Abstract
MicroRNAs (miRNAs) are an emerging class of highly conserved non-coding small RNAs that regulate gene expression at the post-transcriptional level. It is now clear that miRNAs can potentially regulate every aspect of cellular activity, including differentiation and development, metabolism, proliferation, apoptotic cell death, viral infection and tumorigenesis. Recent studies provide clear evidence that miRNAs are abundant in the liver and modulate a diverse spectrum of liver functions. Deregulation of miRNA expression may be a key pathogenetic factor in many liver diseases including viral hepatitis, hepatocellular cancer and polycystic liver diseases. A clearer understanding of the mechanisms involved in miRNA deregulation will offer new diagnostic and therapeutic strategies to treat liver diseases. Moreover, better understanding of miRNA regulation and identification of tissue-specific miRNA targets employing transgenic/knockout models and/or modulating oligonucleotides will improve our knowledge of liver physiology and diseases.
Keywords: MicroRNAs, Non-coding RNAs, Liver, Tumorigenesis, Gene regulation
INTRODUCTION
Genomic studies have demonstrated that many portions of the human genome do not encode conventional protein-coding genes but encode biologically active non-coding RNA species[1]. With the development of small RNA interface techniques over the past decade, it becomes clear that many small RNA molecules could regulate gene and protein expression. One class of such small non-coding RNAs is microRNAs (miRNAs), a group of regulatory RNAs of 19-22 nucleotides involved in control of gene expression at the post-transcriptional level[2]. Recent studies suggest that miRNAs are involved in regulating cell fate (cell death and proliferation), initiation and progression of human cancer, developmental timing, and inflammatory responses[3–6]. Modulation of miRNA expression in vitro as well as in vivo has revealed an important role of miRNAs in liver functions. In this review, we appraise the recent findings on miRNAs in liver physiology and disease. We will also discuss the development and use of miRNA antagonists (antagomirs) to target miRNAs in vivo, which may translate into novel therapeutic strategies for liver disease in the future.
miRNAS ARE REGULATORY NON-CODING SMALL RNAS IMPORTANT TO POST-TRANSCRIPTIONAL GENE REGULATION
miRNAs are endogenous, single-stranded RNA molecules consisting of approximately 22 non-coding nucleotides that regulate target genes[2,3]. miRNA molecules have been identified in over 80 species including those encoded by viral genomes. There are approximately 500-1000 different mammalian miRNA genes; a complete list and details about the nomenclature of the miRNAs can be viewed at Sanger mirBase 10.1 (http://microrna.sanger.ac.uk/sequences/)[7]. Up to 600 miRNAs have been identified in humans[2,3].
Most miRNAs are generated by RNA polymerase II as long primary transcripts (pri-miRNAs) that form a stem-loop structure[8–11]. In the nucleus, pri-miRNAs are processed into 70-100 nucleotide-long hairpin pre-miRNAs by the RNAse III Drosha. These pre-miRNAs are then exported into the cytoplasm by exportin-5[1]. They are then further processed by another RNAse III, Dicer. The resultant approximately 22-nucleotide RNA duplexes contain the mature miRNA and the passenger miRNA strand[12,13]. The mature miRNA can be incorporated into the so-called RNA-induced silencing complex (RISC). This miRNA-mRNA interaction either blocks translation initiation, induces the endonucleolytic cleavage of the target mRNA, or both (Figure 1)[12–14]. In mammal cells, the sequence of the miRNAs loaded in the complex targets the RISC to specific binding sites in the 3’-untranslated region (UTR) of mRNA transcripts. Each mRNA can be regulated by several miRNAs, and one miRNA can recognize several targets[12,13,15]. Intriguingly, miRNAs may also lead to an upregulation of gene expression[16]. However, the exact mechanism is currently unknown but may be the result of direct effects, such as chromatin remodeling, or indirect effects, e.g. suppression of transcriptional repressors. It is believed that expression of 30% of human genes may be regulated by miRNAs[17].
Figure 1.
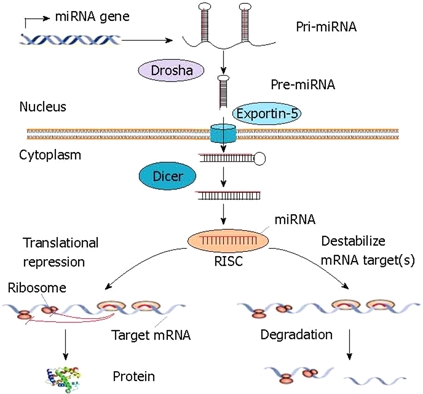
miRNA biogenesis and function in animal cells. Animal genomes have specific genes that encode miRNAs. Primary miRNA transcripts (pri-miRNAs) are processed into precursor miRNA (pre-miRNAs) stem-loops of approximately 60 nucleotides in length by the nuclear RNase III enzyme Drosha. These pre-miRNAs are transported to the cytoplasm via exportin-5 and are further processed by the ribonuclease Dicer. Mature miRNAs are then incorporated in the RNA-induced silencing complex (RISC) and interfere with the regulation of mRNA translation by targeting mRNAs resulting in mRNA degradation or translational repression.
miRNAS ARE ABUNDANT AND FINELY REGULATED IN THE LIVER
One of the first clues of the existence of miRNAs in mammals came from studies on genetic alterations in liver tumors. An unusual transcript, named hcr, was described and characterized as liver-specific, essentially non-coding, specifically nuclear, and processed by endonucleases in one of woodchuck liver tumors investigated in 1989[18]. Later on, the hcr transcript was found to encompass the so-called “pri-miRNA” for miR-122[19]. miRNA-122 was later described as a liver specific miRNA and has been reported in mouse, woodchuck and human livers, in human primary hepatocytes, and in cultured liver derived cells[19,20]. Besides miR-122, many other miRNAs, such as miR-1, miR-16, miR-27b, miR-30d, miR-126, miR-133, miR-143, and the let-7 family, are also abundantly expressed in adult liver tissue. While miR-122 appears as the most highly expressed miRNA in adult liver, miR-92a and miR-483 seem to be more specifically expressed in the fetal liver (Table 1)[21]. Thus, the liver displays a differential miRNA expression profile during its development.
Table 1.
Repertoire of miRNAs in human liver
| Atlas | Adult | Fetal | Atlas | Adult | Fetal | ||
| miR-122 | +++ | +++ | ++ | miR-154 | + | / | / |
| miR-126 | +++ | / | / | miR-193a | + | / | / |
| miR-16 | ++ | ++ | / | miR-193b | + | / | / |
| let-7a | ++ | +++ | / | miR-195 | + | / | / |
| miR-22 | ++ | +++ | + | miR-199a | + | +++ | + |
| miR-125b | ++ | +++ | + | miR-21 | + | +++ | + |
| miR-143 | ++ | +++ | / | miR-223 | + | / | / |
| let-7b | ++ | ++ | / | miR-25 | + | ++ | / |
| miR-99a | ++ | ++ | / | miR-27a | + | ++ | / |
| let-7c | ++ | ++ | / | miR-29b | + | / | / |
| miR-181a | ++ | / | / | miR-29c | + | / | / |
| miR-194 | ++ | +++ | / | miR-3-c | + | +++ | + |
| miR-451 | ++ | / | / | miR-32- | + | / | / |
| miR-3-d | ++ | ++ | / | miR-377 | + | / | / |
| miR-15b | ++ | / | / | miR-378 | + | / | / |
| miR-193a-5p | ++ | / | / | miR-424 | + | / | / |
| miR-24 | ++ | +++ | ++ | miR-425 | + | / | / |
| miR 29a | ++ | / | / | miR-486-5p | + | / | + |
| miR-23b | ++ | + | + | miR-5-5 | + | / | / |
| miR-26b | ++ | / | / | miR-874 | + | / | / |
| miR-27b | ++ | ++ | / | miR-885-5p | + | / | / |
| miR-3-a | ++ | ++ | / | miR-1-7 | Traces | / | / |
| miR-92a | ++ | ++ | +++ | let-7e | Traces | / | / |
| miR-13-a | ++ | / | / | miR-98 | Traces | / | / |
| miR-15a | ++ | ++ | / | miR-3-b | -- | ++ | / |
| miR-186 | ++ | / | / | miR-34 | -- | ++ | / |
| miR-191 | ++ | ++ | / | miR-1-6a | -- | ++ | / |
| miR-26a | ++ | / | ++ | miR-125a | -- | ++ | / |
| miR-28 | ++ | ++ | / | miR-148 | -- | ++ | / |
| miR-1- - | ++ | / | / | miR-149 | -- | ++ | / |
| let-7d | ++ | / | / | miR-189 | -- | ++ | / |
| miR-17 | ++ | / | + | miR-199b | -- | ++ | / |
| miR-185 | ++ | / | / | miR-21- | -- | ++ | / |
| miR-192 | ++ | ++ | / | miR-321 | -- | +++ | / |
| miR-3-e | ++ | / | / | miR-145 | -- | ++ | + |
| miR-381 | ++ | / | / | miR-93 | -- | + | + |
| miR-99b | ++ | / | + | miR-483 | -- | / | +++ |
| miR-1-3 | ++ | + | / | miR-484 | -- | / | + |
| miR-23a | ++ | / | / | miR-485 | -- | / | + |
| let-7f | + | / | / | miR-487 | -- | / | + |
| let-7g | + | / | / | miR-2- | -- | / | ++ |
| miR-139-5p | + | + | / | miR-18 | -- | / | + |
| miR-14- | + | / | / | miR-19b | -- | / | + |
| miR-142 | + | ++ | / | miR-1-6b | -- | / | + |
| miR-144 | + | / | / | miR-345 | -- | / | + |
| miR-151 | + | / | + | miR-41- | -- | / | + |
The minus sign is used to indicate very low to undetectable levels and one plus to three pluses indicate a gradual expression from low levels to very high levels. The slash stands for the miRNAs that were not assayed. This figure is modified from Girard et al[21] with the permission of the authors and the publisher.
The liver contains many cell types including parenchymal cells (i.e. hepatocytes) and “non-parenchymal cells” which include endothelial cells, stellate cells, lymphoid cells, and biliary epithelial cells (cholangiocytes). Each cell type may have completely distinct miRNA expression profiles. However, miRNA expression profile in those cell types, in particular from humans, is not fully tested yet. Current understanding is limited to available human cell lines. For example, a distinct expression profile of miRNAs has been identified in H69 cells, a cell line of SV40 transformed human cholangiocytes[22,23].
The molecular mechanisms underlying transcriptional regulation of miRNA genes in the liver remain largely unknown. Hepatocyte nuclear factor 1α (HNF-1α) is a hepatocyte-enriched transcription factor. Manipulation of HNF-α function through RNA interference causes reciprocal changes in miR-107 expression and thus, may be involved in the regulation of miR-107 transcription in the liver[24]. The myocyte enhancer factor-2 transcription factor can increase the expression of miR-1-2 and miR-133a-1[25]. The transcription factor Myc can up-regulate the expression of miR17-92 cluster and down-regulate several other miRNAs in tumorigenesis[26]. However, whether these transcription factors are also involved in the transcriptional regulation of those miRNAs in the liver is unclear. In addition, decrease of let-7 family expression in human cholangiocytes in response to microbial stimulation appears to be nuclear factor (NF)-κB-dependent[23]. On the other hand, the functional expression of transcription factors can also be regulated by miRNAs, as it has recently been shown for signal transducer and activator of transcription 3 (STAT-3)[22].
miRNAS MAY BE KEY PLAYERS IN THE REGULATION OF LIVER FUNCTIONS
For a comprehensive understanding of miRNA function and potential therapeutic use in liver physiology and disease, identification and validation of miRNA targets are of fundamental importance. Several bioinformatic methods have been developed to predict miRNA targets[7]. Arora and Simpson adapted the ‘ranked ratio (RR)’ described by Yu et al[27] and predicted the genes that are potentially targeted miRNAs in the liver[28]. This RR value is an indicator of the distribution of miRNA target genes within a single mRNA population. A high RR indicates low expression in a greater proportion of target genes and is, therefore, indicative of miRNA expression in that tissue. The RR values for all miRNAs were calculated for the miRNAs in the liver[28] and are shown in Figure 2. Clearly, many mRNAs could be the targets for miRNAs in the liver.
Figure 2.
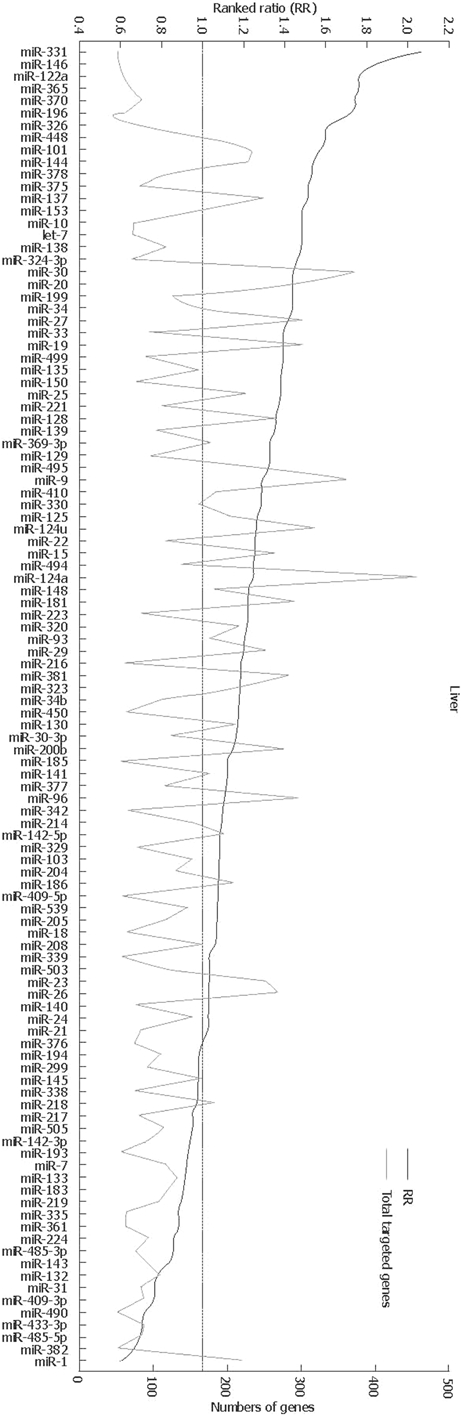
Prediction of targets by miRNAs in the liver. The miRNAs are ordered by RR values (upper-axis). The higher values reflect lower expression of predicted target genes and are, therefore, indicative of miRNA activity. The numbers of genes predicted to be targeted by each miRNA (lower-axis) are indicated by the grey line. This figure is reprinted from Arora and Simpson[28] with the permission of the authors and the publisher.
Validation of those potential targets for each miRNA requires experimental studies both in vitro and in vivo. Several groups have tested the overall importance of miR-122 in the regulation of metabolism in the liver[29,30]. Through an antisense strategy to knock down miR-122, Esau et al[30] observed that several genes which regulate lipid metabolism, specifically the key enzyme phosphomevalonate kinase, were down-regulated. Remarkably, silencing miR-122 in high-fat fed mice resulted in a significant reduction of hepatic steatosis, which was associated with reduced cholesterol synthesis rates and stimulation of hepatic fatty-acid oxidation[30]. Functional inhibition of miR-122 by an “antagomir” resulted in increased expression of several hundred genes including those that are normally repressed in hepatocytes[29]. In addition, Chang et al[20] identified a binding site for miR-122 in the 3’-UTR of the cationic amino acid transporter (CAT-1) mRNA using the Lewis-based miRNA targets approach[31]. Consistently, an inversed pattern of expression of CAT-1 and miR-122 was noted at all stages of liver development. Interestingly, miR-122-induced inhibition through the CAT-1 3’-UTR was efficiently relieved upon amino acid starvation, which validates CAT-1 as a target of miR-122[32].
Several groups, including ourselves, have recently tested the role of miRNAs in the regulation of biliary epithelial immunity in the liver. Human cholangiocytes express let-7 family members, miRNAs with complementarity to TLR4 mRNA. let-7 regulates TLR4 expression via post-transcriptional suppression in cultured human cholangiocytes. Infection of cholangiocytes with Cryptosporidium parvum (C. parvum), a parasite that causes intestinal and biliary disease, results in decreased expression of primary let-7i and mature let-7 in a MyD88/NF-κB-dependent manner. The decreased let-7 expression is associated with C. parvum-induced up-regulation of TLR4 in infected cells[23]. miRNAs may also be involved in cholangiocyte responses to pro-inflammatory cytokines, such as interferon-gamma (IFN-γ), and actively participate in the regulation of biliary inflammatory response in the liver. Specifically, miR-513 regulates B7-H1 translation and mediates IFN-γ-induced B7-H1 expression in human cholangiocytes. B7-H1 (CD274, PD-L1) is a member of the B7 family of costimulatory molecules and plays a critical immunoregulatory role in cell-mediated immune responses. Resting human cholangiocytes express B7-H1 mRNA, but not B7-H1 protein. IFN-γ induces B7-H1 protein expression and alters miRNA expression profile in cholangiocytes. Of those IFN-γ-down-regulated miRNAs, miR-513 has complementarity to the 3’-UTR of B7-H1 and targeting of miR-513 to B7-H1 mRNA results in translational repression, but not B7-H1 mRNA degradation[33].
miRNAS AND LIVER DISEASES
miRNAs and viral hepatitis
Genes encoding miRNAs have also been found in viruses and viral miRNAs have a regulatory effect on their protein-coding genes[34]. This regulatory effect may be beneficiary to the virus toward maintaining its replication, latency and evading the host immune system. Wu and colleagues analyzed the miRNA-encoding potential of the hepatitis B virus (HBV). Using computational approaches, the authors found that HBV putatively encodes only one candidate pre-miRNA. One viral mRNA was found to be targeted by the viral miRNA when they searched the target from viral mRNAs. Thus, HBV has evolved to use viral miRNAs as a means to regulate its own gene expression[35].
miRNAs from the host cells may play a role in building up direct or indirect effect in regulating viral genes[34,36,37]. Hepatitis C virus (HCV) is an enveloped RNA virus of the Flavivirus family, which is capable of causing both acute and chronic hepatitis in humans by infecting liver cells. miRNA-122 has been reported to facilitate the replication of HCV, targeting the viral 5’ non-coding region[38]. Indeed, HCV RNA can replicate in Huh 7 cells, which express miR-122, but not in HepG2 cells, which do not express miR-122. Silencing of miR-122 in hepatocytes resulted in a marked loss of replicating RNAs from HCV[38]. A putative miR-122 binding site in the 50-end of HCV genome was identified, suggesting a direct role of miR-122 in HCV replication[38]. An indirect effect of miR-122 inhibition on HCV regulation via the up-regulation of the cytoprotective enzyme heme-oxygenase 1 (HO-1) and the converse down-regulation of HO-1 repressor Bach1 was also characterized[21]. In addition, HCV replication is associated with an increase in expression of cholesterol biosynthesis genes that are regulated by miR-122[39].
On the other hand, Pedersen et al[40] demonstrated IFN-mediated modulation of the expression of numerous cellular miRNAs in the treatment of hepatocytes infected with HCV. Expression of a total of 30 cellular miRNAs in hepatocytes was influenced by IFN-α/β or IFN-γ. Specifically, eight of the miRNAs (miR-1, miR-30, miR-128, miR-196, miR-296, miR-351, miR-431 and miR-448), having nearly perfect complementarity in their seed sequences with HCV RNA genomes, were up-regulated. Importantly, these miRNAs are capable of inhibiting HCV replication and infection. This has opened the door to our understanding of novel host–defense mechanisms that exist in mammalian cells as well as the antiviral mechanisms employed by interferon.
miRNAs and human hepatocellular cancer (HCC)
Several studies have shown that specific miRNAs are aberrantly expressed in malignant HCC cells or tissues compared to non-malignant hepatocytes or tissue[41–47]. Selected miRNAs such as miR-21, miR-224, miR-34a, miR-221/222, miR-106a, and miR-203 are up-regulated in HCC compared to benign hepatocellular tumors such as adenomas or focal nodular hyperplasia. Certain miRNAs have been noted to be decreased in HCC compared to non-tumoral tissue, such as miR-122a, miR-422b, miR-145, and miR-199a. Murakami et al[48] showed a correlation between miR-222, miR-106a, miR-92, miR-17-5p, miR-20, and miR-18 and the degree of differentiation suggesting an involvement of specific miRNAs in the progression of the disease. Interestingly, the altered expression of some miRNAs was associated with distinctive risk factors, such as miR-96 with hepatitis B virus infection and miR-126* with alcohol use. Further investigations suggested the highly deregulated miR-223 and miR-222 could unequivocally distinguish HCC from adjacent non-tumoral liver, irrespective of viral association[46]. In HCC patients with hepatitis C and liver cirrhosis, miR-122, miR-100, and miR-10a were overexpressed, whereas miR-198 and miR-145 were up to five-fold down-regulated in hepatic tumors compared to normal liver parenchyma[47].
Alteration of expression profile in HCC for some miRNAs may be the consequence of malignant transformation. For other miRNAs, they may play a role in the transformation process. miRNA-122 was reported to be significantly and specifically down-regulated in HCC in humans as well as in rodents[41,42]. Amongst the putative target genes of miR-122 that can be predicted using computational tools, at least three are of interest in tumorigenesis: the gene for N-Myc, which is frequently rearranged in woodchuck liver tumors by woodchuck hepatitis virus[43], the gene referred to as “down-regulated in liver malignancy”[44], and the gene for cyclin G1[45]. In fact, miR-122 was shown to modulate cyclin G1 expression in HCC-derived cell lines, and an inverse correlation between miR-122a and cyclin G1 expression in primary liver carcinomas was further observed[45]. These studies suggest an influence of the down-regulation of miR-122 and the converse expression of cyclin G1 in hepatocarcinogenesis[21].
miRNAs and polycystic liver diseases
The polycystic liver and kidney diseases are a family of disorders with heterogeneous etiologies. Autosomal dominant polycystic kidney disease (ADPKD) is associated with renal and liver cystogenesis that clinically manifests in adulthood, often leading to dialysis and renal transplantation. It is caused by mutations in either of two genes, PKD1 and PKD2, which code for polycystin 1 and polycystin 2, respectively[49]. Autosomal recessive polycystic kidney disease (ARPKD) can present in neonates with massive renal cysts, causing respiratory failure secondary to abdominal competition that subsequently leads to infant demise, although milder forms can present later in life. Proposed mechanisms of disease include ciliary dysfunction, excess cell proliferation, and altered cell-cell or cell-matrix interactions.
Lee and colleagues provide data to support a novel mechanism for cystogenesis involving miRNA. They demonstrate that levels of the miRNA miR15a are decreased in livers of patients with ARPKD and ADPKD, respectively, and congenital hepatic fibrosis as well as in the PKC rat model of ARPKD. This results in increased expression of the cell-cycle regulator Cdc25A, which is a direct target of miR15a, and increased cellular proliferation and cystogenesis in vitro. As a whole, the findings indicate that changes in miRNA expression contribute to the phenotypic changes found in cystic liver disease (Figure 3)[49,50].
Figure 3.
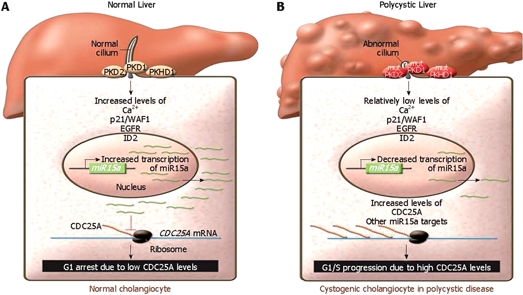
Model of miR15a function in hepatic cystogenesis. A: In normal cholangiocytes, ciliary signaling activates intracellular signaling pathways resulting in miR15a expression and CDC25A repression. Decreased levels of CDC25A then lead to G1 arrest, preventing cyst formation; B: In cholangiocytes of polycystic liver, miR15a level is reduced as a result of mutation of molecules important to the ciliary signaling. Reduction of miR15a results in elevated Cdc25A level, increased cell proliferation, and cyst growth. This figure is reprinted from Chu and Friedman[49] with the permission of the authors and the publisher.
miRNAS FOR THE DIAGNOSIS OF LIVER DISEASES
miRNAs could be of diagnostic significance for many liver diseases but current literature has been focused on tumors in the liver. Hepatocellular tumors comprise diverse benign and malignant neoplasms. Although the phenotypes can be broadly distinguished histologically or immunologically, these tumors can vary widely in their clinical behavior and prognosis. The use of miRNA-based classifications that correlate with etiology, pathogenetic changes, or malignant tendency will enhance molecular diagnosis and enable further definition of these phenotypes. In turn, this may yield clinically useful predictive markers of tumor behavior, as well as identify individual genetic and molecular contributors to tumorigenesis[51,52]. Thus, miRNA profiling studies could be used for defining clinical phenotypes, as well as providing potentially useful molecular diagnostic markers.
Impairments in miRNA functioning seen in cancerogenesis can be used for determination of miRNA expression for diagnostics of tumor origin. Each type of cancer is characterized by a certain profile of miRNA expression[52]. For example, the miRNA expression profiles in malignant hepatocytes differ from those of malignant cholangiocytes. Cluster analysis of miRNA expression profiles in tumors accurately determines not only type of the tissue (e.g. epithelium or hemopoietic system), but also discriminates tumors within the same type of tissue; this may reflect mechanism of transformation[53]. Obviously, evaluation of miRNA profiles can be used for prognosis of the development of tumors[54–56]. Such an approach for tumor diagnostics is very promising. However, it is not widely employed yet due to inadequately developed technology, lack of standards, requirements of very high purity of RNA samples, and not always reproducible results.
miRNAS AS THERAPEUTIC TARGETS FOR LIVER DISEASES
Development of miRNA/RNAi-based therapeutics requires several critical experimental steps, which include: (1) miRNA profiling of disease versus healthy tissue; (2) functional analysis of dyregulated miRNAs; and (3) in vivo studies with the use of different RNAi-based therapeutic methods to dysregulate miRNAs[57]. The success of such strategies for gene therapy will provide clinicians with a larger repertoire that includes miRNA-therapeutic agents. For example, chemically engineered oligonucleotides, termed ‘antagomirs’, have recently been developed and proven to be efficient and specific silencers of endogenous miRNAs in mice[29]. The silencing effect was considerably sustained over time probably because of a long half-life of endogenous miRNAs[12]. In addition, induction of stable loss-of-function phenotypes for specific miRNAs by lentiviral-mediated antagomir expression has recently been described[58].
Over the past several years, strategies based on targeting HBV, and to a lesser extent HCV, by both synthetic and expressed activators of the RNAi pathway have proved efficient to inhibit viral replication both in vitro and in vivo[59,60]. The recent study by Pedersen et al[40] provided great insights into validating sequence-predicted targets of cellular miRNAs within the HCV genome. miRNA-122 antagomir can down-regulate expression of several adult-liver genes[29], providing the potential to generate a new attractive expandable cell source for hepatocyte transplantation that would feature stem/progenitor cell phenotype. In addition, the effect of miR-122 antagomir in high-fat fed mice may be of therapeutic potential to reduce hepatic steatosis[29].
The important breakthrough in the field of hepato-carcinogenesis came from the accurate correlation of alterations in miRNAs with tumor proliferation and differentiation. So far, there have been very limited insights into this characterization. Recent studies by Meng et al[22,61] suggest a role for miRNAs in the influence of interleukin-6 in malignant cholangiocytes. MicroRNA-141, which showed strong overexpression in malignant cholangiocytes, was specifically localized in 12p, a region of known chromosomal aberration in biliary tract cancers[42]. Inhibition of miR-21 sensitized the response of cholangiocarcinoma cell lines to chemotherapy[42]. This observation gives rise to significant hope that miR-21 could serve as a biomarker for drug response in cholangicarcinoma.
CONCLUSION
Study of miRNAs flourished during the decade after their discovery. It is now clear that miRNAs can potentially regulate every aspect of cellular activity, from differentiation and proliferation to apoptosis, and also modulate a large range of physiological processes from developmental timing to organogenesis[1–6]. miRNAs also modulate a diverse spectrum of liver functions with developmental, (patho) physiological, and clinical implications. In the near future, the distinctive signature patterns of miRNA expression associated with liver cancer should allow classification of different stages in tumor progression[21]. Further, creating artificial miRNAs with salutary effects by promoting the expression of beneficial gene products (e.g. tumor-suppressor proteins) or targeting viral genomes (e.g. molecules designed to specifically target HCV-genome sequences) may become part of our patient management and complement chemotherapy and antiviral treatments. Overall, unraveling the regulatory circuits of miRNAs in the liver is a great challenge, but may provide attractive targets for mechanism-based treatment of liver diseases.
Supported by National Institute of Health grant (R01 AI071321) and the Tobacco Settlement Foundation of Nebraska (LB 692)
Peer reviewer: Dr. Bart Rik De Geest, Center for Molecular and Vascular Biology, Katholieke Universiteit Leuven, Campus Gasthuisberg, Herestraat 49, Leuven 3000, Belgium
S- Editor Li LF L- Editor Logan S E- Editor Zheng XM
References
- 1.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- 5.Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Trends Biochem Sci. 2003;28:534–540. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Taganov KD, Boldin MP, Baltimore D. MicroRNAs and immunity: tiny players in a big field. Immunity. 2007;26:133–137. doi: 10.1016/j.immuni.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ying SY, Lin SL. Intronic microRNAs. Biochem Biophys Res Commun. 2005;326:515–520. doi: 10.1016/j.bbrc.2004.10.215. [DOI] [PubMed] [Google Scholar]
- 9.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 13.Zeng Y. Principles of micro-RNA production and maturation. Oncogene. 2006;25:6156–6162. doi: 10.1038/sj.onc.1209908. [DOI] [PubMed] [Google Scholar]
- 14.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 15.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 16.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 17.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 18.Möröy T, Etiemble J, Bougueleret L, Hadchouel M, Tiollais P, Buendia MA. Structure and expression of hcr, a locus rearranged with c-myc in a woodchuck hepatocellular carcinoma. Oncogene. 1989;4:59–65. [PubMed] [Google Scholar]
- 19.Chang J, Provost P, Taylor JM. Resistance of human hepatitis delta virus RNAs to dicer activity. J Virol. 2003;77:11910–11917. doi: 10.1128/JVI.77.22.11910-11917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 21.Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48:648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 22.Meng F, Henson R, Wehbe-Janek H, Smith H, Ueno Y, Patel T. The MicroRNA let-7a modulates interleukin-6-dependent STAT-3 survival signaling in malignant human cholangiocytes. J Biol Chem. 2007;282:8256–8264. doi: 10.1074/jbc.M607712200. [DOI] [PubMed] [Google Scholar]
- 23.Chen XM, Splinter PL, O’Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282:28929–28938. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 25.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci USA. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguda BD, Kim Y, Piper-Hunter MG, Friedman A, Marsh CB. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc Natl Acad Sci USA. 2008;105:19678–19683. doi: 10.1073/pnas.0811166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Z, Jian Z, Shen SH, Purisima E, Wang E. Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos. Nucleic Acids Res. 2007;35:152–164. doi: 10.1093/nar/gkl1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arora A, Simpson DA. Individual mRNA expression profiles reveal the effects of specific microRNAs. Genome Biol. 2008;9:R82. doi: 10.1186/gb-2008-9-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 30.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 33.Gong AY, Zhou R, Hu G, Li X, Splinter PL, O’Hara SP, LaRusso NF, Soukup GA, Dong H, Chen XM. MicroRNA-513 regulates B7-H1 translation and is involved in IFN-gamma-induced B7-H1 expression in cholangiocytes. J Immunol. 2009;182:1325–1333. doi: 10.4049/jimmunol.182.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berkhout B, Jeang KT. RISCy business: MicroRNAs, pathogenesis, and viruses. J Biol Chem. 2007;282:26641–26645. doi: 10.1074/jbc.R700023200. [DOI] [PubMed] [Google Scholar]
- 35.Jin WB, Wu FL, Kong D, Guo AG. HBV-encoded microRNA candidate and its target. Comput Biol Chem. 2007;31:124–126. doi: 10.1016/j.compbiolchem.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh Z, Mallick B, Chakrabarti J. Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res. 2009;37:1035–1048. doi: 10.1093/nar/gkn1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saïb A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 38.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 39.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacob JR, Sterczer A, Toshkov IA, Yeager AE, Korba BE, Cote PJ, Buendia MA, Gerin JL, Tennant BC. Integration of woodchuck hepatitis and N-myc rearrangement determine size and histologic grade of hepatic tumors. Hepatology. 2004;39:1008–1016. doi: 10.1002/hep.20106. [DOI] [PubMed] [Google Scholar]
- 44.Harada H, Nagai H, Ezura Y, Yokota T, Ohsawa I, Yamaguchi K, Ohue C, Tsuneizumi M, Mikami I, Terada Y, et al. Down-regulation of a novel gene, DRLM, in human liver malignancy from 4q22 that encodes a NAP-like protein. Gene. 2002;296:171–177. doi: 10.1016/s0378-1119(02)00855-7. [DOI] [PubMed] [Google Scholar]
- 45.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 46.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 48.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 49.Chu AS, Friedman JR. A role for microRNA in cystic liver and kidney diseases. J Clin Invest. 2008;118:3585–3587. doi: 10.1172/JCI36870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee SO, Masyuk T, Splinter P, Banales JM, Masyuk A, Stroope A, Larusso N. MicroRNA15a modulates expression of the cell-cycle regulator Cdc25A and affects hepatic cystogenesis in a rat model of polycystic kidney disease. J Clin Invest. 2008;118:3714–3724. doi: 10.1172/JCI34922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryazansky SS, Gvozdev VA. Small RNAs and cancerogenesis. Biochemistry (Mosc) 2008;73:514–527. doi: 10.1134/s0006297908050040. [DOI] [PubMed] [Google Scholar]
- 52.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 53.Thum T, Catalucci D, Bauersachs J. MicroRNAs: novel regulators in cardiac development and disease. Cardiovasc Res. 2008;79:562–570. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 54.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 55.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 56.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 57.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 58.Czech MP. MicroRNAs as therapeutic targets. N Engl J Med. 2006;354:1194–1195. doi: 10.1056/NEJMcibr060065. [DOI] [PubMed] [Google Scholar]
- 59.Scherr M, Venturini L, Battmer K, Schaller-Schoenitz M, Schaefer D, Dallmann I, Ganser A, Eder M. Lentivirus-mediated antagomir expression for specific inhibition of miRNA function. Nucleic Acids Res. 2007;35:e149. doi: 10.1093/nar/gkm971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ely A, Naidoo T, Mufamadi S, Crowther C, Arbuthnot P. Expressed anti-HBV primary microRNA shuttles inhibit viral replication efficiently in vitro and in vivo. Mol Ther. 2008;16:1105–1112. doi: 10.1038/mt.2008.82. [DOI] [PubMed] [Google Scholar]
- 61.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]


