Abstract
We present a high-throughput method that enables efficient delivery of biomolecules into cells. The device consists of an array of 96 suspended electrode pairs, where small sample volumes are top-loaded, electroporated and bottom-ejected into 96-well plates. We demonstrate the use of this suspended-drop electroporation (SDE) device to effectively introduce fluorescent dextran, small interfering RNA (siRNA) or cDNA into primary neurons, differentiated neutrophils and other cell types with conventionally low transfection rates.
Delivery of biomolecules such as cDNA, siRNA, proteins and small molecules into living cells is a prerequisite for most cell biology experiments. Even though primary cells and many cell lines serve as important model systems to understand biological processes, effective intracellular delivery of molecules has been a rate-limiting step for cell-based experiments and high-throughput screens. Classical chemical transfection methods based on lipids1 or calcium phosphate precipitation2 are often inefficient or toxic when used for delivering cDNA or other biomolecules into cells such as primary neurons, 3T3-L1 adipocytes, differentiated human promyelocytic leukemia cells (HL-60) or human umbilical vein endothelial cells (HUVEC). Microinjection3 or particle-mediated methods4 produce low yields and are inherently low-throughput. Viral transfections, such as with vaccinia virus or adenovirus5,6, are experimentally difficult to implement and are not suitable for small-molecule or protein transfection.
In contrast, electroporation provides a more general approach for introducing biomolecules into cells7,8 but has so far been of limited use for mammalian cells because of technical problems. For example, relatively large volumes are required in cuvette-type electroporators, which prohibits the use of expensive or precious biomolecules. Commercial strategies that combine electroporation with optimized transfection reagents (that often differ between cell types) are also expensive and typically work only for specific biomolecules such as cDNA or siRNAs (Supplementary Table 1 online). Furthermore, two available commercial multiwell-format electroporation devices that we tested (Ambion siPORTer-96 and Amaxa 96-well shuttle) were difficult to use reproducibly and required more than 10 min for processing a 96-well plate because of a sequential treatment of the samples (Efficient delivery of Dharmacon (SMARTpool) siRNA reagents in difficult-to-transfect cell lines using the amaxa nucleofector 96-well shuttle system. Technical note, Amaxa 9, 8−11; 2007), and relatively slow sample extraction and washing steps (Ovcharenko, D., Jarvis, R., Hunicke-Smith, S., Kelnar, K. & Brown, D. High throughput siRNA delivery in vitro: from cell lines to primary cells. Technical note, Ambion, 12(2); 2005). Both devices were expensive to use because of a need for proprietary buffer kits, and manual steps for sample extraction and cleaning limited the use of these devices for high-throughput experimentation.
Here we present a method of electroporation that is based on an array of suspended electrode pairs that are top-loaded with sample volumes of 10−20 μl; the samples are held between the electrodes by surface tension (Fig. 1a). After electroporation, the sample can be displaced in parallel to the wells below by direct addition of cell-culture medium. We implemented this strategy in a 96-well format and obtained full recovery of the samples without the need of multiple wash steps. The gold coating of the electrode arrays makes them reusable, and standard inexpensive extracellular buffers could be used for different cell types we tested. The top loading and bottom ejection minimizes pipetting and washing, which made it possible to effectively use multichannel robotics (Supplementary Fig. 1 online). We performed a typical 96-well electroporation experiment (Fig. 1a), including ∼10,000 cells per well, in three steps, which required less than a minute; our approach is therefore suitable for high-throughput biomolecule delivery.
Figure 1.
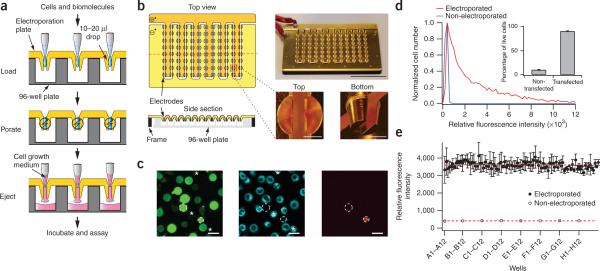
Design of a 96-well–formatted suspended-drop electroporation (SDE) device and its application to deliver small molecules into differentiated HL-60 cells. (a) Sequential loading, electroporation and ejection steps used in the SDE device. (b) The electroporation plate consists of two juxtaposed gold-coated copper pieces with 96 electrodes (1.5 mm apart) that match the layout of a 96-well plate (Supplementary Fig. 2). Scale bars, 5 cm (electroporation plate) and 2 mm (enlarged pictures). (c) Differentiated HL-60 cells were imaged after electroporation with 10-kDa dextran-fluorescein (left) and staining with a live-cell endoplasmic reticulum marker (middle) and propidium iodide (right). Dashed circles and stars highlight dead cells and untranfected cells, respectively. Scale bars, 20 μm. (d) Fluorescence intensity distribution of differentiated HL-60 cells electroporated with dextran-fluorescein and plated in a fibronectin-coated 96-well plate. The fluorescence background intensity was measured from non-electroporated cells (autofluorescence was ∼300 a.u.) and a threshold value of relative fluorescence intensity of 600 was set to separate transfected from non-transfected cells (inset). Single-well electroporation efficiency was 89 ± 3% s.d. (n = 8). (e) Mean fluorescence intensity per well of differentiated HL-60 cells electroporated with dextran-fluorescein and mean fluorescence intensity of untreated cells (error bars, s.d.; n = 3). Dashed lines represent average fluorescence intensity.
The SDE device consists of two machined gold-coated copper pieces with 96 electrodes each (Fig. 1b). These two pieces create an array of 96 vertical electrode pairs that function as suspended electroporation chambers. Each electrode has a surface of 16 mm2 and is separated by 1.5 mm, suitable for sample volumes from 10−20 μl (for construction details, see Supplementary Methods and Supplementary Fig. 2 online). The two gold-coated parts are connected to a power supply that delivers square pulses of tunable time lengths and voltages (Supplementary Methods) for better transfection efficiencies, as had been found earlier8. The power supply and capacitor support currents up to 250 A for several milliseconds. Voltage gradients and pulse lengths used for the 96-well SDE device are not substantially different from those used for single cuvette electroporation.
We tested a range of applications of the SDE device by treating different cell types with small dextran molecules, siRNA and cDNA. As electroporation buffer, we used standard extracellular buffer (pH 7.4 adjusted with HEPES) because it is nontoxic, easily produced and suitable for all mammalian cell types.
We first treated HL-60 cells, a neutrophil-like cell type that is difficult to transfect using existing techniques, which is commonly used as a model for cell migration and chemotaxis9. We electroporated differentiated HL-60 cells using square pulses of 630 V/cm and 7 ms in the presence of 50 μM fluorescein-labeled dextran of 10 kDa (Table 1). We ejected the cells, collected them by centrifugation, resuspended them in fresh growth medium and imaged them (Fig. 1c). Most cells contained sufficient dextran to be visible by fluorescence microscopy (Fig. 1c) with cells ranging from untransfected to weakly transfected to much more highly transfected. Cell death was very low as revealed by live- and dead-cell staining (Fig. 1c). In subsequent experiments, we directly plated electroporated cells in a fibronectin-coated 96-well plate and washed them before imaging to remove non-differentiated cells (Supplementary Methods). The electroporation efficiency of differentiated cells was 90% (Fig. 1d) as determined by comparing the fluorescence intensity distribution of treated versus nontreated cells (Fig. 1d; each condition, ∼2,000 cells). Note that the distribution of the fluorescence intensity per cell is broad and gradually decreases with increasing brightness (Fig. 1d) presumably because biomolecules enter cells by diffusion through multiple transient pores that have variable sizes and opening times7,8. Accordingly, individual cells that have more pores, pores with longer opening times or bigger pore sizes are expected to be brighter because proportionally more fluorescein-labeled dextran diffuses into the cytosol. The resulting electroporation efficiency was reproducible and consistent across the 96 wells (5% s.d. variation over 96 wells; Fig. 1e). This is, to our knowledge, the first demonstration and characterization of a simultaneous electroporation of 96 samples.
Table 1.
Optimized electroporation conditions
| Pulse intensity (V/cm) | Pulse duration (ms) | Pulse number | Transfection efficiencya (%) | Cell deathb (%) | |
|---|---|---|---|---|---|
| HL-60c (cDNAd) | 670 | 20 | 1 | 45 | 40−45 |
| HL-60c (siRNAe) | 630 | 7 | 2 | 90 | 5−10 |
| Hippocampal neurons (cDNAd) | 530 | 15 | 1 | ∼ 50 | 50−60 |
| HUVEC (cDNAd) | 530 | 15 | 1 | 65 | 60−70 |
| 3T3-L1c (cDNAd) | 470 | 20 | 1 | 55 | 40−50 |
| RBL-2H3 (cDNAd) | 670 | 15 | 1 | 45 | 40−45 |
Number of electroporated live cells/number of total live cells.
Determined by comparing the number of live cells between treated versus non-treated samples in which the same amount of cells were pipetted.
Differentiated cells.
Optimized conditions for eGFP cDNA at a concentration of 25−10 ng/μl in extracellular buffer.
Optimized conditions for LMNA siRNA at a concentration of 750−500 nM in extracellular buffer. Data shown for HL-60 cells, hippocampal neurons and HUVECs are averages from n = 8−10 experiments; for 3T3-L1 and RBL-2H3 cells, n = 3.
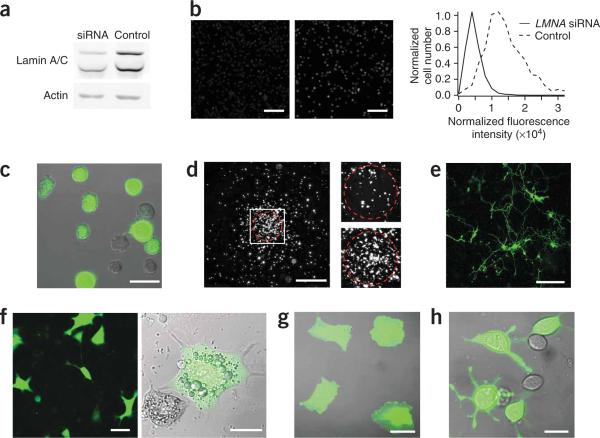
We used the same electroporation conditions to deliver into HL-60 cells siRNA to knock down expression of a nuclear envelope protein, lamin A/C. Three days after treatment, we observed knockdown efficiencies of 70% and 67% of lamin A/C by western blot and immunostaining, respectively (Fig. 2a,b). As all nuclear envelopes had similar fluorescence-staining intensity, knockdown of lamin A/C and electroporation efficiency was consistent across the population (Fig. 2b). We used longer pulse lengths of higher intensities to effectively transfect cDNA encoding enhanced GFP (eGFP) into differentiated HL-60 cells (Fig. 2c) with transfection efficiencies reaching 45% (Table 1). Electroporated cells were physiologically functional; they chemotaxed toward a gradient of chemoattractant N-formyl-methionine-leucine-phenylalanine (fMLP) in an under-agarose assay (Fig. 2d). As a comparison, commercially available electroporation devices did not allow the delivery of siRNA, cDNA and/or dextran into differentiated HL-60 cells as efficiently as the SDE device, which allowed high transfection efficiency combined with low cell death; Supplementary Table 1). Note that transfection efficiency and cell death percentage were correlated (Supplementary Fig. 3 online), and that pulse optimization for the SDE device is also a compromise between cell viability and transfection efficiency. We generally optimized pulse settings to aim for ∼50% cell death (Table 1 and Supplementary Fig. 3).
Figure 2.
cDNA and siRNA transfection of hard-totransfect cells. (a) Western blot of differentiated HL-60 cells electroporated with siRNAs to knock down lamin A/C. (b) Lamin A/C immunostaining of HL-60 differentiated cells treated with LMNA-targeting (left) and non-targeting (middle) siRNA. Scale bars, 200 μm. Fluorescence intensity distributions of lamin A/C staining revealed 67% knockdown (right). (c) Differentiated HL-60 cells electroporated with eGFP cDNA and plated on a fibronectin-coated coverslip. Scale bar, 20 μm. (d) Under-agarose chemotaxis assay of GFP-electroporated HL-60 cells accumulating at the source of the chemoattractant fMLP (red circle) 6 h after exposure to the gradient (left), and enlarged views of the cells before (top right) and after (bottom right) initiation of chemotaxis. Scale bars, 1 mm and 200 μm (close-ups). (e) Rat primary hippocampal neurons 3 d after electroporation of GFP cDNA. Scale bar, 150 μm. (f–h) Differentiated 3T3-L1 adipocytes (f; left, fluorescence micrograph; right, overlay of transmitted light and fluorescence images), HUVEC (g) and RBL-2H3 cells (h) electroporated with eGFP cDNA. Scale bars, 20 μm (f (right), g and h) and 50 μm (f;left).
We also successfully electroporated, with high transfection efficiencies, other physiologically important cell types, including neurons, adipocytes and mast cells using the SDE device. We transfected primary hippocampal neurons with eGFP cDNA (0.2 μg/well) with ∼50% transfection efficiency (Fig. 2e and see Table 1 for electroporation conditions). The transfected neurons had indistinguishable neurite dynamics compared to untransfected neurons, and we observed early synapse formation (Supplementary Fig. 4 online). We similarly electroporated differentiated 3T3-L1 adipocytes, RBL-2H3 tumor mast cells and HUVECs with eGFP (Fig. 2f–h) with transfection efficiencies of 55%, 45% and 65%, respectively (Table 1). Cell death was 40−60% depending on the cell type and the amount of cDNA, which is similar or lower than the amount of cell death seen with commercially available electroporation devices (Supplementary Table 1) and with conventional electroporation8. Higher transfection efficiency could often be reached with the SDE device compared to that with available commercial devices because the several-fold smaller sample volume allows higher relative sample concentrations to be used.
Our study introduces a versatile method to deliver biomolecules into difficult-to-transfect cell types such as differentiated neutrophils and primary cultured neurons. We showed that the SDE device has a several-fold improvement over existing electroporation devices in terms of reduced volume, cost and cycle time, and that it is suitable for high-throughput automated experiments. This opens new possibilities for systems biology experimentation combining relevant cell models with RNA interference or small-molecule screening.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Merkle for machining the SDE device, D. Profitt for building the power supply, A. Hahn and P. Vitorino for careful reading of the manuscript, all the members of the Meyer lab for valuable discussions and advice. This work was financially supported by the Swiss Science Foundation and the US National Institute of Health.
Footnotes
Note: Supplementary information is available on the Nature Methods website.
References
- 1.Felgner PL, et al. Proc. Natl. Acad. Sci. USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jordan M, Schallhorn A, Wurm FM. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaang BK, Kandel ER, Grant SG. Neuron. 1993;10:427–435. doi: 10.1016/0896-6273(93)90331-k. [DOI] [PubMed] [Google Scholar]
- 4.Lo DC, McAllister AK, Katz LC. Neuron. 1994;13:1263–1268. doi: 10.1016/0896-6273(94)90412-x. [DOI] [PubMed] [Google Scholar]
- 5.Moriyoshi K, Richards LJ, Akazawa C, O'Leary DD, Nakanishi S. Neuron. 1996;16:255–260. doi: 10.1016/s0896-6273(00)80044-6. [DOI] [PubMed] [Google Scholar]
- 6.Pettit DL, Koothan T, Liao D, Malinow R. Neuron. 1995;14:685–688. doi: 10.1016/0896-6273(95)90212-0. [DOI] [PubMed] [Google Scholar]
- 7.Kinosita K, Jr, Tsong TY. Nature. 1977;268:438–441. doi: 10.1038/268438a0. [DOI] [PubMed] [Google Scholar]
- 8.Teruel MN, Meyer T. Biophys. J. 1997;73:1785–1796. doi: 10.1016/S0006-3495(97)78209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niggli V. Int. J. Biochem. Cell Biol. 2003;35:1619–1638. doi: 10.1016/s1357-2725(03)00144-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.