Abstract
“Autophagy” is a highly conserved pathway for degradation, by which wasted intracellular macromolecules are delivered to lysosomes, where they are degraded into biologically active monomers such as amino acids that are subsequently re-used to maintain cellular metabolic turnover and homeostasis. Recent genetic studies have shown that mice lacking an autophagy-related gene (Atg5 or Atg7) cannot survive longer than 12 h after birth because of nutrient shortage. Moreover, tissue-specific impairment of autophagy in central nervous system tissue causes massive loss of neurons, resulting in neurodegeneration, while impaired autophagy in liver tissue causes accumulation of wasted organelles, leading to hepatomegaly. Although autophagy generally prevents cell death, our recent study using conditional Atg7-deficient mice in CNS tissue has demonstrated the presence of autophagic neuron death in the hippocampus after neonatal hypoxic/ischemic brain injury. Thus, recent genetic studies have shown that autophagy is involved in various cellular functions. In this review, we introduce physiological and pathophysiological roles of autophagy.
Keywords: Autophagy, Lysosomes, Cathepsins, Autophagic neuron death, Hypoxic/ischemic brain injury
Introduction
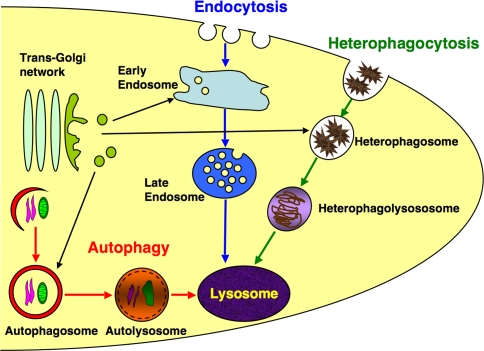
Lysosomes are multifunctional membrane-bound organelles that are present in all mammalian cells. Their internal environment is acidic, with pH ranging from 5.0 to 5.5 and contains various acid hydrolases, while they degrade excess, old, and unneeded intracellular substances and organelles, as well as extracellular materials, into biologically active monomers that are recycled intracellularly. Intracellular trafficking of such macromolecules to lysosomes are mediated by the following processes: endocytosis of cell-surface-receptor proteins with bound ligands, produces early endosomes; heterophagocytosis of large extracellular materials, such as dead cells and bacteria, produces heterophagosomes; and, autophagy of old and unneeded intracellular materials produces autophagosomes. Early endosomes, heterophagosomes, and autophagosomes then receive lysosomal enzymes by fusing with lysosomes or transporting vesicles from the trans-Golgi network (TGN). Degradation begins in these compartments that become subsequently late endosomes/lysosomes, heterophagolysosomes, and autolysosomes, respectively (Fig. 1).
Fig. 1.
Biogenesis of lysosomes: cells execute receptor-mediated endocytosis, heterophagocytosis, and autophagy, forming early endosomes, heterophagosomes, and autophagosomes, respectively. To degrade ingested materials, each structure receives lysosomal enzymes by fusing with transporting vesicles from TGN or lysosomes and becomes late endosomes, heterophagolysosomes, and auto(phago)lysosomes. Before receiving lysosomal enzymes, autophagosomes fuse with endosomes and become amphisomes (Gordon and Seglen 1988) that are not drawn in this diagram
A membrane tissue fraction containing acid hydrolases was first discovered in 1955 by de Duve who called the membrane compartment as a “lysosome” (de Duve et al. 1955). Moreover, the first morphologic description of autophagic processes using electron microscopy was performed by Clark (1957), who noted that the bodies and vacuoles in the basal and apical regions of proximal tubular epithelial cells in infant mouse kidneys were surrounded by dense membranes and contained small canalicular structures, dense lamellar inclusions, and altered mitochondria. Similarly, Ashford and Porter observed the breakdown or hydrolytic process of mitochondria that are sequestered by membrane structures was demonstrated in glucagon-treated hepatocytes (Ashford and Porter 1962). The cellular process by which cytoplasmic organelles such as mitochondria, together with part of the cytoplasm, are sequestered by membrane structures and was termed “autophagy” by de Duve (1963) and de Duve and Wattiaux (1966). The concept of autophagosomes was proposed as prelysosomes that have not yet received acid hydrolases, while the structures become mature as autolysosomes by fusion with primary lysosomes and finally inert residual bodies as postlysosomes (de Duve and Wattiaux 1966).
The early concept of lysosomes and autophagy was established by de Duve and his colleagues (de Duve 1963; de Duve et al. 1955; de Duve and Wattiaux 1966). It was modified by the concept of GERL (Golgi apparatus–endoplasmic reticulum (ER)–lysosomes) in which lysosomal enzymes synthesized in the rough ER are packed in vesicles that are associated with the Golgi apparatus and are transferred to pre-existing lysosomes (Novikoff et al. 1971). Then, the concept of GERL has been improved by understanding the role of the Golgi apparatus and TGN (Griffiths and Simons 1986; Traub and Kornfeld 1997). In particular, the lysosomal system has been developed in relation to endocytosis and the endosomal pathway (Mellman 1996; Mukherjee et al. 1997; van Meel and Klumperman 2008; Kurz et al. 2008; Sandvig et al. 2008).
Unlike studies of the endosomal pathway, most studies of autophagy have examined the morphological aspects of autophagy and experimental conditions that induce autophagy (Arstila and Trump 1968; de Duve and Wattiaux 1966; Dunn 1990a, b; Ericsson 1969a, b; Schworer and Mortimore 1979; Schworer et al. 1981; Yamamoto et al. 1990; Yokota 1993). The origins of isolation membranes, as well as various experimental conditions that induce autophagy, have been areas of intensive research; the isolation membrane of autophagosomes has been suggested to be derived from ER, although isolation and characterization of these compartments would provide evidence about the origin of autophagosomal membranes, which has been a controversial subject (Bolender and Weibel 1973; Dunn 1990a, b; Fengsrud et al. 1995; Kovacs et al. 2000; Mizushima 2005; Ueno et al. 1991; Yokota 1993). Ohsumi and his colleagues have cloned autophagy-related genes from yeast mutants that cannot execute autophagy (Tsukada and Ohsumi 1993) and found a protein conjugation system essential for autophagy and the first mammalian homologues of yeast Atgs (Mizushima et al. 1998a, b). This discovery was the beginning of an era of molecular research in the field of autophagy.
It is well known that autophagy is composed of macroautophagy, microautophagy (Klionsky et al. 2007) and chaperone-mediated autophagy (Massey et al. 2006) that differ in physiological function and the delivery mode to lysosomes. This review is primarily concerned with macroautophagy (hereafter referred to as autophagy). Autophagy is a highly regulated process involving the bulk degradation of cytoplasmic macromolecules and organelles in mammalian cells via the lysosomal system, while it is induced during starvation, differentiation, and normal growth control to maintain homeostasis and survival (Komatsu et al. 2005; Kuma et al. 2004; Shintani and Klionsky 2004). To understand the various roles of autophagy, Mizushima (2005, 2007) proposed the subclassification of autophagy into “induced” and “basal” autophagy; the former produces amino acids in response to starvation, and the latter is important for constitutive turnover of cytoplasmic components. The concept of basal autophagy is based on the fact that the defect of autophagy results in accumulation of cytoplasmic components that are destined to be cleared, cell death, or death of the organism (Hara et al. 2006; Komatsu et al. 2005, 2006, 2007a; Kuma et al. 2004). Induced autophagy is essential for the maintenance of cellular homeostasis and cell survival. However, highly accelerated autophagy may also be involved in pathogenesis of neurodegeneration, such as loss of lysosomal proteinases and of myopathy, such as Pompe disease (Koike et al. 2005; Raben et al. 2007). Moreover, ischemia-induced autophagy contributes to neuron death in mouse brain (Koike et al. 2008). Thus, it is important to further understand how this dual role of autophagy is regulated. In this review, we introduce the physiology and pathophysiology of autophagy based primarily on our recent studies.
What is autophagy?
Morphological aspects of autophagy
As stated above, autophagy is an evolutionarily conserved pathway to lysosomes (Fig. 1). In the process of autophagy, excess, old and unneeded cytoplasmic macromolecules including long-lived proteins and organelles are sequestered by ER-like cisternal structures called the isolation membrane, forming autophagosomes (Reggiori and Klionsky 2005). Autophagosomes further receive lysosomal enzymes by fusing with transporting vesicles from TGN or lysosomes and degradation starts for the turnover and recycling of the cellular constituents (Fig. 2). As evidenced by electron microscopy, autophagosomes are induced to occur in hepatocytes of adult mice after starvation for 24 and 48 h, although it is rather difficult to observe nascent autophagosomes in hepatocytes under physiological conditions (Fig. 3). Typically, autophagosomes detected by electron microscopy in hepatocytes after starvation for 24 h are relatively small (<0.5 mm in diameter) and frequently appear near the bile canaliculi. Nascent autophagosomes that are enwrapped by the ER-like isolation membrane possess part of the cytoplasm, although organelles such as mitochondria and peroxisomes are rarely found within autophagosomes 24 h after the beginning of starvation (Fig. 3). However, relatively larger autophagosomes (approximately 1 to 1.5 mm in diameter) that contain mitochondria and rough ER appear in hepatocytes obtained 48 h later (Fig. 4). It is generally believed that such nascent autophagosomes fuse with lysosomes, although we have never obtained images showing fusion between lysosomes and nascent autophagosomes. Instead of the fusion process of autophagosomes with lysosomes, various types of autophagosomes/autolysosomes can be detected in hepatocytes under starvation conditions; the cisternal space of double membranes shows increased electron density, one part of the double membrane space with a high electron density becomes significantly enlarged, vacuolar structures with single membranes possess membranous structures, and commonly detected lysosomal structures have heterogeneously dense materials (Fig. 4). Thus, maturation of autophagosomes to lysosomes is detectable in hepatocytes of mice starved for 24 and 48 h. Yokota (1993) has shown that the degradation process of excess peroxisomes in rat hepatocytes treated with dioctyl phthalate is rapid and carried out by the autophagic system. In that study he further demonstrated that the isolation membranes enclosing the target organelles are derived from ER. Clearance of mitochondria and peroxisomes has also been observed in isolated hepatocytes (Eskelinen 2005).
Fig. 2.
Diagram showing an autophagic pathway. Excess, old, and unneeded macromolecules, including long-lived proteins and organelles happen to be sequestered and enwrapped completely by the ER-like isolation membrane, the origin of which is unknown, and become autophagosomes that receive lysosomal enzymes by fusing with transporting vesicles from trans-Golgi network or lysosomes. Then degradation starts and autophagosomes become auto(phago)lysosomes. Lysosomes contain acid hydrolases such as cathepsins
Fig. 3.
Electron micrographs of hepatocytes obtained from a mouse housed under starvation conditions for 24 h. Numerous vacuolar structures (arrowheads) are detectable near bile canaliculi (a). Some of these vacuoles are enwrapped by double membranes with morphologically intact cytoplasm (b), and double- or single-membranes with degraded, but morphologically identifiable cytoplasmic materials and structures (c–e). Some vacuoles are encircled by double membranes and a part or whole portion of the intermembrane space is occupied with a dense material (d, e). A lysosome has heterogeneously electron dense materials (f). Bars indicate 1 mm in a and 0.5 μm in b–f
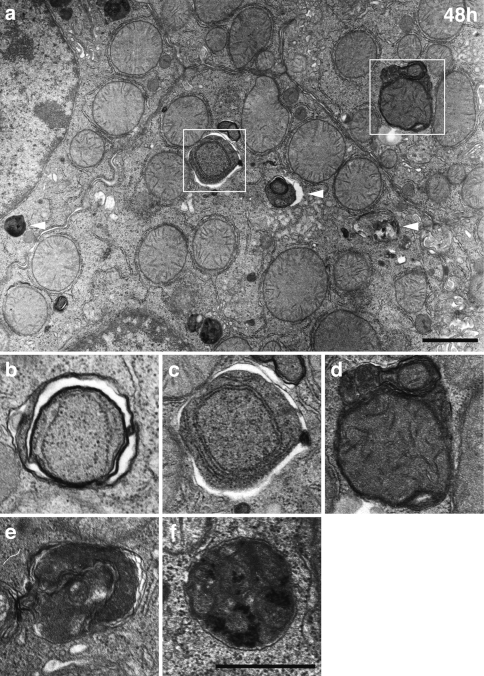
Fig. 4.
Electron micrographs of hepatocytes obtained from a mouse housed under starvation conditions for 48 h. Numerous vacuolar structures (arrowheads) are detectable near bile canaliculi. Vacuolar structures are clearly larger in hepatocytes from mice starved for 48 h (a) than from mice starved for 24 h (see Fig. 3a). Some of these vacuoles are enwrapped by double membranes with morphologically intact cytoplasm (b, c), and single membranes with degraded but morphologically identifiable cytoplasmic materials and structures (d–f). Bars indicate 1 μm in a and 0.5 μm in b–f
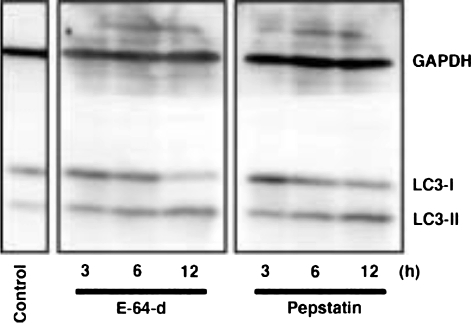
Visualization of autophagosomes and autolysosomes has been intensely investigated, using microtubule-associated protein 1 light chain 3 (LC3), a mammalian homologue of yeast Atg8, as a marker protein of autophagosomes (Kabeya et al. 2000). LC3 that is cleaved near the C-terminal glycine residue by Atg4B soon after being synthesized becomes cytosolic LC3-I (Yoshimura et al. 2006). It is further converted to membrane-bound LC3-II by addition of phosphatidylethanolamine to the C-terminal glycine residue upon induction of autophagy (Kabeya et al. 2000). This modification of LC3 is conducted by the ubiquitin-like conjugation system in which Atg7 and Atg3 act as E1 and E2 enzymes, respectively (Tanida et al. 2001). As evidenced by western blotting, LC3-I is converted to LC3-II in PC12 cells, a rat pheochromocytoma cell line, when they are incubated in the absence of serum (Ohsawa et al. 1998; Uchiyama 2001). This tendency is clearly detected when the cells are cultured in the presence of cysteine and/or aspartic proteinase inhibitors, such as E-64-d and/or pepstatin A (Fig. 5). Positive immunostaining for LC3 becomes punctated in the cytoplasm (Fig. 6). However, it is difficult to show membrane-bound LC3-II on the autophagosomal membrane using ordinary immunoelectron microscopy. We have clearly demonstrated that LC3 is localized on the membrane of autophagic structures in neurons of cathepsin D-deficient mouse brains by immunoelectron microscopy using the sodium dodecyl sulphate-digested freeze-fracture replica labeling (SDS-FRL) technique (Koike et al. 2005) (Fig. 7).
Fig. 5.
Western blotting for LC3. PC12 cells were incubated in the absence of serum and in the presence of a cysteine proteinase inhibitor, E-64-d, or an aspartic proteinase inhibitor, pepstatin A, for 3, 6 or 12 h (h). Lysates from E-64-d-treated, pepstatin A-treated, and control untreated (before serum-free culturing) PC12 cells at each time point were subjected to western blotting. Protein bands immunoreactive for LC3 are detected at molecular weights of 18 and 16 kDa, which correspond to membrane-bound LC3-II and cytosolic LC3-I, respectively. The LC3-II form increases with time after the start of serum-free culturing, indicative of progression of the autophagic process
Fig. 6.
Immunostaining for LC3 in PC12 cells. Cells were cultured in the absence of serum and in the presence of E-64-d, a cysteine proteinase inhibitor, and/or pepstatin A, an aspartic proteinase inhibitor, for 3 or 6 h (h). Cells that were cultured in the presence of serum and in the absence of serum without inhibitors were used as control. Punctate signals for LC3 are distinct in cells treated with inhibitors. Bar indicates 10 μm
Fig. 7.
Immunoelectron microscopy for LC3 using the SDS-FRL method. Immunogold particles indicating LC3 are specifically present on the granule membranes. Bar indicates 0.5 μm
GFP-LC3 transgenic mice, engineered to monitor autophagosome formation, have been used in many studies of autophagy (Mizushima et al. 2003). Using the mice under starvation conditions, Mizushima et al. (2003) have shown by fluorescence microscopy that cup-shaped, ring-shaped, and punctated structures appear in various tissue cells including hepatocytes, cardiac myocytes, and skeletal myocytes. Similar to the results of electron microscopy, autophagosomes with ring-shaped structures were larger than 1 μm (Mizushima et al. 2003). More recently, methods for monitoring autophagic processes have been introduced by Klionsky et al. (2008).
Molecular aspects of autophagy
The first genetic screen for autophagy mutants in yeast was performed by Ohsumi and his colleagues (1993), who identified the first (Atg1) (Matsuura et al. 1997) and, recently, the 31st autophagy-related gene (Atg31) (Kabeya et al. 2007). In yeast autophagy, many of these 31 Atgs participate in initiation of autophagosome formation and gather one spot near the vacuolar membrane—the preautophagosomal structure (PAS) (Kim et al. 2002; Suzuki et al. 2001). Among 31 Atg proteins, the 18 that are involved in autophagosome formation are called AP-Atg proteins (Kabeya et al. 2007; Klionsky et al. 2003; Suzuki et al. 2007): Atg1 to 10, Atg12 to 14, Atg16 to 18, Atg29, and Atg31. The roles of these gene products, together with their mammalian homologues, are summarized in Table 1. Until recently, structures corresponding to PAS have not yet been found in mammals; however, considering the fact that membrane dynamics during autophagic process are conserved from yeast to mammals, mammalian homologous proteins of yeast Atg proteins that are localized to PAS must be essential for initiation of autophagosome formation. Further studies are required to resolve these questions.
Table 1.
AP-Atg proteins
| ATG | Characteristics |
|---|---|
| 1 | Protein kinase |
| 2 | Interact with Atg9 |
| 3 | Phosphatidylethanolamine (PE) conjugation enzyme to Atg8 like E2 |
| 4 | Cysteine protease processing C-terminal end of Atg8 and phosphatidylethanolamine (PE) deconjugation enzyme from Atg8-II |
| 5 | E3 like activity for Atg8 conjugation system in corporation with Atg12 |
| 6 (Beclin) | Bcl-2 binding protein and a component of PI3 kinase complexes |
| 7 | Atg8 or atg12 activating enzyme like E1 |
| 8 (LC3, GABARAP, GATE-16) | Ubiquitin-like protein conjugated to PE and marker for autophagosome |
| 9 | Only membrane protein among atg genes |
| 10 | Atg5 conjugation enzyme to Atg12 like E2 |
| 12 | Ubiquitin-like protein conjugated to Atg5 |
| 13 | Activation of Atg1 |
| 14 | Associate with PI3-Kinase complex I |
| 16 | Interact with Atg12-Atg5 complex |
| 17 | Activation of Atg1, dispensable for Cvt vesicle formation |
| 18 | Interact with Atg9 |
| 29 | PAS organization via interaction with Atg17, dispensable for Cvt vesicle formation |
| 31 | PAS organization via interaction with Atg17 |
Autophagy in disease
Defects in autophagy machinery
According to Kuma et al. (2004), the mice lacking Atg5 cannot survive more than 12 h after birth, during which time they encounter the first, and probably most severe, period of starvation during their lifespan. Kuma et al. (2004) concluded that the nutrient supply derived from neonatal autophagy is essential for survival, although the presence of potential suckling defects may partially account for the natural death of mice deficient in Atg5. Similar phenotypes have been confirmed in mice deficient in Atg7, which is essential for ATG conjugation systems and autophagosome formation in mice (Komatsu et al. 2005). Conditional knockout mice of Atg7 in the liver or central nervous system (CNS) tissue have also been produced by Tanaka and his colleagues (Komatsu et al. 2005, 2006). As for phenotypes of these mice, it has been shown that ubiquitin aggregates accumulate in the cytoplasm of hepatocytes or neurons (Figs. 8, 9). Various abnormalities were observed in the livers of conditionally Atg7-deficient (Atg7Flox/Flox: Mx1-Cre) animals: concentric membrane structures that are continuous to rough ER; accumulation of peroxisomes; and, deformed mitochondria in the cytoplasm of hepatocytes, resulting in hepatomegaly 90 days after injection of pIpC (Komatsu et al. 2005). In Atg7-deficient mice specifically in CNS tissue, loss of cerebral and cerebellar cortical neurons occurs and ubiquitin aggregates accumulate in neuronal perikarya and axons (Figs. 8, 9), leading to neurodegeneration, abnormal neurological signs, and death (Komatsu et al. 2005, 2006). Ubiquitin aggregates were detected in Atg7-deficient brains in which proteasomal function was normal. Until these studies, it has been believed that autophagy is a non-selective degradation pathway upon nutrient deprivation. However, as stated above, accumulation of ubiquitin aggregates occurs in Atg7- or Atg5-deficient CNS neurons, in which nutrient must be supplied constantly from other organs even under starvation conditions. These findings indicate that the constitutive/basal autophagy plays an essential role in the elimination of unfavorable proteins. At present, it remains largely unknown why ubiquitin aggregates occur in Atg7- or Atg5-deficient neurons (see the next session). It is clear that the ubiquitin-proteasomal pathway is responsible for selective clearance of structurally aberrant proteins. Simultaneously, it is also worthy to note that ubiquitination may function as a signal for selective clearance of some kinds of proteins in CNS neurons. There are many neurodegenerative diseases in which ubiquitin aggregates accumulate in CNS neurons, but, to date, autophagy-related genes have not been implicated in the etiology of these neurodegenerative diseases. However, mice whose CNS neurons are unable to execute autophagy may be useful for studying the pathogenesis of neurodegenerative diseases such as Huntington’s disease, Parkinson’s disease, amyotrophic lateral sclerosis (ALS), and peripheral neuropathies, although huntingtin, α-synuclein, superoxide dismutase, and peripheral myelin protein 22, respectively, are believed to be involved in the pathogenesis of these diseases (Ciechanover and Brundin 2003).
Fig. 8.
Immunostaining for ubiquitin (Ubi) in the liver of control (Atg7flox/flox) (a) and Atg7flox/flox; Mx1-Cre (b) mice. Mice were injected with pIpg once a week and killed 16 days after the first injection. Positive staining for ubiquitin is intensely detected in coarse granules in Atg7-deficient hepatocytes but not in the control ones. Bar indicates 20 μm
Fig. 9.
Immunostaining for ubiquitin (Ubi) in the cerebral cortex of control (Atg7flox/flox) (a) and Atg7flox/flox; Nestin-Cre (b) mice at 4 weeks of age. Positive signals for ubiquitin are intensely detected in cortical neurons of an Atg7-deficient brain, but not in the control cortical neurons. Bar indicates 10 μm
It has also been shown that specific ablation of an essential autophagy gene, Atg7, in Purkinje cells initially causes cell-autonomous, progressive dystrophy (manifested by axonal swellings) and degeneration of axon terminals (Komatsu et al. 2007b). Moreover, Komatsu et al. (2007b) have suggested that the autophagy protein, Atg7, is required for membrane trafficking and turnover in the axons, while impairment of axonal autophagy, a possible mechanism of axonopathy, is associated with neurodegeneration. On the other hand, loss of Atg5, specifically in Purkinje cells plays an important role in the maintenance of axonal morphology and membrane structures, and its loss of function leads to axonal swelling, followed by progressive neurodegeneration in mammalian neurons (Nishiyama et al. 2007). We have also noted the importance of basal autophagy in axons of CNS tissue, since the accumulation of nascent autophagosomes is detected in axons of the corpus callosum in mice deficient in lysosomal cathepsin D or doubly deficient in cathepsins B and L (Koike et al. 2005).
Autophagy in lysosome storage disorders due to cathepsin deficiency
Lysosomal cathepsins B, L, and D (CB, CL, and CD) are representative cysteine and aspartic proteinases in lysosomes and major proteinases in CNS neurons. The most common inherited neurodegenerative disease in childhood is neuronal ceroid-lipofuscinosis (NCL), which is categorized as a lysosomal storage disorder and pathologically characterized by the accumulation of proteolipids, such as subunit c of mitochondrial ATP synthase and sphingolipid activator proteins in the lysosomes of neurons (Fearnley et al. 1990; Hall et al. 1991; Kominami et al. 1992; Palmer et al. 1989). We have shown that the CNS neurons in CD-deficient and CB/CL-double deficient mice show a new form of lysosomal accumulation disease with a phenotype resembling NCLs and subunit c accumulates in lysosomes of the affected neurons (Koike et al. 2000, 2003, 2005; Nakanishi et al. 2001). Morphological hallmarks of NCL neurons are granular osmiophilic deposits (GRODs) and fingerprint profiles that can be seen in these mutant mouse neurons. CB and subunit c are detected in GRODs of CD-deficient neurons (Koike et al. 2000), indicating that the GRODs and fingerprint profiles are lysosomes. We have demonstrated that immunosignals for subunit c are granular in the neuronal perikarya of CD-deficient and CB/CL-double deficient mouse brains, while the localization pattern of LC3 is similar to that of subunit c in the neuronal perikarya and fibrous in dendrites of cerebral and cerebellar cortical neurons (Fig. 10) (Koike et al. 2005). Electron microscopy of these mutant mouse brains showed that double membrane-bound vacuoles (AV) containing part of the cytoplasm are frequently detected in CNS neurons of CD-deficient and CB/CL-double deficient mice that are near their terminal stages of the disease (Fig. 11). According to Dunn (1994), autophagosomes undergo stepwise maturation by fusing with endosomes and/or lysosomes, while two types of AVs in this maturation process are distinguishable by electron microscopy (Dunn 1990b; Liou et al. 1997): nascent or immature AVs encircled by endoplasmic reticulum (ER)-like membrane saccules contain part of the morphologically-intact cytoplasm (AVi); and, mature AVs encircled by a single membrane possess degraded but morphologically identifiable cytoplasmic materials and structures (AVd) (Fig. 4). It is interesting that lysosome-like structures such as dense bodies, GROD-like inclusions and autophagosome-like structures themselves are often surrounded, along with part of the cytoplasm, by membrane saccules in CD-deficient and CB/CL-deficient neurons (Fig. 11). This observation indicates that autophagosome formation occurs frequently in these mutant neurons. Morphometric analysis of these lysosomal structures in CD-deficient neurons demonstrates that the volume densities of GRODs and AVs (AVi and AVd) increase with days after birth. More interestingly, half of the AVi counted possess GRODs, and 20% of GRODs are detected in AVi. These data suggest that the presence of GROD-like inclusions and AVs with undigested materials such as GROD-like inclusions may be a potent inducer of autophagy in neuronal cells.
Fig. 10.
Immunostaining for LC3 in cathepsin D-deficient (CD−/−) (b) and littermate control (CD+/−) (a) mouse cerebral cortexes at P23. Positive signals for LC3 are intensely detected in granules of cortical neurons deficient in CD, while they are fibrilar in dendrites of both control and mutant neurons. Bar indicates 20 μm
Fig. 11.
Electron micrographs of cerebral cortical neurons in mouse brains deficient in cathepsin D (CD−/−) at 3,523 and doubly deficient in cathepsins B and L (CB−/−CL−/−) at P13. Granular osmiophilic deposits (GRODs) abundantly accumulate in neuronal perikarya of both mutant mouse brains. GRODs in the neurons are frequently enwrapped together with part of the cytoplasm by double-membrane structures. Bars indicate 1 μm
Although different from loss of lysosomal cathepsins, increased autophagosome formation has also been found in various tissue cells of LAMP2-deficient mice (Tanaka et al. 2000). In such mice, autophagic degradation of long-lived proteins is severely impaired in hepatocytes, while cardiac myocytes are ultrastructurally abnormal and heart contractility is severely reduced, indicating that LAMP2 is critical for autophagy. The deficiency of LAMP2 in humans has been shown to cause Danon’s disease that is associated with the accumulation of autophagic material in striated myocytes (Tanaka et al. 2000). Moreover, since subunit c of mitochondrial ATP synthase accumulates in the lysosomes of NCL neurons, Cao et al. (2006) hypothesized that autophagy, a pathway that regulates mitochondrial turnover, might be impaired in CLN neurons (Cao et al. 2006). Therefore, they produced knock-in mice of Cln3, and found that, in homozygous knock-in mice, the autophagy marker LC3-II is increased, and mammalian target of rapamycin is down-regulated. Moreover, they detected immature isolated autophagic vacuoles and lysosomes from homozygous knock-in mice. Thus, impairment of lysosomal functions due to loss of lysosomal cathepsins and LAMP2, and knock-in of CLN3 facilitates autophagosome formation, resulting in lysosomal storage disorder.
Until recently, however, it remains largely unknown what signaling is essential for autophagosome formation. As stated above, in conditional Atg7-knock-out mice specifically in liver or CNS tissue, numerous ubiquitinated aggregates are detected in the cytosol of hepatocytes or CNS neurons with the presence of functional proteasomes (Hara et al. 2006; Komatsu et al. 2005, 2006), indicating that protein ubiquitination may serve as a signal to the autophagic process in addition to the proteasomal pathway. The presence of ubiquitin aggregates is one of the common pathological characteristics of neurodegenerative diseases, including lysosomal storage disorders (Ardley et al. 2005; Settembre et al. 2008; Zhan et al. 1992). Moreover, LC3, a marker of autophagosomes that is localized on both outer and inner membranes of autophagosomes (Kabeya et al. 2000), has been proposed to function as a receptor for selective substrate, a multifunctional protein, p62/A170/SQSTM1 (p62) (Bjørkøy et al. 2005) that mediates diverse signaling pathways including cell stress, survival, and inflammation (Moscat et al. 2006; Wooten et al. 2006). Since the p62 protein can bind a large number of proteins through its multiple protein–protein interaction motifs (Moscat et al. 2006), including both LC3 and ubiquitin (Komatsu et al. 2007a), the ubiquitin- and LC3-binding protein “p62” regulates the formation of protein aggregates and is removed by autophagy (Komatsu et al. 2007a). In fact, p62 is degraded in lysosomes through autophagy, and accumulates in autophagy-deficient cells (Komatsu et al. 2007a; Nakai et al. 2007; Wang et al. 2006). Moreover, according to Komatsu et al. (2007), genetic ablation of p62 suppresses the appearance of ubiquitin-positive protein aggregates in hepatocytes and neurons, indicating that p62 plays an important role in inclusion body formation, although the pathologic process associated with autophagic deficiency is cell type-specific.
Cell death and autophagy
Based on observations of ultrastructural changes in metamorphosis-related degeneration of insect intersegmental muscles, Beaulaton and Lockshin have shown that autophagy is responsible for selective degradation of mitochondria, glycogen particles, ribosomes, and other organized sarcoplasmic structures, but not for dissolution of myofilaments that appear to be independent of lysosomal activity (Beaulaton and Lockshin 1977). Therefore, they suggested that lysosomal hydrolases are liberated into the sarcoplasm, leading to destruction of myofibrils and cell death. Recent understanding concerning the degradation of myofilaments is that it is conducted by the ubiquitin proteasomes system in cooperation with calpain (Goll et al. 2007). As Beaulation and Lockshin (1977) observed the degradation of intersegmental muscular components shortly after ecdysis to the moth by electron microscopy, the proteolysis by the autophagy/lysosomal system in cooperation with the ubiquitin–proteasome system and calpain serves the adaptive function of providing amino acids for the non-feeding adult insect. Therefore, it may be difficult to speculate that excess autophagy induces metamorphosis-related cell death of intersegmental muscles.
Since Peter Clarke (1990) categorized physiological neuron death into three types that can be detected in CNS tissue during development—apoptotic, autophagic, and non-lysosomal vesiculate—numerous studies have shown the presence of autophagy-related cell death in various CNS tissues, peripheral tissues, and cultured cells (Bursch 2001; Canu et al. 2005; Isahara et al. 1999; Ohsawa et al. 1998; Shibata et al. 1998; Telbisz et al. 2002; Uchiyama 2001; Yu et al. 2004). Since 3-methyladenin (3-MA) was identified as an inhibitor of autophagy (Gordon and Seglen 1982; Seglen and Gordon 1982), cell death that is accompanied by autophagy and inhibited by 3-MA is suggested to be autophagic cell death (Bursch et al. 1996; Uchiyama 2001). L298 fibroblastic cells are known to die by apoptosis when treated with tumor necrosis factor, ceramide, oxidants, or irradiation. Such apoptotic cell death is mediated by activation of the caspase cascade. However, Z-VAD-fmk (benzyloxycarbonylvalyl-alanyl-aspartic acid (O-methyl)-fluoro-methylketone), a pan-caspase inhibitor, does not prevent this apoptosis of L298 cells (Fiers et al. 1999). The treatment with Z-VAD-fmk also induces death in various cell lines, such as U937 monocytoid cells, RAW 264.7 macrophage cells, and primary mouse macrophages. Yu et al. (2004) have shown that Z-VAD-fmk-induced cell death is prevented by 3-MA or wortmannin. Subsequently, they tried to find ways to directly inhibit autophagy, because 3-MA and wortmannin are general inhibitors of phosphatidylinositol-3 (PI3) kinase, which may affect both autophagy and non-apoptotic cell death. Thus, they found that mRNA knockdown of Atg7 or Beclin-1, which are essential for autophagy, prevent Z-VAD-fmk-induced cell death. Therefore, potent inhibitors of apoptosis, like Z-VAD-fmk, may have the unanticipated effects on autophagic cell death (Yu et al. 2004).
As discussed above, autophagy is induced during starvation, differentiation, and normal growth control to maintain homeostasis and survival (Komatsu et al. 2005; Kuma et al. 2004; Shintani and Klionsky 2004). However, it is also involved in neurodegenerative disorders (Chu 2006; Koike et al. 2005; Nixon 2006; Zhu et al. 2007). In fact, autophagy is highly induced in CA1 pyramidal neurons of gerbil hippocampus after brief forebrain ischemia and such damaged pyramidal neurons undergo delayed neuronal death (Nitatori et al. 1995). Induction of autophagy assessed using LC3 as an autophagic marker has also been shown in neonatal and adult mouse cortex, hippocampus, and striatum after hypoxic/ischemic (H/I) brain injury (Adhami et al. 2006, 2007; Koike et al. 2008; Uchiyama et al. 2008; Zhu et al. 2005, 2006). To answer the question of whether autophagy is neuroprotective or anti-neuroprotective in the execution of neuron death after H/I injury, we analyzed this H/I injury-induced neuron death using autophagy-deficient neonatal mice, caspase-3-deficient and caspase activated DNase (CAD)-deficient mice (Koike et al. 2008). Our data can be summarized into three primary conclusions. First, H/I injury-induced pyramidal neuron death in the neonatal hippocampus occurs in both caspase 3-dependent and caspase 3-independent manners. In this model, the caspase 3-independent neuron death is accompanied by DNA ladder formation that is mediated by an unknown DNase other than CAD. Second, H/I injury induces autophagy in neonatal hippocampal pyramidal neurons, while this pyramidal neuron death is prevented by Atg7 deficiency. Finally, pyramidal neuron death in the adult hippocampus after H/I injury is caspase-independent and accompanied by autophagosome formation. In particular, morphological features of these degenerating adult neurons resemble the features of type 2 neuron death, as defined by Clarke (1990). Our data provide direct evidence for autophagy-induced neuron death following neonatal mouse H/I brain injury in animals that cannot execute autophagy, specifically in CNS tissue (Koike et al. 2008). Even though we obtained these results, using mice deficient in Atg7 specifically in CNS tissue, it remains largely unknown how loss of Atg7 prevents H/I injury-induced pyramidal neuron death in the neonatal hippocampus (Koike et al. 2008; Uchiyama et al. 2008). It is therefore very important to understand the mechanism of how pyramidal neurons regulate the two opposite downstream effects of autophagy, survival and death, after H/I insult.
Concluding remarks
This review summarized the physiological and pathophysiological aspects of autophagy. Both “constitutive or basic” and “induced” types of autophagy are very important for the maintenance of cellular metabolism. In particular, genetic studies have contributed to the understanding of multifunctional aspects of autophagy. Although, we have described our recent study of autophagic neuron death after H/I brain injury, it remains largely unexplained why autophagy-deficient neurons are resistant to H/I insult. Selective and non-selective enwrapping mechanisms during autophagosome formation and the origin of isolation membranes are also largely unknown. Further studies are required to resolve these physiological and pathological problems.
Acknowledgments
The authors wish to thank Ms. K. Ikeue, K. Ohta, A. Koseki and K. Isahara for their technical assistance. The studies introduced here are supported by Grant-in-Aid for Scientific Research from Japan Society for the Promotion of Science (16GS0315) from the Ministry of Education, Science, Sports and Culture, Japan.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- AP-Atg proteins
Autophagosome formation-Atg protein;
- AV
Autophagic vacuoles
- CNS
Central nervous system
- Cvt
Cytoplasm-to-vacuole targeting
- ER
Endoplasmic reticulum
- GERL
Golgi apparatus endoplasmic reticulum lysosomes
- GFP
Green fluorescent protein
- LC3
Microtubule associated protein 1 light chain 3
- Macroautophagy
Autophagy
- 3-MA
3-Methyladenine
- PE
Phosphatidylethanolamine
- SDS-FRL
Sodium dodecyl sulphate-digested freeze-fracture replica labeling
- TGN
trans-Golgi network
- Z-VAD-fmk
Benzyloxycarbonylvalyl-alanyl-aspartic acid (O-methyl)-fluoro-methylketone
References
- Adhami F, Liao G, Morozov YM, Schloemer A, Schmithorst VJ, Lorenz JN, Dunn RS, Vorhees CV, Wills-Karp M, Degen JL, Davis RJ, Mizushima N, Rakic P, Dardzinski BJ, Holland SK, Sharp FR, Kuan CY (2006) Cerebral ischemia-hypoxia induces intravascular coagulation and autophagy. Am J Pathol 169:566–583 [DOI] [PMC free article] [PubMed]
- Adhami F, Schloemer A, Kuan CY (2007) The roles of autophagy in cerebral ischemia. Autophagy 3:42–44 [DOI] [PubMed]
- Ardley HC, Hung CC, Robinson PA (2005) The aggravating role of the ubiquitin–proteasome system in neurodegeneration. FEBS Lett 579:571–576 [DOI] [PubMed]
- Arstila AU, Trump BF (1968) Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. Am J Pathol 53:687–733 [PMC free article] [PubMed]
- Ashford TP, Porter KR (1962) Cytoplasmic components in hepatic cell lysosomes. J Cell Biol 12:198–202 [DOI] [PMC free article] [PubMed]
- Beaulaton J, Lockshin RA (1977) Ultrastructural study of the normal degeneration of the intersegmental muscles of Anthereae polyphemus and Manduca sexta (Insecta, Lepidoptera) with particular reference of cellular autophagy. J Morphol 154:39–57 [DOI] [PubMed]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171:603–614 [DOI] [PMC free article] [PubMed]
- Bolender RP, Weibel ER (1973) A morphometric study of the removal of phenobarbital-induced membranes from hepatocytes after cessation of threatment. J Cell Biol 56:746–761 [DOI] [PMC free article] [PubMed]
- Bursch W (2001) The autophagosomal–lysosomal compartment in programmed cell death. Cell Death Differ 8:569–581 [DOI] [PubMed]
- Bursch W, Ellinger A, Kienzl H, Torok L, Pandey S, Sikorska M, Walker R, Hermann RS (1996) Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis 17:1595–1607 [DOI] [PubMed]
- Canu N, Tufi R, Serafino AL, Amadoro G, Ciotti MT, Calissano P (2005) Role of the autophagic-lysosomal system on low potassium-induced apoptosis in cultured cerebellar granule cells. J Neurochem 92:1228–1242 [DOI] [PubMed]
- Cao Y, Espinola JA, Fossale E, Massey AC, Cuervo AM, MacDonald ME, Cotman SL (2006) Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J Biol Chem 281:20483–20493 [DOI] [PubMed]
- Chu CT (2006) Autophagic stress in neuronal injury and disease. J Neuropathol Exp Neurol 65:423–432 [DOI] [PMC free article] [PubMed]
- Ciechanover A, Brundin P (2003) The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron 40:427–446 [DOI] [PubMed]
- Clark SLJ (1957) Cellular differentiation in the kidneys of newborn mice studied with the electron microscope. J Biophys Biochem Cytol 3:349–364 [DOI] [PMC free article] [PubMed]
- Clarke PG (1990) Developmental cell death: morphological diversity and multiple mechanisms. Anat Embryol (Berl) 181:195–213 [DOI] [PubMed]
- de Duve C (1963) Lysosomes. J. & A. Churchill, London
- de Duve C, Wattiaux R (1966) Functions of lysosomes. Annu Rev Physiol 28:435–492 [DOI] [PubMed]
- de Duve C, Pressman BC, Gianetto R, Wattiaux R, Appelmans F (1955) Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J 60:604–617 [DOI] [PMC free article] [PubMed]
- Dunn WA Jr. (1990a) Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol 110:1923–1933 [DOI] [PMC free article] [PubMed]
- Dunn WA Jr. (1990b) Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J Cell Biol 110:1935–1945 [DOI] [PMC free article] [PubMed]
- Dunn WA Jr. (1994) Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol 4:139–143 [DOI] [PubMed]
- Ericsson JL (1969a) Studies on induced cellular autophagy. I. Electron microscopy of cells with in vivo labelled lysosomes. Exp Cell Res 55:95–106 [DOI] [PubMed]
- Ericsson JL (1969b) Studies on induced cellular autophagy. II. Characterization of the membranes bordering autophagosomes in parenchymal liver cells. Exp Cell Res 56:393–405 [DOI] [PubMed]
- Eskelinen EL (2005) Maturation of autophagic vacuoles in Mammalian cells. Autophagy 1:1–10 [DOI] [PubMed]
- Fearnley IM, Walker JE, Martinus RD, Jolly RD, Kirkland KB, Shaw GJ, Palmer DN (1990) The sequence of the major protein stored in ovine ceroid lipofuscinosis is identical with that of the dicyclohexylcarbodiimide-reactive proteolipid of mitochondrial ATP synthase. Biochem J 268:751–758 [DOI] [PMC free article] [PubMed]
- Fengsrud M, Roos N, Berg T, Liou W, Slot JW, Seglen PO (1995) Ultrastructural and immunocytochemical characterization of autophagic vacuoles in isolated hepatocytes: effects of vinblastine and asparagine on vacuole distributions. Exp Cell Res 221:504–519 [DOI] [PubMed]
- Fiers W, Beyaert R, Declercq W, Vandenabeele P (1999) More than one way to die: apoptosis, necrosis and reactive oxygen damage. Oncogene 18:7719–7730 [DOI] [PubMed]
- Goll DE, Neti G, Mares SW, Thompson VF (2007) Myofibrillar protein turnover: the proteasome and the calpains. J Anim Sci doi:10.2527/jas.2007-0395 [DOI] [PubMed]
- Gordon PB, Seglen PO (1982) 6-substituted purines: a novel class of inhibitors of endogenous protein degradation in isolated rat hepatocytes. Arch Biochem Biophys 217:282–294 [DOI] [PubMed]
- Gordon PB, Seglen PO (1988) Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun 151:40–47 [DOI] [PubMed]
- Griffiths G, Simons K (1986) The trans Golgi network: sorting at the exit site of the Golgi complex. Science 234:438–443 [DOI] [PubMed]
- Hall NA, Lake BD, Dewji NN, Patrick AD (1991) Lysosomal storage of subunit c of mitochondrial ATP synthase in Batten’s disease (ceroid-lipofuscinosis). Biochem J 275(Pt 1):269–272 [DOI] [PMC free article] [PubMed]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885–889 [DOI] [PubMed]
- Isahara K, Ohsawa Y, Kanamori S, Shibata M, Waguri S, Sato N, Gotow T, Watanabe T, Momoi T, Urase K, Kominami E, Uchiyama Y (1999) Regulation of a novel pathway for cell death by lysosomal aspartic and cysteine proteinases. Neuroscience 91:233–249 [DOI] [PubMed]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728 [DOI] [PMC free article] [PubMed]
- Kabeya Y, Kawamata T, Suzuki K, Ohsumi Y (2007) Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem Biophys Res Commun 356:405–410 [DOI] [PubMed]
- Kim J, Huang WP, Stromhaug PE, Klionsky DJ (2002) Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem 277:763–773 [DOI] [PMC free article] [PubMed]
- Klionsky DJ, Cregg JM, Dunn WA Jr., Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y (2003) A unified nomenclature for yeast autophagy-related genes. Dev Cell 5:539–545 [DOI] [PubMed]
- Klionsky DJ, Cuervo AM, Seglen PO (2007) Methods for monitoring autophagy from yeast to human. Autophagy 3:181–206 [DOI] [PubMed]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA Jr., Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovacs AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, Lopez-Otin C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Melendez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Munz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nurnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Talloczy Z, Tanaka K, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcategui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL (2008) Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4:151–175 [DOI] [PMC free article] [PubMed]
- Koike M, Nakanishi H, Saftig P, Ezaki J, Isahara K, Ohsawa Y, Schulz-Schaeffer W, Watanabe T, Waguri S, Kametaka S, Shibata M, Yamamoto K, Kominami E, Peters C, von Figura K, Uchiyama Y (2000) Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J Neurosci 20:6898–6906 [DOI] [PMC free article] [PubMed]
- Koike M, Shibata M, Ohsawa Y, Nakanishi H, Koga T, Kametaka S, Waguri S, Momoi T, Kominami E, Peters C, Figura K, Saftig P, Uchiyama Y (2003) Involvement of two different cell death pathways in retinal atrophy of cathepsin D-deficient mice. Mol Cell Neurosci 22:146–161 [DOI] [PubMed]
- Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, Gotow T, Peters C, von Figura K, Mizushima N, Saftig P, Uchiyama Y (2005) Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease). Am J Pathol 167:1713–1728 [DOI] [PMC free article] [PubMed]
- Koike M, Shibata M, Tadakoshi M, Gotoh K, Komatsu M, Waguri S, Kawahara N, Kuida K, Nagata S, Kominami E, Tanaka K, Uchiyama Y (2008) Inhibition of autophagy prevents hippocampal pyramidal neuron death after hypoxic-ischemic Injury. Am J Pathol 172:454–469 [DOI] [PMC free article] [PubMed]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169:425–434 [DOI] [PMC free article] [PubMed]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884 [DOI] [PubMed]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K (2007a) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131:1149–1163 [DOI] [PubMed]
- Komatsu M, Wang QJ, Holstein GR, Friedrich VL Jr., Iwata J, Kominami E, Chait BT, Tanaka K, Yue Z (2007b) Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci USA 104:14489–14494 [DOI] [PMC free article] [PubMed]
- Kominami E, Ezaki J, Muno D, Ishido K, Ueno T, Wolfe LS (1992) Specific storage of subunit c of mitochondrial ATP synthase in lysosomes of neuronal ceroid lipofuscinosis (Batten’s disease). J Biochem (Tokyo) 111:278–282 [DOI] [PubMed]
- Kovacs AL, Rez G, Palfia Z, Kovacs J (2000) Autophagy in the epithelial cells of murine seminal vesicle in vitro. Formation of large sheets of nascent isolation membranes, sequestration of the nucleus and inhibition by wortmannin and 3-ethyladenine. Cell Tissue Res 302:253–261 [DOI] [PubMed]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N (2004) The role of autophagy during the early neonatal starvation period. Nature 432:1032–1036 [DOI] [PubMed]
- Kurz T, Terman A, Gustafsson B, Brunk UT (2008) Lysosomes in iron metabolism, ageing and apoptosis. Histochem Cell Biol doi:10.1007/s00418-008-0394-y [DOI] [PMC free article] [PubMed]
- Liou W, Geuze HJ, Geelen MJ, Slot JW (1997) The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J Cell Biol 136:61–70 [DOI] [PMC free article] [PubMed]
- Massey AC, Zhang C, Cuervo AM (2006) Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol 73:205–235 [DOI] [PubMed]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y (1997) Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192:245–250 [DOI] [PubMed]
- Meel E van, Klumperman J (2008) Imaging and imagination: understanding the endo-lysosomal system. Histochem Cell Biol doi:10.1007/s00418-008-0384-0 [DOI] [PMC free article] [PubMed]
- Mellman I (1996) Endocytosis and molecular sorting. Annu Rev Cell Dev Biol 12:575–625 [DOI] [PubMed]
- Mizushima N (2005) The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ 12(suppl 2):1535–1541 [DOI] [PubMed]
- Mizushima N (2007) Autophagy: process and function. Genes Dev 21:2861–2873 [DOI] [PubMed]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y (1998a) A protein conjugation system essential for autophagy. Nature 395:395–398 [DOI] [PubMed]
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y (1998b) A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem 273:33889–33892 [DOI] [PubMed]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y (2003) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15:1101–1111 [DOI] [PMC free article] [PubMed]
- Moscat J, Diaz-Meco MT, Albert A, Campuzano S (2006) Cell signaling and function organized by PB1 domain interactions. Mol Cell 23:631–640 [DOI] [PubMed]
- Mukherjee S, Ghosh RN, Maxfield FR (1997) Endocytosis. Physiol Rev 77:759–803 [DOI] [PubMed]
- Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K (2007) The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med 13:619–624 [DOI] [PubMed]
- Nakanishi H, Zhang J, Koike M, Nishioku T, Okamoto Y, Kominami E, von Figura K, Peters C, Yamamoto K, Saftig P, Uchiyama Y (2001) Involvement of nitric oxide released from microglia-macrophages in pathological changes of cathepsin D-deficient mice. J Neurosci 21:7526–7533 [DOI] [PMC free article] [PubMed]
- Nishiyama H, Fukaya M, Watanabe M, Linden DJ (2007) Axonal motility and its modulation by activity are branch-type specific in the intact adult cerebellum. Neuron 56:472–487 [DOI] [PMC free article] [PubMed]
- Nitatori T, Sato N, Waguri S, Karasawa Y, Araki H, Shibanai K, Kominami E, Uchiyama Y (1995) Delayed neuronal death in the CA1 pyramidal cell layer of the gerbil hippocampus following transient ischemia is apoptosis. J Neurosci 15:1001–1011 [DOI] [PMC free article] [PubMed]
- Nixon RA (2006) Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends Neurosci 29:528–535 [DOI] [PubMed]
- Novikoff PM, Novikoff AB, Quintana N, Hauw JJ (1971) Golgi apparatus, GERL, and lysosomes of neurons in rat dorsal root ganglia, studied by thick section and thin section cytochemistry. J Cell Biol 50:859–886 [DOI] [PMC free article] [PubMed]
- Ohsawa Y, Isahara K, Kanamori S, Shibata M, Kametaka S, Gotow T, Watanabe T, Kominami E, Uchiyama Y (1998) An ultrastructural and immunohistochemical study of PC12 cells during apoptosis induced by serum deprivation with special reference to autophagy and lysosomal cathepsins. Arch Histol Cytol 61:395–403 [DOI] [PubMed]
- Palmer DN, Martinus RD, Cooper SM, Midwinter GG, Reid JC, Jolly RD (1989) Ovine ceroid lipofuscinosis. The major lipopigment protein and the lipid-binding subunit of mitochondrial ATP synthase have the same NH2-terminal sequence. J Biol Chem 264:5736–5740 [PubMed]
- Raben N, Roberts A, Plotz PH (2007) Role of autophagy in the pathogenesis of Pompe disease. Acta Myol 26:45–48 [PMC free article] [PubMed]
- Reggiori F, Klionsky DJ (2005) Autophagosomes: biogenesis from scratch? Curr Opin Cell Biol 17:415–422 [DOI] [PubMed]
- Sandvig K, Torgersen ML, Raa HA, Deurs B van (2008) Clathrin-independent endocytosis: from nonexinsting to an extreme degree of complexity. Histochem Cell Biol doi:10.1007/s00418-007-0376-5 [DOI] [PMC free article] [PubMed]
- Schworer CM, Mortimore GE (1979) Glucagon-induced autophagy and proteolysis in rat liver: mediation by selective deprivation of intracellular amino acids. Proc Natl Acad Sci USA 76:3169–3173 [DOI] [PMC free article] [PubMed]
- Schworer CM, Shiffer KA, Mortimore GE (1981) Quantitative relationship between autophagy and proteolysis during graded amino acid deprivation in perfused rat liver. J Biol Chem 256:7652–7658 [PubMed]
- Seglen PO, Gordon PB (1982) 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA 79:1889–1892 [DOI] [PMC free article] [PubMed]
- Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, de Pablo R, Tacchetti C, Rubinsztein DC, Ballabio A (2008) A block of autophagy in lysosomal storage disorders. Hum Mol Genet 17:119–129 [DOI] [PubMed]
- Shibata M, Kanamori S, Isahara K, Ohsawa Y, Konishi A, Kametaka S, Watanabe T, Ebisu S, Ishido K, Kominami E, Uchiyama Y (1998) Participation of cathepsins B and D in apoptosis of PC12 cells following serum deprivation. Biochem Biophys Res Commun 251:199–203 [DOI] [PubMed]
- Shintani T, Klionsky DJ (2004) Autophagy in health and disease: a double-edged sword. Science 306:990–995 [DOI] [PMC free article] [PubMed]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 20:5971–5981 [DOI] [PMC free article] [PubMed]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y (2007) Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12:209–218 [DOI] [PubMed]
- Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P (2000) Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406:902–906 [DOI] [PubMed]
- Tanida I, Tanida-Miyake E, Ueno T, Kominami E (2001) The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem 276:1701–1706 [DOI] [PubMed]
- Telbisz A, Kovacs AL, Somosy Z (2002) Influence of X-ray on the autophagic-lysosomal system in rat pancreatic acini. Micron 33:143–151 [DOI] [PubMed]
- Traub LM, Kornfeld S (1997) The trans-Golgi network: a late secretory sorting station. Curr Opin Cell Biol 9:527–533 [DOI] [PubMed]
- Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333:169–174 [DOI] [PubMed]
- Uchiyama Y (2001) Autophagic cell death and its execution by lysosomal cathepsins. Arch Histol Cytol 64:233–246 [DOI] [PubMed]
- Uchiyama Y, Koike M, Shibata M (2008) Autophagic neuron death in neonatal brain ischemia/hypoxia. Autophagy 4:1–5 [DOI] [PubMed]
- Ueno T, Muno D, Kominami E (1991) Membrane markers of endoplasmic reticulum preserved in autophagic vacuolar membranes isolated from leupeptin-administered rat liver. J Biol Chem 266:18995–18999 [PubMed]
- Wang QJ, Ding Y, Kohtz DS, Mizushima N, Cristea IM, Rout MP, Chait BT, Zhong Y, Heintz N, Yue Z (2006) Induction of autophagy in axonal dystrophy and degeneration. J Neurosci 26:8057–8068 [DOI] [PMC free article] [PubMed]
- Wooten MW, Hu X, Babu JR, Seibenhener ML, Geetha T, Paine MG, Wooten MC (2006) Signaling, polyubiquitination, trafficking, and inclusions: Sequestosome 1/p62’s role in neurodegenerative disease. J Biomed Biotechnol 2006:62079 [DOI] [PMC free article] [PubMed]
- Yamamoto A, Masaki R, Tashiro Y (1990) Characterization of the isolation membranes and the limiting membranes of autophagosomes in rat hepatocytes by lectin cytochemistry. J Histochem Cytochem 38:573–580 [DOI] [PubMed]
- Yokota S (1993) Formation of autophagosomes during degradation of excess peroxisomes induced by administration of dioctyl phthalate. Eur J Cell Biol 61:67–80 [PubMed]
- Yoshimura K, Shibata M, Koike M, Gotoh K, Fukaya M, Watanabe M, Uchiyama Y (2006) Effects of RNA interference of Atg4B on the limited proteolysis of LC3 in PC12 cells and expression of Atg4B in various rat tissues. Autophagy 2:200–208 [DOI] [PubMed]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ (2004) Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 304:1500–1502 [DOI] [PubMed]
- Zhan SS, Beyreuther K, Schmitt HP (1992) Neuronal ubiquitin and neurofilament expression in different lysosomal storage disorders. Clin Neuropathol 11:251–255 [PubMed]
- Zhu C, Wang X, Xu F, Bahr BA, Shibata M, Uchiyama Y, Hagberg H, Blomgren K (2005) The influence of age on apoptotic and other mechanisms of cell death after cerebral hypoxia-ischemia. Cell Death Differ 12:162–176 [DOI] [PubMed]
- Zhu C, Xu F, Wang X, Shibata M, Uchiyama Y, Blomgren K, Hagberg H (2006) Different apoptotic mechanisms are activated in male and female brains after neonatal hypoxia-ischaemia. J Neurochem 96:1016–1027 [DOI] [PubMed]
- Zhu JH, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT (2007) Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am J Pathol 170:75–86 [DOI] [PMC free article] [PubMed]