Abstract
Aldo-keto reductase family 1 member B10 (AKR1B10) is overexpressed in human hepatocellular carcinoma, lung squamous carcinoma, and lung adenocarcinoma in smokers. Our recent studies have showed that AKR1B10 plays a critical role in the growth and proliferation of cancer cells by detoxifying reactive carbonyls and regulating fatty acid biosynthesis. However, little is known about the regulatory mechanisms of AKR1B10 expression. In this study, we determined the structure of AKR1B10 gene and characterized its promoter. The results demonstrated that AKR1B10 consists of 10 exons and 9 introns, stretching approximately 13.8 kb. A 5′-RACE study determined the transcriptional start site of AKR1B10 at 320 bp upstream of the ATG translational start codon. A TATA-like (TAATAA) and a CAAT box are present from −145 to −140 bp and −193 to −190 bp upstream of the transcriptional start site, respectively. Motif analysis recognized multiple putative oncogenic and tumor suppressor protein binding sites in the AKR1B10 promoter, including c-Ets-1, C/EBP, AP-1, and p53, but osmolytic response elements were not found. A -4,091 bp of the 5′-flanking fragment of the AKR1B10 gene was capable of driving GFP and luciferase reporter gene expression in HepG2 cells derived from human hepatocellular carcinoma; progressive 5′-deletions revealed that a −255 bp fragment possesses full promoter activity.
Keywords: AKR1B10, Aldose reductase-like-1, ARL-1, promoter, and gene structure
1. Introduction
Aldo-keto reductase family 1 member B10 (AKR1B10), also known as aldose reductase-like-1 (ARL-1), is a novel protein identified from human hepatocellular carcinoma (HCC) (Cao et al., 1998; Hyndman and Flynn, 1998). This protein belongs to the aldo-keto reductase superfamily, a protein cluster implicated in osmolytic regulation, carbonyl detoxification, cellular carcinogenesis, and cancer therapeutics (Ko et al., 1997; Lee et al., 2001; Crosas et al., 2003; Hyndman et al., 2003; Jin et al., 2006). AKR1B10 is primarily expressed in the adrenal gland, colon and small intestine with low levels in the liver, thymus, prostate and testis (Cao et al., 1998; Hyndman and Flynn, 1998), but overexpressed in 54% of HCC, 84.4% of lung squamous cell carcinoma, and 29.2% of lung adenocarcinoma in smokers, potentially serving as a diagnostic and/or prognostic marker (Cao et al., 1998; Fukumoto et al., 2005; Penning, 2005).
AKR1B10 is a monomeric enzyme that efficiently catalyzes the reduction of carbonyls with NADPH as a co-enzyme (Cao et al., 1998; Gallego et al., 2007). This reaction converts highly reactive aldehydic and ketonic groups into hydroxy groups, protecting cells against carbonyl toxicity. Our recent studies have demonstrated that targeting expression of AKR1B10 gene in colorectal cancer cells (HCT-8) and transformed human embryonic kidney cells (293T) significantly affected the cell proliferation, clonogenic growth, and susceptibility to reactive carbonyls, such as acrolein and crotonaldehyde (Yan et al., 2007; Zu et al., 2007). AKR1B10 also shows strong enzymatic activity toward all-trans-retinal, 9-cis-retinal, and 13-cis-retinal, converting them to retinols (Crosas et al., 2003). This reaction may diminish cellular retinoic acid, a signaling molecule regulating cell proliferation and differentiation (Dragnev et al., 2000; Penning, 2005).
In H-ras transformed human mammary epithelial cells and colorectal cancer cells (HCT-8), AKR1B10 affects de novo synthesis of fatty acids by mediating acetyl-CoA carboxylase-α (ACCA) degradation through the ubiquitination-proteasome pathway (Ma et al., 2008). ACCA is a rate-limiting enzyme in long chain fatty acid synthesis, catalyzing malonyl-CoA formation by ATP-dependent carboxylation of acetyl-CoA (Witters et al., 1994; Zang et al., 2005). Long chain fatty acids are the building blocks of biomembranes and the precursors of lipid second messengers, thus being critical to cell proliferation, migration, signal transduction, and intracellular trafficking (Manes et al., 1999; Simons and Toomre, 2000; Rouquette-Jazdanian et al., 2002; Swinnen et al., 2004; Swinnen et al., 2006). In breast cancer cells, ACCA knockdown induced by small interfering RNA resulted in cell cycle arrest and apoptosis (Chajes et al., 2006). Therefore, AKR1B10 may play a critical role in the development and progression of cancer through detoxifying intracellular cytotoxic carbonyls, mediating retinal metabolism, and regulating fatty acid biosynthesis. However, the regulatory mechanisms of AKR1B10 expression remain unclear. This study determined AKR1B10 gene structure and characterized its promoter, aiding in the understanding of its expression regulation.
2. Materials and Methods
2.1. Cell culture
HepG2 (human hepatocellular carcinoma) cells were purchased from American Type Culture Collection (Manassas, VA) and maintained in RMPI 1640 medium (Hyclone, UT) containing 10% fetal bovine serum, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C, 5% CO2.
2.2. 5′-RACE (rapid amplification of cDNA ends)
Total RNA was extracted using Trizol® reagent (Invitrogen, CA) from HepG2 cells and hepatocellular carcinoma tissues (collected via Co-operative Human Tissue Network sponsored by National Cancer Institute) as previously described (Cao et al., 2005). Contaminated genomic DNA was removed by DNase I at 37°C for 30 min in 200 μl mixture containing 20 μg total RNA, 20 U RNase-free DNase I, and 50 U RNase inhibitor (Invitrogen, CA), followed by incubation with proteinase K (2.0 mg/ml in 0.5% SDS) and purification with phenol/chloroform extraction. The first strand cDNA was synthesized at 42°C for 1 hour in 40 μl mixture consisting of 10 pmol gene specific primer (refer to Figure 2), 200 μM dNTP, 10 μg total RNA, 8 μl 5 × reaction buffer, 4 μl 0.1 M DTT, and 400 U Superscript II® retrotranscriptase (RTase). The RNA strand was removed by RNase H (0.5 U/μl, final) at 55°C for 10 min, and cDNA was purified with GLASSMAX DNA isolation spin cartridge. Poly (dC) tail was added by terminal dNTP transferase (TdT, Invitrogen, CA) at 37°C for 10 min in reaction mixture composed of 10 U TdT and 200 μM dCTP, followed by heat inactivation at 65°C for 15 min. The dC-tailed cDNA was amplified in 50 μl PCR mix containing 5 μl dC-tailed cDNA, 10 pmol gene specific primer, 10 pmol anchor primer (Invitrogen, CA), 200 μM dNTP, and 2.5 U Taq DNA polymerase at cycling conditions: 94°C/30 sec, 45°C/30 sec, and 72°C/45 sec for 35 cycles. PCR products were examined in 1.0% agarose gel and subcloned using TA Cloning® kit (Invitrogen, CA). Plasmid DNAs were purified from 5 colonies each and submitted for sequencing analysis in the Center for Genetic Medicine at Northern University, Chicago, IL.
Figure 2. Transcriptional start site of AKR1B10 gene.

The 5′-ends of AKR1B10 transcripts were determined by 5′-RACE as described in the Section 2.2. PCR products (upper) and DNA sequenced (lower) are exhibited. The transcriptional start site is marked as +1. ATG translational start codon is bolded and primers used in 5′-RACE are underlined and orientated by arrows. RACE, rapid amplification of cDNA ends; H, HepG2 cells; T, hepatocellular carcinoma tissues; and M, DNA molecular marker.
2.3. AKR1B10 gene structure
AKR1B10 gene structure was determined by blasting its cDNA (Cao et al., 1998) with genomic sequence in human genome database of National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/), following the GT-AG rule (Wieben, 2003).
2.4. Cloning of AKR1B10 promoter and construction of reporter gene vectors
A 4,411 bp fragment, containing 320 bp of 5′-untranslated region and 4,091 bp of 5′-flanking region, was amplified by PCR using human genomic DNA extracted from HepG2 cells as template and cloned into pGlow-TOPO vector (Invitrogen, CA), creating pAKR1B10/Glow-TOPO vector that expresses green fluorescent protein (GFP). Thereafter, AKR1B10 promoter DNA fragment was released by KpnI and BglII and inserted into pGL4 (Promega, WI), constructing luciferase reporter vectors. Progressive 5′-end deletions of AKR1B10 promoter were performed by enzyme digestion, where available, or PCR amplification and subcloning. Table I summarizes the primers used in the construction of 5′-deletion vectors. Constructs were verified by DNA sequencing.
Table I.
Primer sequences used for promoter subcloning
| Primer | Sequences | Location |
|---|---|---|
| P1 | 5′- CTAATCTGTCACCTTGGAGG - 3′ | −4,091 bp |
| P2 | 5′- CTAAAAAAGATATCCCTTCTCACTGATTC - 3′ | −2,067 bp |
| P2341 | 5′- AGGACCTCTG AACAACTGCG TG - 3′ | −1,776 bp |
| P3056 | 5′- GAAGTATAAGATTTTTCACTCATAG - 3′ | −1,061 bp |
| P3862 | 5′- CCCTACCTTCCAACTTTTGGCTG - 3′ | −255 bp |
| P320a (R)* | 5′- GAAT CATTTCTGCA CCAACC - 3′ | +320 bp |
Reverse primer, used to pair with all forward primers for amplification.
2.5. Transient transfection and luciferase activity assays
HepG2 cells (5 × 104/well) were plated in 24-well plates and incubated overnight. AKR1B10 promoter-GFP (1.0 μg) or promoter-luciferase vectors (0.9 μg) plus CMV-driven β-galactosidase plasmid (0.1 μg, as an internal control) were transfected into HepG2 cells using ExGen 500 reagent (Fermentas, MD) at 3.3 μl ExGen 500 plus 1 μg plasmid DNA per well. The Glow-TOPO or pGL4 empty vector was used as a negative control. Forty-eighty hours after transfection, cells were examined under fluorescent microscopy (for GFP) or harvested by scrapers, rinsed with PBS and lysed in 30 μl passive lysis buffer (Promega, WI). Luciferase activity was determined using luciferase reporter assay system (Promega, WI); and β-galactosidase activity was measured at 37°C for 5–10 min in 300 μl mixture containing 15 μl cell lysates and 150 μl 2 × β-galactosidase assay buffer (120 mM Na2HPO4 (pH 7.5), 2 mM MgCl2, 100 mM β-mercaptoethanol, and 1.33 mg/ml ortho-nitrophenyl-β-galactoside). Reactions were stopped by the addition of 500 μl of 1 M Na2CO3; and enzymatic products, chromophore o-nitrophenol, were detected at OD420. Promoter activity was calibrated by dividing luciferase activity over β-galactosidase activity and expressed as fold over the pGL4 vector control.
2.6. Statistical analysis
Statistical analysis was performed using Student’s t test with INSTAT statistical analysis package (Graph Pad Software, CA). Significance was defined as p < 0.05.
3. Results
3.1. AKR1B10 gene structure
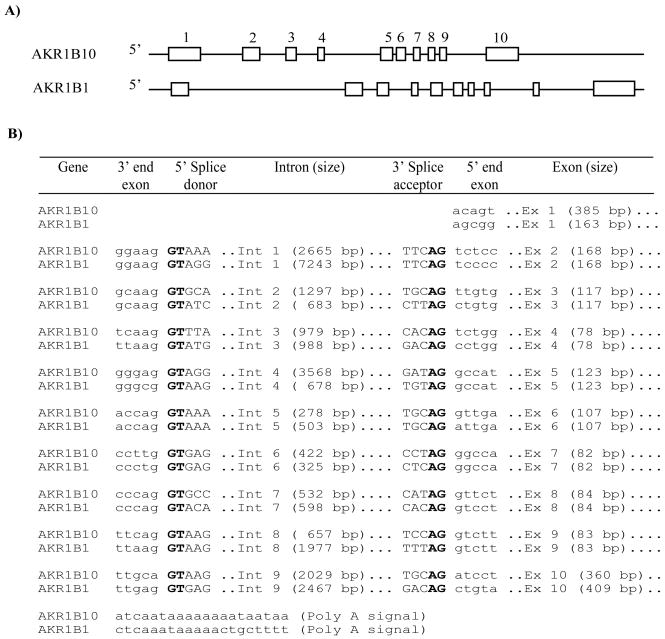
The structure of AKR1B10 gene was determined using cDNA sequence (Cao et al., 1998) and NCBI human genome database. As shown in Figure 1, AKR1B10 consists of 10 exons and 9 introns. Exons range in length from 78–385 bp; and the first in-frame ATG codon is located in exon 1. Exon 10 encodes the -COOH terminus of AKR1B10 protein and contains a translational stop codon TGA and a 3′-untranslated region. A polyadenylation signal, AATAAA, is located 297 bp downstream of the TGA codon. AKR1B10 introns have typical donor and acceptor sites, following the GT-AG rule; the whole AKR1B10 gene spans approximately 13.8 kb. These in silico data are consistent with the variant with 1590 bp in the NCBI AceView web site (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/) (Thierry-Mieg and Thierry-Mieg, 2006). AKR1B10 gene structure is highly identical to AKR1B1, which stretches about 16.8 kb; exons 2–9 of these two genes are exactly same in length (Figure 1), indicating their evolutionary relationship. AKR1B10 gene is located at 7q33 while AKR1B1 is at 7q35 (NCBI database).
Figure 1. Gene structures of AKR1B10 and AKR1B1.
A) Scaled representation of AKR1B10 and AKR1B1 genes. Open boxes indicate the exons numbered on the top. B) Nucleotide sequences in intron-exon borders. AKR1B10 and AKR1B1 genes both contain ten exons and nine introns following a typical GT/AG (bolded) rule at donor and acceptor sites.
3.2. Transcriptional start site and regulatory motifs of AKR1B10 promoter
To characterize the AKR1B10 promoter, we first determined its transcriptional start site. 5′-Ends of AKR1B10 transcripts were amplified by 5′-RACE to determine the DNA sequences. As shown in Figure 2, the transcriptional start site of AKR1B10 was mapped 320 bp upstream of the ATG translational start codon. The AKR1B10 mRNA from HepG2 cells and HCC tissues exhibited a same transcription site.
A motif analysis of the AKR1B10 promoter was performed using Motif Search Software (Bioinformatics Center, JP; http://motif.genome.jp/). As shown in Figure 3, putative TATA-like (TAATAATT) and CAAT (CAATGA) boxes are located at −145 to −140 bp and −193 to −190 bp upstream of the transcriptional start site, respectively. Other important putative promoter elements include a C/EBP binding site at −168 to −155 bp, an AP-1 element at −222 to −212 bp, a SP1 site at −333 to −324 bp, a CHOP-C/EBP-α site at −385 to −373 bp, a p53 binding motif at −666 to −657 bp, and an AP4 site at −793 to −784 bp. In addition, a putative c-Ets-1 binding site overlaps with a C/EBP consensus element at −99 to −80 bp. Osmolytic response elements were not found in this AKR1B10 promoter region.
Figure 3. Putative motifs of the 5′-flanking region of AKR1B10 gene.
Important putative motifs are identified and underlined as indicated. The transcriptional start site is indicated as +1; the complex microsatellite is boxed; and the translational start codon is bolded.
Interestingly, a complex microsatellite (TTCC)2(TCCC)5(TCCT)7 (−604 to −549 bp) is present in a C and T enriched repeat region (−656 to −502 bp) of the AKR1B10 promoter. Preliminary studies (unpublished data) suggested that this highly polymorphic complex microsatellite may associate with the prognosis of colorectal cancer.
3.3. Promoter activity of 5′-flanking fragment of AKR1B10 gene
To characterize the promoter activity, a 4,411 bp fragment, containing 320 bp of 5′-untranslated region and 4,091 bp of the 5′-flanking fragment of the AKR1B10 gene, was subcloned into pGlow-TOPO vector to drive GFP expression. Figure 4 shows GFP expression in HepG2 cells, driven by this 4,411 bp fragment. To quantitatively measure the promoter activity, this 5′-fragment and its progressive 5′-deletions were used to transiently drive luciferase reporter gene expression in HepG2 cells. The CMV-driven β-galactosidase vector was co-transfected as an internal control. Data indicated that a −255 bp fragment, containing most of the putative motifs identified, showed full promoter activity in the HepG2 cells; a further deletion to −21 bp completely abolished the promoter activity, suggesting −255 to −21 bp of the 5′-flanking region of the AKR1B10 gene contains essential promoter components required for basal promoter activity (Figure 5). A significant decrease of promoter activity was observed in the −1,061 bp promoter-luciferase construct, indicating the presence of a repressor element between −1,061 and −804 bp.
Figure 4. Promoter activity of the 5′-flanking region of AKR1B10 gene.
HepG2 cells were transfected with pAKR1B10/Glow-TOPO and empty pGlow-TOPO vectors as described in the Section 2.5. Green fluorescent proteins were examined under fluorescent microscopy with excitation at 395 nm and emission at 509 nm (10 x objectives).
Figure 5. Progressive deletions and activity of AKR1B10 promoter.
AKR1B10 promoter-luciferase vectors with progressive 5′-deletions were constructed and transiently transfected into HepG2 cells as described in the Section 2.4. CMV-driven β-galactosidase vector was used as an internal control to correct the transfection efficiency. Lines to the left indicate AKR1B10 promoter linked to luciferase (open box). Bars on the right denote luciferase activity, expressed by fold over empty vector after corrected for β-galactosidase activity.
4. Discussion
AKR1B10 is overexpressed in human cancers and promotes the growth and proliferation of cancer cells, thus being critical to the development and progression of cancer (Cao et al., 1998; Fukumoto et al., 2005; Yan et al., 2007; Zu et al., 2007; Ma et al., 2008). This study characterized the gene structure and promoter region of the AKR1B10 and identified a novel complex microsatellite in its promoter region.
AKR1B10 possesses more than 70% amino acid sequence identity to and overlapping substrate specificity with AKR1B1 (Cao et al., 1998). This study revealed a highly similar gene structure and very close chromosome loci between these two genes, suggesting their evolutionary relationship. However, the 5′-flanking region of the AKR1B10 gene, which showed promoter activity in our studies, does not appear to have common features with AKR1B1 (Wang et al., 1993). Unlike AKR1B1, the AKR1B10 promoter does not have a typical TATA box although a TAATAA sequence is present from −145 to −140 bp upstream of the transcription start point. The lack of a typical TATA consensus sequence is a feature of the genes with a housekeeping function, such as N-ras and transforming growth factor-α (Hagiwara et al., 1991). In addition, osmotic regulatory elements, which regulate AKR1B1 response to environmental osmolytic changes (Ko et al., 1997), were not found in the AKR1B10 promoter, reflecting its distinct expression pattern (Cao et al., 1998). However, some important promoter elements were indeed recognized in the AKR1B10 promoter, including: (a) an AP-1 consensus element, which responds to transcription factors composed of heterodimeric proteins from c-Fos, c-Jun, ATF, and JPD families (Troen et al., 2004; Bossis et al., 2005); (b) consensus motifs for C/EBP and CHOP-C/EBP-α, the transcription factors involved in a broad cellular responses, such as proliferation, growth and differentiation (Koshiishi et al., 2008); and (c) a c-Ets-1 proto-oncogenic transcription factor and a p53 tumor suppressor protein binding site (Muller-Tiemann et al., 1998; Yamaguchi et al., 2007). Considering the upregulation of the AKR1B10 gene in some types of cancer tissues, one might postulate that the expression of AKR1B10 is regulated directly by these oncogenic and tumor suppressor products.
A complex microsatellite (TTCC)2(TCCC)5(TCCT)7, was found in a C and T enriched repetitive region (−656 to −502 bp) in the AKR1B10 promoter. We have expanded the studies on its polymorphism in colorectal cancer, normal adjacent tissues, and normal donors. Preliminary data (unpublished) indicates that the polymorphism of this microsatellite may associate with colorectal cancer prognosis, being a potential biomarker.
In summary, we characterized the AKR1B10 gene structure and promoter and recognized a number of important putative promoter elements that warrant further evaluations to fully understand the regulation of the AKR1B10 expression. The similarities of the AKR1B10 and AKR1B1 gene structures suggest their evolutionary relationship, but the distinct promoter elements and tissue distributions of AKR1B10 and AKR1B1 transcripts suggest their difference in biological function that merits further investigation.
5. Conclusion
AKR1B10 gene, composed of 10 exons and 9 introns, is structurally identical to AKR1B1, but demonstrates distinct promoter components. The putative oncogenic and tumor suppressor protein binding sites in the AKR1B10 promoter may regulate its tumor-specific expression, warranting a further study. A complex microsatellite is present in the AKR1B10 promoter region, which may be a potential biomarker. A −255 bp of 5′-flanking fragment contains most of important promoter elements and possesses the full promoter activity, representing a targeting region for further elucidation of AKR1B10 expression regulation in the cancer cells.
Acknowledgments
This work was supported in part by American Cancer Society grant (RSG-04-031-01-CCE) and National Cancer Institute (CA122327 and CA122622).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bossis G, Malnou CE, Farras R, Andermarcher E, Hipskind R, Rodriguez M, Schmidt D, Muller S, Jariel-Encontre I, Piechaczyk M. Down-regulation of c-Fos/c-Jun AP-1 dimer activity by sumoylation. Mol Cell Biol. 2005;25:6964–79. doi: 10.1128/MCB.25.16.6964-6979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Fan ST, Chung SS. Identification and characterization of a novel human aldose reductase-like gene. J Biol Chem. 1998;273:11429–35. doi: 10.1074/jbc.273.19.11429. [DOI] [PubMed] [Google Scholar]
- Cao D, Leffert JJ, McCabe J, Kim B, Pizzorno G. Abnormalities in uridine homeostatic regulation and pyrimidine nucleotide metabolism as a consequence of the deletion of the uridine phosphorylase gene. J Biol Chem. 2005;280:21169–75. doi: 10.1074/jbc.M412343200. [DOI] [PubMed] [Google Scholar]
- Chajes V, Cambot M, Moreau K, Lenoir GM, Joulin V. Acetyl-CoA carboxylase alpha is essential to breast cancer cell survival. Cancer Res. 2006;66:5287–94. doi: 10.1158/0008-5472.CAN-05-1489. [DOI] [PubMed] [Google Scholar]
- Crosas B, Hyndman DJ, Gallego O, Martras S, Pares X, Flynn TG, Farres J. Human aldose reductase and human small intestine aldose reductase are efficient retinal reductases: consequences for retinoid metabolism. Biochem J. 2003;373:973–9. doi: 10.1042/BJ20021818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragnev KH, Rigas JR, Dmitrovsky E. The retinoids and cancer prevention mechanisms. Oncologist. 2000;5:361–8. doi: 10.1634/theoncologist.5-5-361. [DOI] [PubMed] [Google Scholar]
- Fukumoto S, Yamauchi N, Moriguchi H, Hippo Y, Watanabe A, Shibahara J, Taniguchi H, Ishikawa S, Ito H, Yamamoto S, Iwanari H, Hironaka M, Ishikawa Y, Niki T, Sohara Y, Kodama T, Nishimura M, Fukayama M, Dosaka-Akita H, Aburatani H. Overexpression of the aldo-keto reductase family protein AKR1B10 is highly correlated with smokers’ non-small cell lung carcinomas. Clin Cancer Res. 2005;11:1776–85. doi: 10.1158/1078-0432.CCR-04-1238. [DOI] [PubMed] [Google Scholar]
- Gallego O, Ruiz FX, Ardevol A, Dominguez M, Alvarez R, de Lera AR, Rovira C, Farres J, Fita I, Pares X. Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10. Proc Natl Acad Sci U S A. 2007;104:20764–9. doi: 10.1073/pnas.0705659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara K, Stenman G, Honda H, Sahlin P, Andersson A, Miyazono K, Heldin CH, Ishikawa F, Takaku F. Organization and chromosomal localization of the human platelet-derived endothelial cell growth factor gene. Mol Cell Biol. 1991;11:2125–32. doi: 10.1128/mcb.11.4.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyndman D, Bauman DR, Heredia VV, Penning TM. The aldo-keto reductase superfamily homepage. Chem Biol Interact. 2003;143–144:621–31. doi: 10.1016/s0009-2797(02)00193-x. [DOI] [PubMed] [Google Scholar]
- Hyndman DJ, Flynn TG. Sequence and expression levels in human tissues of a new member of the aldo-keto reductase family. Biochim Biophys Acta. 1998;1399:198–202. doi: 10.1016/s0167-4781(98)00109-2. [DOI] [PubMed] [Google Scholar]
- Jin J, Krishack PA, Cao D. Role of aldo-keto reductases in development of prostate and breast cancer. Front Biosci. 2006;11:2767–73. doi: 10.2741/2006. [DOI] [PubMed] [Google Scholar]
- Ko BC, Ruepp B, Bohren KM, Gabbay KH, Chung SS. Identification and characterization of multiple osmotic response sequences in the human aldose reductase gene. J Biol Chem. 1997;272:16431–7. doi: 10.1074/jbc.272.26.16431. [DOI] [PubMed] [Google Scholar]
- Koshiishi C, Park HM, Uchiyama H, Tanaka Y. Regulation of expression of the mouse adiponectin gene by the C/EBP family via a novel enhancer region. Gene. 2008 doi: 10.1016/j.gene.2008.07.039. [DOI] [PubMed] [Google Scholar]
- Lee KW, Ko BC, Jiang Z, Cao D, Chung SS. Overexpression of aldose reductase in liver cancers may contribute to drug resistance. Anticancer Drugs. 2001;12:129–32. doi: 10.1097/00001813-200102000-00005. [DOI] [PubMed] [Google Scholar]
- Ma J, Yan R, Zu X, Cheng JM, Rao K, Liao DF, Cao D. Aldo-keto reductase family 1 B10 affects fatty acid synthesis by regulating the stability of acetyl-CoA carboxylase-alpha in breast cancer cells. J Biol Chem. 2008;283:3418–23. doi: 10.1074/jbc.M707650200. [DOI] [PubMed] [Google Scholar]
- Manes S, Mira E, Gomez-Mouton C, Lacalle RA, Keller P, Labrador JP, Martinez AC. Membrane raft microdomains mediate front-rear polarity in migrating cells. Embo J. 1999;18:6211–20. doi: 10.1093/emboj/18.22.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Tiemann BF, Halazonetis TD, Elting JJ. Identification of an additional negative regulatory region for p53 sequence-specific DNA binding. Proc Natl Acad Sci U S A. 1998;95:6079–84. doi: 10.1073/pnas.95.11.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penning TM. AKR1B10: a new diagnostic marker of non-small cell lung carcinoma in smokers. Clin Cancer Res. 2005;11:1687–90. doi: 10.1158/1078-0432.CCR-05-0071. [DOI] [PubMed] [Google Scholar]
- Rouquette-Jazdanian AK, Pelassy C, Breittmayer JP, Cousin JL, Aussel C. Metabolic labelling of membrane microdomains/rafts in Jurkat cells indicates the presence of glycerophospholipids implicated in signal transduction by the CD3 T-cell receptor. Biochem J. 2002;363:645–55. doi: 10.1042/0264-6021:3630645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–9. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Swinnen JV, Brusselmans K, Verhoeven G. Increased lipogenesis in cancer cells: new players, novel targets. Curr Opin Clin Nutr Metab Care. 2006;9:358–65. doi: 10.1097/01.mco.0000232894.28674.30. [DOI] [PubMed] [Google Scholar]
- Swinnen JV, Heemers H, van de Sande T, de Schrijver E, Brusselmans K, Heyns W, Verhoeven G. Androgens, lipogenesis and prostate cancer. J Steroid Biochem Mol Biol. 2004;92:273–9. doi: 10.1016/j.jsbmb.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7 Suppl 1:S121–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troen G, Nygaard V, Jenssen TK, Ikonomou IM, Tierens A, Matutes E, Gruszka-Westwood A, Catovsky D, Myklebost O, Lauritzsen G, Hovig E, Delabie J. Constitutive expression of the AP-1 transcription factors c-jun, junD, junB, and c-fos and the marginal zone B-cell transcription factor Notch2 in splenic marginal zone lymphoma. J Mol Diagn. 2004;6:297–307. doi: 10.1016/S1525-1578(10)60525-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Bohren KM, Gabbay KH. Characterization of the human aldose reductase gene promoter. J Biol Chem. 1993;268:16052–8. [PubMed] [Google Scholar]
- Wieben ED. Primer on medical genomics. Part VII: The evolving concept of the gene. Mayo Clin Proc. 2003;78:580–7. doi: 10.4065/78.5.580. [DOI] [PubMed] [Google Scholar]
- Witters LA, Widmer J, King AN, Fassihi K, Kuhajda F. Identification of human acetyl-CoA carboxylase isozymes in tissue and in breast cancer cells. Int J Biochem. 1994;26:589–94. doi: 10.1016/0020-711x(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Yamaguchi E, Nakayama T, Nanashima A, Matsumoto K, Yasutake T, Sekine I, Nagayasu T. Ets-1 proto-oncogene as a potential predictor for poor prognosis of lung adenocarcinoma. Tohoku J Exp Med. 2007;213:41–50. doi: 10.1620/tjem.213.41. [DOI] [PubMed] [Google Scholar]
- Yan R, Zu X, Ma J, Liu Z, Adeyanju M, Cao D. Aldo-keto reductase family 1 B10 gene silencing results in growth inhibition of colorectal cancer cells: Implication for cancer intervention. Int J Cancer. 2007;121:2301–6. doi: 10.1002/ijc.22933. [DOI] [PubMed] [Google Scholar]
- Zang Y, Wang T, Xie W, Wang-Fischer YL, Getty L, Han J, Corkey BE, Guo W. Regulation of acetyl CoA carboxylase and carnitine palmitoyl transferase-1 in rat adipocytes. Obes Res. 2005;13:1530–9. doi: 10.1038/oby.2005.188. [DOI] [PubMed] [Google Scholar]
- Zu X, Yan R, Robbins S, Krishack PA, Liao DF, Cao D. Reduced 293T cell susceptibility to acrolein due to aldose reductase-like-1 protein expression. Toxicol Sci. 2007;97:562–8. doi: 10.1093/toxsci/kfm033. [DOI] [PubMed] [Google Scholar]