Bacteria secrete and respond to small chemical signals, or autoinducers, in a cell density-dependent process known as quorum sensing (QS).1 As the number of cells, and consequently autoinducer concentration, increases, bacterial populations coordinate their gene expression to behave as a unified group. This coordinated effort of bacteria might result in deleterious effects for humans, as certain bacteria use QS to regulate the formation of biofilms and the secretion of virulence factors. As a result, the modulation of QS has emerged as a therapeutic target of considerable interest.2,3
One class of autoinducers is AI-2, derived from the precursor (S)-4,5-dihydroxy-2,3-pentanedione (DPD), and the gene encoding the DPD synthase, LuxS, has been identified in over 55 bacterial species.4 QS systems based on this autoinducer have been shown to regulate bioluminescence in Vibrio harveyi, virulence and biofilm formation in Vibrio cholera, AI-2 transport and virulence expression in Salmonella typhimurium, and mixed-species biofilm development in oral pathogens.5,6 Illustrated in this final example is the fact that AI-2 QS, in contrast to other QS systems that are used for communication between members of the same species, serves as a mechanism of interspecies communication.4 Thus, the development of agonists or antagonists for this system would have implications for broad range QS modulation.
Despite the wide distribution of the AI-2 synthase LuxS, the discrete structures of DPD-based autoinducers and their respective receptor proteins have only been identified in two species: V. harveyi and S. typhimurium. These two signals are distinct despite the fact that both are derived from DPD and rapidly interconvert in solution: S. typhimurium responds to the R-tetrahydroxytetrahydrofuran (R-THMF) form of DPD, whereas V. harveyi responds to the S-THMF borate diester form of DPD (Supporting Information, Figure S1).4 This lack of structural information has been detrimental to the development of agonists and antagonists of AI-2-based QS, and reports of modulators of this system remain limited. Several reports have identified weak and partial agonists,7-9 while reports of antagonists remain largely limited to one class of natural products.10,11 Because of this dearth of information regarding the modulation of AI-2 QS, there is no solid rationale for the design of new ligands. Herein, we report the discovery of a class of synergistic compounds toward the QS of V. harveyi, as well as a remarkable switch in the biological transmission of AI-2-based QS in S. typhimurium stemming from the addition of methylene groups to the C1 position of DPD.
As a first step toward the discovery of new modulators of AI-2-based QS, we created a panel of C1-substituted DPD analogues according to Scheme 1. The synthesis is founded upon previously reported routes for the construction of DPD but modified for the introduction of alkyl groups at the C1 position.12,13 Synthesis began from known aldehyde 1,13 which was converted in one step to the terminal alkyne 2.14 Alkyne 2 served as the branch point for construction of the various alkylated DPD analogues. Toward this end, alkyne 2 was treated with butyl lithium and various alkyl iodides, the products of which were then oxidized to compounds of type 4. To obtain the final compounds, the protected diketones were incubated in aqueous acid to reveal 5a-g (Table 1).
Scheme 1. Synthesis of C1-Substituted DPD Analoguesa.
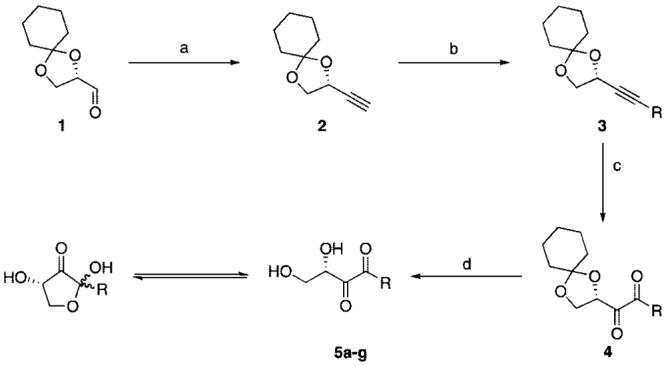
a Conditions: (a) LDA, TMSCHN2, -78 °C, 6 h, 71%; (b) n-BuLi, R-I, -78 °C to room temp, 12 h, 40-75%; (c) RuO2, NaIO4, MeCN, CCl4, H2O, 15 min. (see Table 1 for yields); (d) pH 1.5, 99%.
Table 1.
C1-Substituted Analogues Synthesized via Scheme 1
| compound | R | name | yield [%] |
|---|---|---|---|
| 5a | CH3 | DPD | 70 |
| 5b | CH2CH3 | ethyl-DPD | 53 |
| 5c | CH2CH2CH3 | propyl-DPD | 64 |
| 5d | CH2(CH2)2CH3 | butyl-DPD | 58 |
| 5e | CH2(CH2)4CH3 | hexyl-DPD | 61 |
| 5f | CH2CH2(C6H5) | phenyl-DPD | 44 |
| 5g | CH2(CH2)3N3 | azidobutyl-DPD | 36 |
The series of C1-substituted compounds (Scheme 1) was evaluated for modulation of QS in two established biological assays: induction of β-galactosidase activity in S. typhimurium and bioluminescence production in V. harveyi. These two assays were selected for biological evaluation because our analogues were designed from the AI-2 signals employed by both species as well as because of the rapid readout provided by each assay.
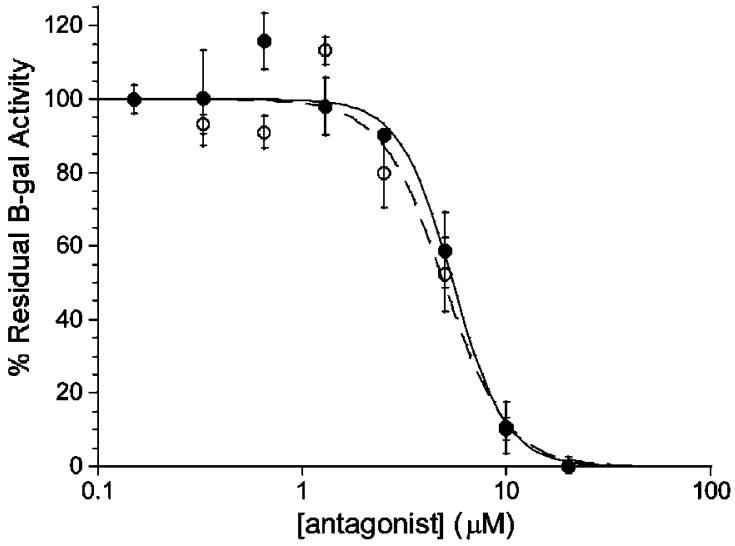
The screens performed for (ant)agonistic activity in S. typhimurium were measured via a β-galactosidase activity assay.15 Assays were conducted using S. typhimurium strain Met844, a δluxS strain with a lacZ-lsr fusion, and assays were performed in the absence of DPD (agonist assay) or in the presence of 50 μM DPD (antagonist assay). The lacZ fusion, which encodes for the biosynthesis of β-galactosidase under lsr promoter control allows for the monitoring of AI-2-dependent lsr activation.16 No agonists were uncovered from the screening, but in the presence of 50 μM DPD, all compounds were found to act as antagonists of AI-2-based QS (Table 2). Notably, the propyl-substituted (5c) and butyl-substituted (5d) analogues were potent inhibitors with IC50 values 10-fold below the concentration of the natural DPD signal, placing these two analogues among the most potent inhibitors of QS relative to the concentration of natural autoinducer.3,17,18 Importantly, none of these compounds affected the growth of S. typhimurium, implicating their role in the specific antagonism of AI-2 QS. Also notable is the activity of the azidobutyl-DPD, which may be used in the identification of unknown AI-2 receptor proteins using the recently developed tag-free approach, utilizing click chemistry, to protein identification.19 Intriguingly, addition or deletion of a single methyl group between DPD (5a), ethyl-DPD (5b), and propyl-DPD (5c), completely alters the biological activity of these substances ranging from the natural substrate to no activity to a potent antagonist.
Table 2.
Summary of QS Modulation by C1-Alkylated DPD Analogues
| compound | IC50 in S. typhimurium assay (μM)a | fold-activation in V. harveyi assayb |
|---|---|---|
| ethyl-DPD (5b) | > 50 | 6.30 ± 0.72 |
| propyl-DPD (5c) | 5.30 ± 0.43 | 7.69 ± 0.30 |
| butyl-DPD (5d) | 5.04 ± 0.61 | 6.05 ± 0.93 |
| hexyl-DPD (5e) | 24.9 ± 5.4 | 2.74 ± 0.24 |
| phenyl-DPD (5f) | > 50 | 1.81 ± 0.12 |
| azidobutyl-DPD (5g) | 20.3 ± 1.3 | 7.44 ± 0.77 |
Assay performed in the presence of 50 μM DPD and varying concentrations of test compound.
Assay performed in the presence of 1 μM DPD and 25 μM compound. Luminescence was measured after 8 h.
To explore an expanded role of these compounds as modulators of QS, we evaluated their effects on the QS of V. harveyi in a bioluminescence assay.20 Modulation of bioluminescence was examined using V. harveyi MM32 cells (ATCC BAA-1121, ΔluxS, ΔluxN), a cell line incapable of producing luminescence either through the acylhomoserine lactone pathway or AI-2 pathways in the absence of exogenous DPD. Although V. harveyi responds to a borate diester form of DPD, boric acid was not added during these assays, as the presence of boric acid itself induces QS activity, rendering V. harveyi less sensitive to different concentrations of DPD. Additionally, V. harveyi does respond to DPD without the addition of boric acid.12,13,20 Thus, the test compounds were evaluated for agonist activity, but only the ethyl-DPD exhibited weak agonistic activity (50-fold less active than natural DPD, data not shown). However, when the test compounds were incubated with V. harveyi and 1 μM DPD to monitor antagonism, a synergistic effect was observed (Table 2). This synergistic activity was observed across the entire series of analogues, with ethyl-DPD (5b) and butyl-DPD (5d) exhibiting greater than 6-fold activation, and propyl-DPD (5c) and azidobutyl-DPD (5g) exhibiting at least 7-fold activation over 1 μM DPD. Interestingly, these compounds were inactive in the absence of DPD, leading to the hypothesis that these analogues are interacting with the AI-2 receptor protein LuxP in a manner productive only in the presence of natural DPD.
To lend credence to the use of these compounds in in vivo settings, we have examined the effects of DPD and the corresponding C1-substituted DPD analogues against a mouse leukemic monocyte macrophage cell line (RAW 264.7) using an XTT based in vitro toxicology assay kit (Sigma). The panel of compounds, including DPD itself, were found to be nontoxic toward mammalian cells as cells retained at least 90% viability in the presence of 50 μM compound (Supporting Information, Table S1). These results, coupled with the differential activity of these compounds in the two reporter assays, make these analogues candidates for the study and inhibition of AI-2-based QS in vivo.
In conclusion, we have shown how a panel of alkyl-substituted DPD analogues can elicit strikingly different biological effects in two different species with known AI-2 QS systems: V. harveyi and S. typhimurium. This difference in activity could not be predicted solely on the basis of the crystal structures of the AI-2 signals nor the receptor proteins but rather was revealed through chemical synthesis and the exploration of bacterial phenotypes with the use of a series of structurally related compounds. In sum, these findings validate, in principle, our approach to the design of DPD-based analogues for the modulation of AI-2-based QS. Although it remains to be seen if these compounds will affect other species that respond to AI-2, this class of compounds nevertheless represents a logical starting point for the development of broad range modulators of QS and identification of unknown AI-2 receptor proteins.
Supplementary Material
Figure 1.
IC50 curves of propyl-DPD (closed symbols) and butyl-DPD (open symbols) for the inhibition of β-galactosidase activity in S. typhimurium.
Acknowledgment
We gratefully acknowledge Prof. Bonnie Bassler (Princeton University) for providing us with Met844 and Dr. Michael Meijler (Ben Gurion University) and Dr. Tobin Dickerson for insightful conversations. We also thank the Skaggs Institute for Chemical Biology and Sanofi-Aventis Graduate Fellowship (C.A.L.) for funding.
Footnotes
Supporting Information Available: Experimental procedures, spectral data, and biological protocols. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Fuqua WC, Winans SC, Greenberg EP. J. Bacteriol. 1994;176:269–75. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Smith KM, Bu Y, Suga H. Chem. Biol. 2003;10:563–71. doi: 10.1016/s1074-5521(03)00107-8. [DOI] [PubMed] [Google Scholar]
- (3).Geske GD, O'Neill JC, Miller DM, Mattmann ME, Blackwell HE. J. Am. Chem. Soc. 2007;129:13613–25. doi: 10.1021/ja074135h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Waters CM, Bassler BL. Annu. Rev. Cell Dev. Biol. 2005;21:319–46. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- (5).Rickard AH, Palmer RJ, Jr., Blehert DS, Campagna SR, Semmelhack MF, Egland PG, Bassler BL, Kolenbrander PE. Mol. Microbiol. 2006;60:1446–56. doi: 10.1111/j.1365-2958.2006.05202.x. [DOI] [PubMed] [Google Scholar]
- (6).Surette MG, Miller MB, Bassler BL. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1639–44. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chan WC, Coyle BJ, Williams P. J. Med. Chem. 2004;47:4633–41. doi: 10.1021/jm0400754. [DOI] [PubMed] [Google Scholar]
- (8).Lowery CA, McKenzie KM, Qi L, Meijler MM, Janda KD. Bioorg. Med. Chem. Lett. 2005;15:2395–8. doi: 10.1016/j.bmcl.2005.02.069. [DOI] [PubMed] [Google Scholar]
- (9).Raffa RB, Iannuzzo JR, Levine DR, Saeid KK, Schwartz RC, Sucic NT, Terleckyj OD, Young JM. J. Pharmacol. Exp. Ther. 2005;312:417–23. doi: 10.1124/jpet.104.075150. [DOI] [PubMed] [Google Scholar]
- (10).Ren D, Bedzyk LA, Ye RW, Thomas SM, Wood TK. Biotechnol. Bioeng. 2004;88:630–42. doi: 10.1002/bit.20259. [DOI] [PubMed] [Google Scholar]
- (11).Ni N, Choudhary G, Li M, Wang B. Bioorg. Med. Chem. Lett. 2008;18:1567–72. doi: 10.1016/j.bmcl.2008.01.081. [DOI] [PubMed] [Google Scholar]
- (12).Meijler MM, Hom LG, Kaufmann GF, McKenzie KM, Sun C, Moss JA, Matsushita M, Janda KD. Angew. Chem. Int. Ed. 2004;43:2106–8. doi: 10.1002/anie.200353150. [DOI] [PubMed] [Google Scholar]
- (13).Semmelhack MF, Campagna SR, Federle MJ, Bassler BL. Org. Lett. 2005;7:569–72. doi: 10.1021/ol047695j. [DOI] [PubMed] [Google Scholar]
- (14).Feldman KS, Cutarelli TD, Di Florio R. J. Org. Chem. 2002;67:8528–37. doi: 10.1021/jo026337w. [DOI] [PubMed] [Google Scholar]
- (15).De Keersmaecker SC, Varszegi C, van Boxel N, Habel LW, Metzger K, Daniels R, Marchal K, De Vos D, Vanderleyden J. J. Biol. Chem. 2005;280:19563–8. doi: 10.1074/jbc.M412660200. [DOI] [PubMed] [Google Scholar]
- (16).Taga ME, Miller ST, Bassler BL. Mol. Microbiol. 2003;50:1411–27. doi: 10.1046/j.1365-2958.2003.03781.x. [DOI] [PubMed] [Google Scholar]
- (17).Defoirdt T, Miyamoto CM, Wood TK, Meighen EA, Sorgeloos P, Verstraete W, Bossier P. Environ. Microbiol. 2007;9:2486–95. doi: 10.1111/j.1462-2920.2007.01367.x. [DOI] [PubMed] [Google Scholar]
- (18).Smith KM, Bu Y, Suga H. Chem. Biol. 2003;10:81–9. doi: 10.1016/s1074-5521(03)00002-4. [DOI] [PubMed] [Google Scholar]
- (19).Speers AE, Cravatt BF. Chembiochem. 2004;5:41–7. doi: 10.1002/cbic.200300721. [DOI] [PubMed] [Google Scholar]
- (20).Taga ME. Curr. Protoc. Microbiol. 2005;1C:1–18. doi: 10.1002/9780471729259.mc01c01s00. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.