAAGCCAGC-3′; ITD-R: 5′-CTTTCAGCATTTTGACGGCAACC-3′. After PCR amplification, 1μl product and 0.5μl of 1:1 diluted 500 HD Rox size standard (Applied Biosystems, Foster City, CA, USA) were added to 13μl formamide and analyzed using the ABI Prism 310 Genetic Analyzer (Applied Biosystems) with an extended run time of 30 minutes. Patients negative for the ITD mutation will have a single 332bp fragment. For patients positive for an ITD mutation, an additional fragment ranging from 350 to 554bp will be present.
NRAS mutational screening
Screening for mutations in exons 1 and 2 of NRAS was carried out using direct genomic DNA sequencing. The following primer sets were used: 1F: 5′-GGCCGATATTAATCCGGTGT-3′; 1R: 5′-TGGGTAAAGATGATCCGACA-3′; 2F: 5′-GCAATTTGAGGGACAAACCA-3′; 2R: 5′-TGGTAACCTCATTTCCCCATA-3′. PCR conditions: 94°C for 4 minutes, 30 cycles of 94°C for 30 seconds, 51°C for 30 seconds, and 72°C for 30 seconds; one cycle at 72°C for 5 minutes. Following PCR amplification, product was prepared for direct sequencing using standard conditions and analyzed using the 3100-Avant Genetic Analyzer (Applied Biosystems).
c-Cbl mutational screening
To screen patients for mutations in c-Cbl, direct genomic DNA sequencing was performed. Exons 7, 8, and 9 from genomic DNA were amplified using the following primer sets: 7F: 5′-ACACCACGTTGCCCTTTTAG-3′, 7R: 5′-GTCAATGGGTTCCAATGAAT-3′; 8F: 5′-GGACCCAGACTAGATGCTTTCT-3′, 8R: GAAAATACATTTCCTAGAGATCAAAAA-3′; 9F: 5′-CTGGCTTTTGGGGTTAGGTT-3′, 9R: 5′-TCGTTAAGTGTTTTACGGCTTT-3′. PCR reaction conditions were as follows: 94°C for 4 minutes, 30 cycles of 94°C for 30 seconds, 48.5°C for 30 seconds, and 72°C for 30 seconds; one cycle at 72°C for 5 minutes. The PCR product was then prepared for sequencing under standard conditions.
RESULTS
Detection of UPD using 250K SNP-A
To define acquired somatic UPD and distinguish this lesion from germ-line, non-clonal regions of homozygosity, we first performed SNP-A analysis on a control cohort consisting of 116 healthy internal controls. Areas of homozygosity were defined as regions spanning >400 consecutive homozygous SNPs as detected by the AsCNAR (Allele-specific Copy Number Analysis using anonymous References) algorithm, which allows for the detection of abnormal clones comprising 20-30% of the total cell population.(15, 19) The criteria of >400 homozygous SNPs is specific for our 250K array data and was chosen based upon the uneven distribution of SNP probes on the array. Certain regions of the genome are not sufficiently covered by the 250K array due to relatively long stretches of LOH definable by only a few SNPs.(19) This provides a greater risk of technical artifacts. Thus, the longer the series of homozygous SNPs, the higher the probability the LOH is genuine. 400 sequential SNPs was deemed to be of sufficient length to account for these regions. While such an approach has a greater risk of generating false negative results, it is also more likely to eliminate technical artifacts. However, this criterion is not necessary for the 6.0 array due to the much higher, more uniform density of the SNP probes on the array (see below).
Using this criteria, and through the application of CNAG v3.0 software, segmental regions of homozygosity were identified in 3.5% (4/116) of the internal control cohort, comparable to previous reports.(20) SNP-A analysis of CD3+ lymphocytes in these controls confirmed the germ-line derivation of these regions. Moreover, we identified a similar percentage (4.9%) of patients harboring germ-line regions of homozygosity in the CEU HapMap cohort.(13) The average size of these regions in both control groups was 10.3Mba (4.2Mba-22.4Mba) and occurred in small areas on chromosomes 2, 3, 4, 6, 9, 13, and 15 (Supplemental Table A). In general, regions of homozygosity detected in controls were all short, interstitial fragments that did not regularly overlap with commonly affected regions of UPD seen in patients. Any potential gene candidates located in regions of homozygosity found in controls were subsequently excluded in patient analyses.
Potentially pathogenic UPDs in patients were chosen based on a rational algorithm of exclusion compared to controls. All UPD regions smaller than 20Mba (the median size of lesions found in controls + 2X S.D.) were excluded from analysis unless they extended to the telomere. The rationale for the latter is based on our analysis of control cases (none had UPD extending to the telomere) and the observation of frequent extension of UPD to the telomere in JAK2 V617F cases with UPD9p.(5, 8) While such stringent criteria may be associated with a higher false negative rate, in the interest of minimizing false positives, we feel that this method is the best method to remove any non-pathogenic regions in patient samples. This exclusion principle was substantiated by running CD3+ lymphocyte fractions from patients showing regions of UPD similar to those seen as controls as well as those with longer, telomeric UPD (N=18). In agreement with findings from the control cohort described above, small interstitial regions of UPD identified in patients using CD3+ comparison were found to be germ-line while longer, telomeric regions of UPD were all clonal. Comparison of 250K results of both blood and bone marrow samples from the same patients showed identical results each time (N=6).(21)
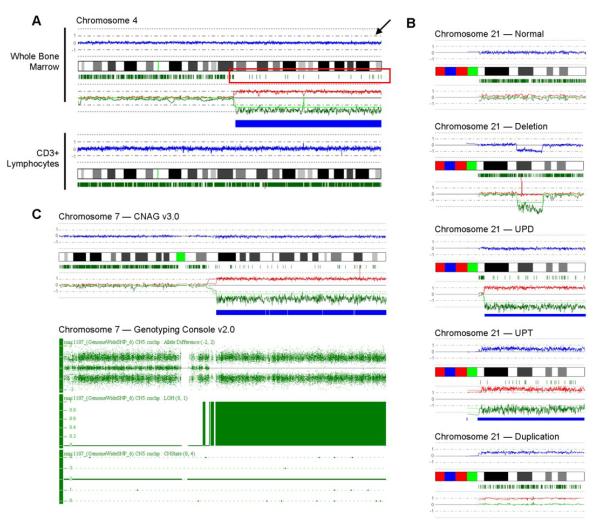
Of the 301 total patients in our cohort, 44 had CMML or AML evolving from CMML, 140 had MDS or AML evolving from MDS, 37 had MDS/MPDu or AML evolving from MDS/MPDu, 14 had MPD or AML evolving from MPD, and 66 had primary AML (Table 1). Across diseases tested, UPD is a frequent type of chromosomal lesion, occurring in 18% of MDS cases (25/140), 37% of MPD cases (8/14, particularly UPD9p in those positive for the JAK2 V617F mutation), and 24% (16/66) of all primary AML cases; however, there appears to be a slightly higher frequency of segmental UPD in patients with MDS/MPDu and CMML, occurring in 38% (14/37) and 48% (21/44) of patients respectively. The average size of UPD in patients was 52.4Mba. Copy number changes and UPD were identified using CNAG v3.0 software. In contrast to deletions, which show clear loss of copy number (CN) signal intensity, UPD is characterized by drastic loss of heterozygous loci but normal, diploid CN (Figure 1B). In one patient with AML, a distinct area of LOH was accompanied by hyperploid CN consistent with the presence of uniparental trisomy 21 (Figure 1B). In many patients (N=95), we have confirmed the presence of segmental LOH detected on 250K arrays, including UPD, by repeated analysis using new, ultra-high density Affymetrix 6.0 arrays and Genotyping Console v2.0 software (Figure 1C). Of 95 patients tested, 21 had previously-identified UPD. There was remarkable concordance between the 6.0 array and the 250K arrays. All UPD regions detected by 6.0 arrays were detected by the 250K; however, it should be noted that because of the greater resolution of the 6.0 array, which allowed for the identification of smaller regions of homozygosity in controls, the average size of UPD detected in controls was reduced. Therefore, if our same “cut-off” criteria method was applied, the minimal size of pathogenic lesions would be decreased from 20Mba to approx. 10Mba for 6.0 arrays.
Table 1.
Characteristics of patients
| Diagnosis | Sub-group | N | % of total with abnormal SNP-A |
% of total with UPD |
% of those with abnormal SNP-A having UPD |
|
|---|---|---|---|---|---|---|
| Total (N=301) |
MDS (N=140) |
RA/RARS/RCMD/5q | 67 | 63 | 18 | 29 |
| RAEB I/II | 28 | 79 | 14 | 18 | ||
| Secondary AML | 45 | 67 | 20 | 30 | ||
| MDS/MPD (N=81) |
MDS/MPDu | 29 | 66 | 34 | 53 | |
| CMML | 27 | 74 | 41 | 55 | ||
| Secondary AML□ | 25 | 88 | 60 | 68 | ||
| MPD (N=14) |
n/a | 7 | 100 | 57 | 57 | |
| Secondary AML | 7 | 86 | 57 | 67 | ||
| pAML (N=66) |
n/a | 66 | 56 | 24 | 43 | |
Samples from 116 healthy individuals were used as controls; n/a, not applicable; pAML, primary acute myeloid leukemia
Includes patients who evolved to AML from both MDS/MPDu and CMML
Figure 1. Detecting acquired, segmental UPD using 250K SNP-A technology.
(A) SNP-A “karyograms” of both whole bone marrow cells and CD3+ lymphocytes in one patient demonstrate the somatic nature of acquired UPD. In this representative sample, chromosome (ch.) 4 is displayed. The blue line (indicated by the arrow) represents average copy number (CN) signal intensity of SNPs on the array chip. In this instance, there are no CN variations, and thus the blue line does not deviate from normal diploid CN. The green marks below the idiogram represent heterozygosity at particular DNA loci. In the region of UPD seen in whole bone marrow cells, drastic reduction of heterozygous loci denotes the region of UPD (red box). Remaining green marks in the region of UPD delineate the presence of non-clonal cells in the sample. The green and red lines represent the AsCNAR algorithm and show the relative signal intensity for individual homologs. In CD3+ sorted lymphocytes, a normal ch. 4 is seen. (B) SNP-A analysis of chromosome 21 in 1 healthy control and in 4 patients demonstrates the various types of lesions observed: deletions, UPD, duplications, and in a unique patient, uniparental trisomy (UPT). (C) Comparison of Affymetrix 250K and 6.0 arrays in the detection of UPD7q for one patient. CNAG v3.0 analysis (top) shows clear UPD of ch. 7 both by loss of heterozygous loci and allelic imbalance. Repeated testing on the 6.0 array and analysis using Genotyping Console v2.0 software (bottom) confirms the 250K SNP-A findings. Note that the Genotyping Console output includes Allele Difference, LOH, and CN variation (CNState) plots. The Allele Difference graph represents the genotypes for each individual SNP. Dots with a value of 1 represent SNPs with an “AA” genotype, while those with a value of -1 represent SNPs with a “BB” genotype. Dots at 0 represent heterozygous SNPs (“AB”). Complete loss of all “AB” SNPs indicates copy-neutral LOH. This is further demonstrated by both the LOH and CNState graphs, which show no loss in copy number but clear LOH.
Acquired, segmental UPD serves as a molecular marker for known gene mutations
Previous studies in AML have demonstrated that regions of acquired UPD detected by SNP-A may correlate with areas that harbor genes implicated in disease pathogenesis.(12) One could hypothesize that acquired UPD leading to homozygosity of a pre-existing pathogenic mutation provides an already mutant clone with an additional growth advantage. Examples of recurrent UPD regions identified in our cohort that include genes previously associated with homozygosity are presented in Figure 2. For example, in 10/11 patients, including 4/37 patients with MDS/MPDu (11%) and 6/14 patients with MPD or AML evolved from MPD (43%), we found UPD9p coinciding with homozygous JAK2 V617F mutations (Figure 2). In addition, we also identified 7/143 patients (5%) with either primary or secondary AML harboring segmental UPD in the region of FLT3. All 7 of these patients were positive for a biallelic FLT3-ITD mutation (Figure 2).
Figure 2. Acquired UPD detected by 250K SNP-A may serve as a molecular marker for mutated genes in patients.
Topographical maps show regions of UPD in individual patients on chromosomes 9, 13 and 1 (Left). Yellow bars corresponding to the idiogram represent the regions affected for each patient. Individual diagnoses are listed to the right of the depicted lesion (disease sub-types are indicated in parentheses; for those patients who transformed to secondary AML, the initial presentation is given in parentheses). Red lines on the idiogram represent the physical location of the JAK2, FLT3 and c-MPL genes respectively. To the right: mutational screening of JAK2, FLT3, NRAS and c-MPL in patients with segmental UPD in the region of each gene confirms their homozygous status (note that sequencing analysis of NRAS exon 2 in patient 31 appears heterozygous due to the presence of non-clonal cells in the sample). Cartoons to the left of the maps represent the diploid chromosome set and identify the minimally overlapping region on each chromosome along with representative candidate genes in the region, regardless if the segment overlaps with known gene mutations or not. Gel images have been cropped and enhanced.
In addition to JAK2 and FLT3, we tested for a homozygous c-KIT mutation in patient 45 who had UPD in the region of 4q12 (Figure 3), and c-MPL mutations in 7 patients with UPD1p. While patient 45 had wild-type c-KIT, we did identify homozygous c-MPL W515L mutations (1p34.2) in 2 of the 7 patients with UPD1p in this region (Figure 2). In patients 30 and 31, who were both negative for a c-MPL mutation, we hypothesized that given the localization of their lesions and their history of CMML, they may instead harbor homozygous NRAS (1p13.2) mutations.(22) Direct genomic DNA sequencing of NRAS exons 1 and 2 in these two patients revealed missense mutations at codon 13 (G13V) in patient 30, and at codon 61 (Q61H) in patient 31.
Figure 3. Genome-wide scan of overlapping UPD in patients.
Topographical maps show all overlapping regions of UPD occurring in greater than 2 patients. Gray bars corresponding to the idiogram represent the regions affected for each patient. Individual diagnoses are listed to the right of the depicted lesion. Cartoons to the left of the maps represent the diploid chromosome set and identify the minimally overlapping region on the chromosome along with representative candidate genes in each region.
Other previously reported gene mutations (not investigated in this study) occurring in biallelic configuration as a result of acquired UPD have included WT1, CEBPA, RUNX1 and NF1.(12, 23) Thus, segmental UPD detected by SNP-A may serve as a clear molecular marker for potentially pathogenic homozygous gene mutations.
Subsequently, we hypothesized that the presence of biallelic mutations may have prognostic significance. Previous studies have shown that hemizygosity for the FLT3-ITD mutation due to deletion of the remaining wild-type allele has a negative impact on overall survival.(24) Therefore, as an example, we tested whether duplication of a FLT3-ITD mutation due to acquired UPD would also have prognostic impact. We screened all primary and secondary AML patients for FLT3-ITD mutations on whom DNA was available. 23/127 patients (18%) were positive for a FLT3-ITD mutation, 7 of which occurred in homozygous configuration due to acquired UPD. When survival outcomes of AML patients heterozygous for a FLT3-ITD mutation (N=16) were compared to those with a homozygous FLT3-ITD mutation (N=7), Kaplan-Meier analysis revealed a significant detrimental effect on overall survival (p=0.0011) (Supplemental Figure A).
Overlapping segmental UPD in patients reveals other regions which may harbor pathogenic gene mutations
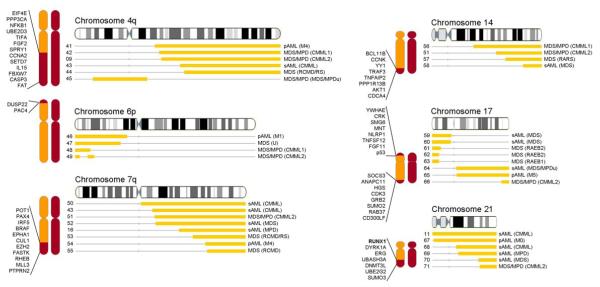
In our cohort of 301, we have identified several regions of the genome commonly affected by UPD that may also harbor genes implicated in disease pathogenesis. Minimally overlapping regions of UPD found in greater than 2 patients on any chromosome are defined in Figures 2 & 3. Chromosomes frequently affected by UPD include 1p (N=11), 4q (N=6), 7q (N=8), 9p (N=11), 13 (N=7), 17 (N=8), and 21 (N=6). 83% of these lesions occurred in patients with an otherwise normal SNP-A karyotype (data not shown). The chromosome arm most often affected was 11q, occurring in 12/301 patients, 6 of which had MDS/MPDu, CMML, or related disorders by clinical phenotype (Figure 4A). Certain lesions were associated with specific disease sub-types. For instance, all patients with UPD17p had advanced stages of MDS or AML evolving with MDS, perhaps consistent with a p53 mutation.(25) Likewise, all patients with UPD17q had MDS/MPD or abnormal monocytic features, including CMML and M5 primary AML (Figure 3). A similar scenario occurs in patients with UPD4q. Perhaps not surprisingly, all but one patient with UPD21 have either primary or secondary AML, consistent with a potential RUNX1 mutation.(22) However, greater variation exists on chromosomes 7q and 14q, where a broader range of patients harbor UPD in these regions. This is consistent with the clinical spectrum of chromosome 7q deletions in MDS, AML, and MPD.
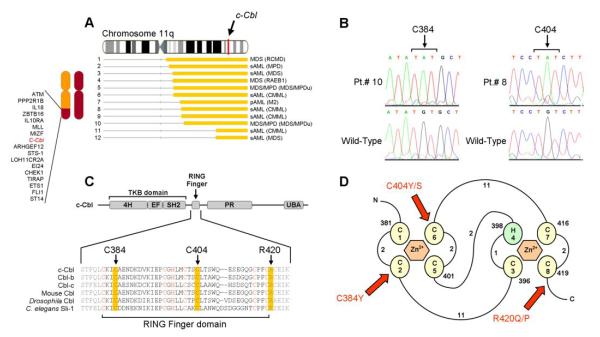
Figure 4. Identification of unique, missense mutations in c-Cbl (11q23.3).
(A) Map of individual UPD lesions on chromosome 11q and the location of the c-Cbl gene. (B) Direct genomic DNA sequencing of exon 8 in c-Cbl reveals the presence of missense mutations. Each base pair change occurs in a homozygous state due to the copy-neutral LOH and results in the substitution of cysteine residues for tyrosine or serine residues at positions 384 (in patients 9 and 10) and 404 (in patients 6, 8, and 12). (C) Schematic representation showing the major domains of mammalian c-Cbl, primarily the tyrosine kinase binding (TKB) domain, RING finger, proline-rich (PR) region, and ubiquitin-associated domain (UBA). Also shown is the amino acid sequence spanning the RING finger domain of mammalian c-Cbl along with homologs Cbl-b, Cbl-c, and those found in Mus musculus, Drosophila, and C. elegans (Sli-1). Critical cysteine/histidine residues that make up the RING finger domain are indicated in red. Residues affected by mutations are highlighted in yellow. (D) Schematic of the prototypical RING finger domain found in c-Cbl (adapted from (39) (Fig1)) C’s denote cysteine residues; H, histidine residues and are numbered in the order with which they occur in the domain. Curved lines represent amino acid chains and are accompanied by numbers which denote their lengths. Red arrows indicate the affected residues in patients positive for the mutations and are accompanied by the resulting amino acid substitutions.
Identification of point mutations in c-Cbl (11q23.3) using SNP-A
Given the prevalence of UPD on chromosome 11q, we screened for candidate genes located in this region in an attempt to identify novel mutations. Candidate genes were chosen using a simple algorithm based on physical location and function of the encoded protein. Genes were first considered according to their location within minimally affected regions of UPD. Next, genes involved with leukemia, cell signaling, cell cycle regulation, or apoptosis were selected (particularly receptor tyrosine kinases or those linked to tyrosine kinase activity). Finally, genes already known to be involved with disease processes were isolated. In this manner, potential gene candidates were minimized from a list of many to a small few.
Among several candidate genes on 11q, including ATM, MLL, and STS1, was c-Cbl, a gene which encodes an E3 ubiquitin ligase involved in the ubiquitylation and degradation of active protein tyrosine kinase receptors.(26) Oncogenic mutations of c-Cbl occurring in a highly-conserved alpha-helix linker joining an SH2 tyrosine kinase receptor binding domain with a RING domain have been previously identified in cell lines and in a small number of patients with primary AML (5/162 in all);(27-30) however, to our knowledge, no mutations occurring in other c-Cbl protein domains and/or in other hematologic malignancies have been found thus far. Therefore, c-Cbl seemed a logical target for sequence analysis.
Among our UPD11q cohort, all 12 patients harbored UPD11q in the region of the c-Cbl gene (Figure 4A). Direct genomic sequencing of c-Cbl in these patients revealed the presence of 3 unique missense mutations, all occurring within or directly adjacent to the RING finger domain, which gives c-Cbl its ubiquitylation activity.(31) In total, 7/12 patients were affected (Table 2). One mutation, occurring in 2/7 patients, resulted in the substitution of an arginine residue with either glutamine or proline at position 420 (R420Q/P). R420Q, lying just outside the RING domain, has been previously identified in 1/150 AML patients;(30) however, we also found two additional, newly-identified missense mutations, both affecting the cysteine residues of the RING finger in the remaining 5 patients (Figures 4B and 4C). The cysteine residues, which bind zinc ions that form the zinc-chelating loops responsible for accommodating contacts with UbcH7, give c-Cbl its ubiquitylation activity. In 2/5 patients, residue 384 was altered by substitution of a tyrosine. In the other 3 patients, residue 404 was altered by substitution of either a tyrosine (in 1 patient) or serine (in 2 patients) (Figure 4D). The presence of each somatic mutation was confirmed by bidirectional DNA sequencing of multiple isolates and comparison against CD3+ sorted lymphocytes when available.
Table 2.
Characteristics of patients with c-Cbl mutations
| Pt. | Age | Sex | Initial presentation |
Current dx. |
Current BM blast % |
Metaphase karyotype | SNP-A karyotype | c-Cbl mutation |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Gain | Loss | UPD | ||||||||
| 2 | 61 | F | MPD (IMF) | sAML | 10 | 46,XX,del(13)(q12q22) [15]/46,XX[5] |
N | 13q13.2q31.1 | 9p13.3pter 11q12.3qter |
R420Q |
| 4 | 49 | F | MDS-RAEB1 | n/c | 7 | 47,XX,+21[20] | 21 | N | 6p21.32p22.1 11q13.1qter |
R420P |
| 6 | 78 | M | CMML2 (MP)□ | sAML | 60 | 46,XY[20] | 8 | N | 2p22.3p23.3 11q13.2qter |
C404S |
| 8 | 54 | F | CMML1 (MP)□ | sAML | 60 | 46,X,-X[17]/46,XX[3] | N | N | 11q13.4qter | C404Y |
| 9 | 69 | M | CMML2 (MD)□ | sAML | 50 | 46,XY[20] | 21q11.1 | 7q21.1 | 4q21.21qter 11q13.4qter |
C384Y |
| 10 | 82 | M | MDS/MPDu | n/c | 0 | 46,XY[20] | N | 3p14.2, 7p21.2 | 11q13.5qter | C384Y |
| 12 | 44 | F | MDS-RAEB2 | sAML | 14 | 46,XX[20] | 15q13.3 | N | 11q22.3qter | C404S |
Given the preponderance of c-Cbl mutations in patients who exhibit MDS/MPD features, we tested for the presence of monoallelic mutations in all patients with a diagnosis of MDS/MPD, including those negative for UPD11q; however, in 40 such patients screened, no c-Cbl mutations were identified.
DISCUSSION
SNP-A technology, with its improved resolution and ability to detect uniparental disomy, increases the overall detection rate of chromosomal abnormalities and complements metaphase cytogenetics (MC) in the delineation of chromosomal lesions associated with hematological malignancies. Recently, we and others have shown that in MDS, MDS/MPD and MDS-derived AML, previously unrecognized chromosomal defects detected by SNP-A have similar impact on prognostic parameters including overall survival as those lesions detected by MC.(7, 9) In addition to its potential clinical implications, SNP-A karyotyping is also an excellent investigative tool, facilitating delineation of invariant chromosomal defects and, in turn, corresponding genes which may be implicated in the pathogenesis of hematological malignancies.
Using 250K SNP-A and CNAG v3.0 software, we have identified in a large cohort of patients with MDS, MDS/MPD, MPD, and AML, a significant proportion of UPD, particularly in MDS/MPD. Often times, overlapping regions of UPD correlated with the presence of homozygous gene mutations, including FLT3-ITD in 7/7 cases with UPD13q, accounting for 30% of all cases positive for a FLT3-ITD mutation (N=23), and JAK2 V617F mutations in all MDS/MPD and MPD cases characterized by UPD9p. In addition, we identified c-MPL or NRAS point mutations in 4 patients with UPD1p. c-MPL mutations were associated with MDS/MPD, and we believe that this is the first report of biallelic c-MPL or NRAS mutations occurring as a result of acquired UPD. Although not formally tested here, one could also hypothesize that, in accordance with previous studies,(11, 12) there are homozygous gene mutations affecting either RUNX1 or CEBPA in those patients with acquired UPD on chromosomes 21 (N=6) or 19 (N=2) respectively.
Systematic mapping of copy-neutral LOH has allowed for the identification of other frequently shared areas of UPD that are currently not attributable to any known gene mutations in myeloid malignancies. Identification of novel mutations in these regions may explain the pathophysiology of those individual cases affected. Examples of these commonly affected areas include segments on chromosomes 4q, 6p, 7q, 14, 17 and 11q. Various genes potentially involved in the pathogenesis of leukemia are located in these regions including EIF4E, CCNA2 and FGF2 on 4q, DUSP22 and PAC4 on 6p, BRAF, CUL1, and EZH2 on 7q, TNFAIP2 and AKT1 on 14, and p53 and GRB2 on 17.
When we examined patients with UPD11q, we noted several common clinical phenotypic trends, including history of MDS/MPD, the presence of monocytic blasts or increased numbers of differentiated monocytes, reticulin fibrosis and propensity to AML transformation. Monoallelic mutations in the alpha-helix linker domain of c-Cbl disrupt binding of c-Cbl to E2 ubiquitin-conjugating enzymes and thus, deregulate proper ubiquitylation and degradation of phosphorylated protein tyrosine kinase receptors in animal models(32) and in a small number of patients with AML.(27-30) We hypothesized that mutated, dysfunctional c-Cbl could lead to deregulated activity along proliferative transduction pathways consistent with the phenotype of MDS/MPD. For example, other TK receptors including CSF1R, PDGFRβ and EGFR have all been described to be inactivated by members of the Cbl ubiquitin ligase family either directly or via binding to Grb2.(33-37)
In addition to 2 patients harboring biallelic R420Q/P mutations previously reported, we identified new, biallelic mutations of c-Cbl in 5 additional patients which cause single amino acid substitutions affecting the highly-conserved cysteine residues at positions 384 and 404 of the RING domain. Analysis of non-clonal T-cells from these cases confirmed the acquired nature of these mutations. However, unlike previous reports, sequencing targeted to identify RING and linker domain monoallelic mutations in heterozygous configuration did not identify patients with monoallelic c-Cbl. Based on our observation of only biallelic mutations in our MDS/MPD patients, we hypothesize that the pathogenic role of c-Cbl mutations may differ in AML versus MDS/MPD. Alternatively or additionally, the discrepancy between our findings and those previously reported in AML may be complicated by technical problems which arise when distinguishing between true monoallelic mutations in one clone and a mixture of abnormal clonal cells harboring biallelic lesions (e.g., due to UPD) and nonclonal, wild-type cell contributions. We have shown that patients with c-Cbl mutations all have corresponding UPD11q; perhaps SNP-A applied to the previously described AML patients would also reveal UPD11q.
Patients with c-Cbl mutations showed a great deal of diversity with regard to clinical phenotype (Table 2). While c-Cbl mutations have been previously linked to primary AML only,(29, 30) all patients positive for c-Cbl mutations in this study had developed from a previous hematologic disease. In addition, some patients with c-Cbl mutations have additional chromosomal aberrations and/or genetic mutations that could modify the resultant clinical phenotype. For example, one patient (Patient 2, Table 2) was JAK2 V617F+. Another two patients (Patients 6 and 12, Table 2) were NPM1+. In addition, related genes Cbl-b (3q13.11) and Cbl-c (19q13.31) may harbor mutations with pathogenetic significance which could explain other cases of myeloid malignancies at the molecular level. These results serve to illustrate that advanced myeloid malignancies likely have many cooperating genetic and epigenetic lesions, and that high resolution SNP arrays provide insight into sorting out affected molecular pathways.
Systematic analysis of UPD also provides many important clues as to the pathogenesis that may operate in MDS, MDS/MPD, or AML. We have noted that certain chromosomes are as frequently affected by UPD as deletion (e.g. 7 and 21). However, UPD may suggest a different mechanism and type of gene affected (e.g., duplication of an activating mutation) compared to deletion, which can lead to haploinsufficiency or “unmasking” of a pre-existing inactivating mutation in a tumor suppressor gene. This may explain why some chromosomes are more frequently affected by deletion than UPD. For example, while we have identified 35 cases with deletions involving the minimally affected region on chromosome 5q, only 1 example of UPD was found in the corresponding area, suggesting that affected genes in this region do not provide an additional selection advantage if duplicated.
It should also be noted, however, that not all UPD is likely to be pathogenic but may simply serve as a marker of clonality and/or underlying previous DNA damage.(1) To enable analysis of as many samples as possible and determine clinical applicability of SNP-A, most patient samples were run using whole bone marrow and/or blood. Most often the admixture of non-clonal with clonal cells is not an issue in samples with advanced MDS, AML and CMML as the percentages of non-clonal cells are low. However, in low grade MDS, it is possible that the admixture of clonal cells with non-clonal cells may be much lower and thus, some UPD may have been undetected by our analysis.
In sum, our investigations provide a proof of principle that delineation of shared areas of UPD identified by SNP-A may facilitate the identification of genes affected by pathogenic mutations and potentially explain clinical phenotype. Segmental UPD appears to be a common clonal lesion in myeloid malignancies. As more sensitive techniques and study of additional myeloid malignancy cases accrue, comparison of individual UPD lesions with current clinical, molecular, genetic, and cytogenetic prognostic schemes in larger cohorts of patients will likely provide additional important diagnostic, prognostic, and translational biomarkers.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH R01 HL082983(JPM), DOD 903687 (MAM), U54 RR019391 (JPM, MAS), K24 HL077522 (JPM) and a charitable donation from Robert Duggan Cancer Research Fund.
Footnotes
All authors declare no financial or any other conflicts of interest.
Reference List
- 1.Robinson WP. Mechanisms leading to uniparental disomy and their clinical consequences. Bioessays. 2000;22:452–9. doi: 10.1002/(SICI)1521-1878(200005)22:5<452::AID-BIES7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 2.Jiang YH, Bressler J, Beaudet AL. Epigenetics and human disease. Annu.Rev.Genomics Hum.Genet. 2004;5:479–510. doi: 10.1146/annurev.genom.5.061903.180014. [DOI] [PubMed] [Google Scholar]
- 3.Kralovics R, Guan Y, Prchal JT. Acquired uniparental disomy of chromosome 9p is a frequent stem cell defect in polycythemia vera. Exp.Hematol. 2002;30:229–36. doi: 10.1016/s0301-472x(01)00789-5. [DOI] [PubMed] [Google Scholar]
- 4.Baxter EJ, Scott LM, Campbell PJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–61. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 5.Kralovics R, Passamonti F, Buser AS, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–90. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 6.Levine RL, Wadleigh M, Cools J, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–97. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 7.Mohamedali A, Gaken J, Twine NA, et al. Prevalence and prognostic significance of allelic imbalance by single-nucleotide polymorphism analysis in low-risk myelodysplastic syndromes. Blood. 2007;110:3365–73. doi: 10.1182/blood-2007-03-079673. [DOI] [PubMed] [Google Scholar]
- 8.Gondek LP, Dunbar AJ, Szpurka H, McDevitt MA, Maciejewski JP. SNP Array Karyotyping Allows for the Detection of Uniparental Disomy and Cryptic Chromosomal Abnormalities in MDS/MPD-U and MPD. PLoS.ONE. 2007;2:e1225. doi: 10.1371/journal.pone.0001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gondek LP, Tiu R, O’Keefe CL, et al. Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD, and MDS-derived AML. Blood. 2008;111:1534–42. doi: 10.1182/blood-2007-05-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths M, Mason J, Rindl M, et al. Acquired isodisomy for chromosome 13 is common in AML, and associated with FLT3-itd mutations. Leukemia. 2005;19:2355–8. doi: 10.1038/sj.leu.2403988. [DOI] [PubMed] [Google Scholar]
- 11.Ferrara JL, Cooke KR, Pan L, Krenger W. The immunopathophysiology of acute graft-versus-host-disease. Stem Cells. 1996;14:473–89. doi: 10.1002/stem.140473. [DOI] [PubMed] [Google Scholar]
- 12.Fitzgibbon J, Smith LL, Raghavan M, et al. Association between acquired uniparental disomy and homozygous gene mutation in acute myeloid leukemias. Cancer Res. 2005;65:9152–4. doi: 10.1158/0008-5472.CAN-05-2017. [DOI] [PubMed] [Google Scholar]
- 13.Mingari MC, Moretta A, Moretta L. Regulation of KIR expression in human T cells: a safety mechanism that may impair protective T-cell responses 16. Immunol.Today. 1998;19:153–7. doi: 10.1016/s0167-5699(97)01236-x. [DOI] [PubMed] [Google Scholar]
- 14.Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–18. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto G, Nannya Y, Kato M, et al. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays. Am.J.Hum.Genet. 2007;81:114–26. doi: 10.1086/518809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nannya Y, Sanada M, Nakazaki K, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–9. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- 17.Ye S, Dhillon S, Ke X, Collins AR, Day IN. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001;29:E88. doi: 10.1093/nar/29.17.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collett K, Eide GE, Arnes J, et al. Expression of enhancer of zeste homologue 2 is significantly associated with increased tumor cell proliferation and is a marker of aggressive breast cancer. Clin.Cancer Res. 2006;12:1168–74. doi: 10.1158/1078-0432.CCR-05-1533. [DOI] [PubMed] [Google Scholar]
- 19.Maciejewski JP, Mufti GJ. Whole genome scanning as a cytogenetic tool in hematologic malignancies. Blood. 2008;112:965–74. doi: 10.1182/blood-2008-02-130435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ting JC, Ye Y, Thomas GH, Ruczinski I, Pevsner J. Analysis and visualization of chromosomal abnormalities in SNP data with SNPscan. BMC.Bioinformatics. 2006;7:25. doi: 10.1186/1471-2105-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gondek LP, Tiu R, O’Keefe CL, et al. chromosomal lesions and UPD detected by SNP-A in MDS, MPD/MDS and MDS-derived AML. Blood. 2007 doi: 10.1182/blood-2007-05-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renneville A, Roumier C, Biggio V, et al. Cooperating gene mutations in acute myeloid leukemia: a review of the literature. Leukemia. 2008 doi: 10.1038/leu.2008.19. [DOI] [PubMed] [Google Scholar]
- 23.Flotho C, Steinemann D, Mullighan CG, et al. Genome-wide single-nucleotide polymorphism analysis in juvenile myelomonocytic leukemia identifies uniparental disomy surrounding the NF1 locus in cases associated with neurofibromatosis but not in cases with mutant RAS or PTPN11. Oncogene. 2007;26:5816–21. doi: 10.1038/sj.onc.1210361. [DOI] [PubMed] [Google Scholar]
- 24.Whitman SP, Archer KJ, Feng L, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a cancer and leukemia group B study. Cancer Res. 2001;61:7233–9. [PubMed] [Google Scholar]
- 25.Kaneko H, Misawa S, Horiike S, Nakai H, Kashima K. TP53 mutations emerge at early phase of myelodysplastic syndrome and are associated with complex chromosomal abnormalities. Blood. 1995;85:2189–93. [PubMed] [Google Scholar]
- 26.Joazeiro CA, Wing SS, Huang H, et al. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–12. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 27.Andoniou CE, Thien CB, Langdon WY. Tumour induction by activated abl involves tyrosine phosphorylation of the product of the cbl oncogene. EMBO J. 1994;13:4515–23. doi: 10.1002/j.1460-2075.1994.tb06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thien CB, Langdon WY. Tyrosine kinase activity of the EGF receptor is enhanced by the expression of oncogenic 70Z-Cbl. Oncogene. 1997;15:2909–19. doi: 10.1038/sj.onc.1201468. [DOI] [PubMed] [Google Scholar]
- 29.Caligiuri MA, Briesewitz R, Yu J, et al. Novel c-CBL and CBL-b ubiquitin ligase mutations in human acute myeloid leukemia. Blood. 2007;110:1022–4. doi: 10.1182/blood-2006-12-061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sargin B, Choudhary C, Crosetto N, et al. Flt3-dependent transformation by inactivating c-Cbl mutations in AML. Blood. 2007;110:1004–12. doi: 10.1182/blood-2007-01-066076. [DOI] [PubMed] [Google Scholar]
- 31.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–9. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 32.Thien CB, Walker F, Langdon WY. RING finger mutations that abolish c-Cbl-directed polyubiquitination and downregulation of the EGF receptor are insufficient for cell transformation. Mol.Cell. 2001;7:355–65. doi: 10.1016/s1097-2765(01)00183-6. [DOI] [PubMed] [Google Scholar]
- 33.Swaminathan G, Tsygankov AY. The Cbl family proteins: ring leaders in regulation of cell signaling. J Cell Physiol. 2006;209:21–43. doi: 10.1002/jcp.20694. [DOI] [PubMed] [Google Scholar]
- 34.Lee PS, Wang Y, Dominguez MG, et al. The Cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. EMBO J. 1999;18:3616–28. doi: 10.1093/emboj/18.13.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonita DP, Miyake S, Lupher ML, Jr., Langdon WY, Band H. Phosphotyrosine binding domain-dependent upregulation of the platelet-derived growth factor receptor alpha signaling cascade by transforming mutants of Cbl: implications for Cbl’s function and oncogenicity. Mol.Cell Biol. 1997;17:4597–610. doi: 10.1128/mcb.17.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levkowitz G, Waterman H, Zamir E, et al. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–74. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun J, Pedersen M, Bengtsson S, Ronnstrand L. Grb2 mediates negative regulation of stem cell factor receptor/c-Kit signaling by recruitment of Cbl. Exp.Cell Res. 2007;313:3935–42. doi: 10.1016/j.yexcr.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 38.Bennett JM, Catovsky D, Daniel MT, et al. The chronic myeloid leukaemias: guidelines for distinguishing chronic granulocytic, atypical chronic myeloid, and chronic myelomonocytic leukaemia. Proposals by the French-American-British Cooperative Leukaemia Group. Br.J Haematol. 1994;87:746–54. doi: 10.1111/j.1365-2141.1994.tb06734.x. [DOI] [PubMed] [Google Scholar]
- 39.Kosarev P, Mayer KF, Hardtke CS. Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-4-research0016. RESEARCH0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.