Abstract
AIM: To develop a method of labeling and micro-dissecting mouse Kupffer cells within an extraordinarily short period of time using laser capture microdissection (LCM).
METHODS: Tissues are complex structures comprised of a heterogeneous population of interconnected cells. LCM offers a method of isolating a single cell type from specific regions of a tissue section. LCM is an essential approach used in conjunction with molecular analysis to study the functional interaction of cells in their native tissue environment. The process of labeling and acquiring cells by LCM prior to mRNA isolation can be elaborate, thereby subjecting the RNA to considerable degradation. Kupffer cell labeling is achieved by injecting India ink intravenously, thus circumventing the need for in vitro staining. The significance of this novel approach was validated using a cholestatic liver injury model.
RESULTS: mRNA extracted from the microdissected cell population displayed marked increases in colony-stimulating factor-1 receptor and Kupffer cell receptor message expression, which demonstrated Kupffer cell enrichment. Gene expression by Kupffer cells derived from bile-duct-ligated, versus sham-operated, mice was compared. Microarray analysis revealed a significant (2.5-fold, q value < 10) change in 493 genes. Based on this fold-change and a standardized PubMed search, 10 genes were identified that were relevant to the ability of Kupffer cells to suppress liver injury.
CONCLUSION: The methodology outlined herein provides an approach to isolating high quality RNA from Kupffer cells, without altering the tissue integrity.
Keywords: Kupffer cells, India ink, Laser capture microdissection, Bile duct ligation, DNA microarray
INTRODUCTION
Kupffer cells, resident tissue macrophages that line the liver sinusoids, play a key role in modulating inflammation in a number of experimental models of liver injury[1]. Since Kupffer cells represent only a small portion of the entire liver cell population, greatly outnumbered by the parenchymal cells, Kupffer cell isolation faces major technical obstacles. Initial Kupffer cell preparations were heavily contaminated with other cell types or, if sufficiently purified, were only a small fraction of the total cell number and therefore not truly representative. Preparation improved drastically with the introduction of techniques that involved perfusion of the liver with collagenase and pronase[2].
After enzymatic digestion of the liver, the non-parenchymal liver cells are purified by density gradient centrifugation or centrifugal elutriation[3]. Neither of these methods is able to separate Kupffer cells from other non-parenchymal cell types since cell size and density exhibit considerable overlap[4]. Furthermore, Kupffer cell purification can be achieved by adherence to plastic[5], or positive selection using specific antibodies and magnetic beads[6].
Laser capture microdissection (LCM) circumvents many of the limitations inherent in conventional isolation methods. LCM was created at the National Institutes of Health, Bethesda, MD, USA and further developed by Arcturus Engineering, Inc., Mountain View, CA, USA[7]. The principals of LCM entail overlaying the tissue section with a transparent ethylene vinylacetate thermoplastic film. At the point of interest, the film is melted onto the tissue surface with a laser integrated with the microscope optics and then removed, capturing sample areas as small as 3-5 μm from intact tissue sections[8].
For the analysis of gene expression, an important challenge for LCM remains the specific and rapid labeling of the target cell population, thus minimizing the degradation of RNA. In this regard, classic immunohistochemical staining methods for visualizing Kupffer cells are not generally applicable. In the present study, a well-established and efficient method using India ink to label Kupffer cells in vivo was described[9]. Kupffer cells are mainly located in the periportal areas, where they have ready access to pathogens and particulate antigens entering the liver with portal-venous blood[10,11]. In contrast to liver sinusoidal endothelial cells (LSECs), which mainly ingest soluble materials via pinocytosis, Kupffer cells take up particulate material via phagocytosis[12]. It is relevant to note, therefore, that while colloidal gold ≤ 100 nm diameter particle size is internalized almost exclusively by LSECs, colloidal carbon is taken up primarily by Kupffer cells. This apparent contradiction is explained by the fact that blood platelets bind carbon and the platelet-carbon complexes are subsequently phagocytosed by Kupffer cells[9]. Importantly, the phagocytic capacity of Kupffer cells is not altered by the ingestion of these complexes[13].
The ability to isolate carbon-labeled Kupffer cells from intact tissue sections using LCM offers an attractive approach to studying gene expression under diverse conditions. The aim of this study was to validate this approach and to apply it to an animal model of cholestatic liver injury.
MATERIALS AND METHODS
Animals
Wild-type female, C57BL/6J mice were purchased from The Jackson Laboratories (Bar Harbor, ME, USA) and used at 8-12 wk of age. The animals were treated in accordance with NIH publications entitled “Principles for Use of Animals” and “Guide for the Care and Use of Laboratory Animals.” The mice were housed in well-ventilated rooms maintained at 22°C and an alternating 12-h light and dark cycle; food and water were provided ad libitum.
Common bile duct ligation
Ligation of the common bile duct was performed as previously described[6]. The abdomen of mice under deep anesthesia was disinfected with 70% ethanol. A midline upper abdominal incision was made and the abdominal wall was retracted. The common bile duct was identified, isolated and double-ligated with #4-0 braided silk sutures and divided in between. The fascia and skin of the midline abdominal incision were closed with #6-0 braided silk sutures. Control mice underwent sham operations in which the common bile duct was exposed, but not ligated.
Kupffer cell staining
Frozen, 6-μm thick liver sections were cut with a cryostat (Leica, Wetzlar, Germany). Slides were warmed to room temperature for 30 min, fixed by immersion in ice cold acetone (4°C) for 5 min, and air dried for 30 min. After rinsing the slides three times in PBS to remove the tissue freezing matrix, non-specific binding was blocked with 5% normal rabbit serum and 1% BSA for 60 min. This step was followed by 15 min of avidin blocking and 15 min of biotin blocking (Vector Laboratories, Burlingame, CA, USA). After each step, the slides were rinsed in PBS. The slides were stained first with a 1/10 dilution of rat IgG2a anti-mouse F4/80 (a pan-specific macrophage marker; eBioscience, San Diego, CA, USA) monoclonal antibody for 60 min. Subsequently, the slides were washed three times for 10 min with PBS and then incubated for 45 min on a shaker with a 1/50 dilution of biotinylated goat anti-rat polyclonal antibody (Vector Laboratories). After rinsing the slides, pre-diluted streptavidin-Cy3 (Invitrogen, Carlsbad, CA, USA) was applied to the tissue sections and incubated for 30 min; the slides were then dried and mounted with Fluoromount-G (SouthernBiotech, Birmingham, AL, USA). All the steps were performed at room temperature.
Processing of the liver tissue and LCM
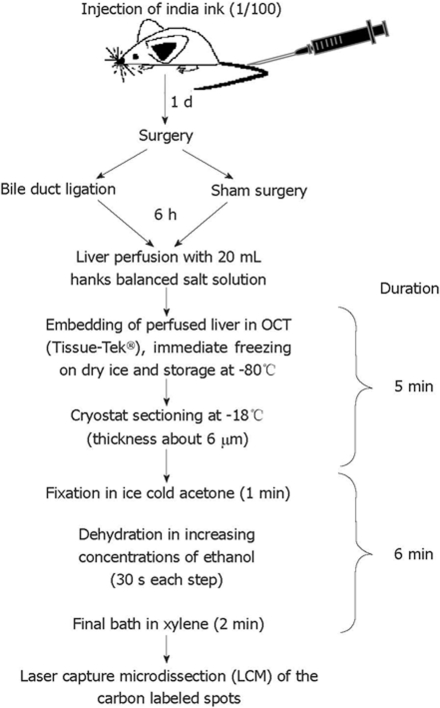
Mice were inoculated iv via the tail vein with 200 μL India ink diluted 1:100 in saline 1 d prior to surgery. Immediately following euthanasia, the livers were perfused in situ with 20 mL Hank’s buffered salt solution to eliminate blood cells, and dissected. Tissue wedges were flash frozen in Tissue-Tek® Optimum Cutting Temperature™ (OCT) compound and stored at -80°C. Cryostat sections (6 μm) were prepared under RNase-free conditions at -20°C, fixed immediately in acetone for 2 min at 4°C, dehydrated by sequential immersion for 30 s in 75%, 95% and 100% ethanol, immersed for 2 min in xylene, and then air dried. For each cutting session, new blades, single use staining jars, and fresh (RNAse free) solutions were used.
LCM was performed using an AutoPix Automated Laser Capture Microdissection System equipped with an infrared diode laser according to the protocols and methods provided by the manufacturer (Arcturus Engineering, Santa Clara, CA, USA). Air-dried slides were placed under the microscope of the LCM system and one drop of xylene was applied to visualize the carbon-containing Kupffer cells (Figure 1). A static image was taken and, after the xylene evaporated, the liver sections were overlaid with thermoplastic membranes mounted on transparent, CapSure Macro LCM Caps (Arcturus). The carbon-labeled (Kupffer) cells were captured by laser activation (12-15 μm laser spot size, 60-80 mW power, 3.0 ms duration) and focal melting of the membrane. At least 300 cells were captured on each cap, and three different caps (equivalent to approximately 1000 cells) were prepared for each sample.
Figure 1.
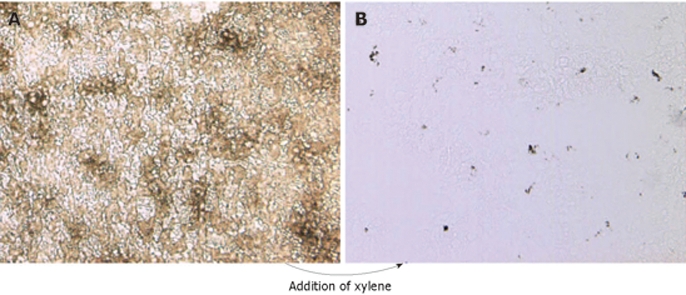
India-ink-positive cells revealed in tissue section overlaid with xylene. A non-stained liver section derived from a mouse inoculated iv with India ink was not mounted (A), or was overlaid with xylene (B).
RNA extraction and purification
Cells captured on thermoplastic membranes were immersed in Lysis Buffer (RLT) (Qiagen Inc., Valencia, CA, USA). Subsequently, the RNA was extracted, purified using an RNeasy Micro Kit (Qiagen), and quantified using Lab-on-Chip technology (Agilent Technologies, Palo Alto, CA, USA). Intact 18S and 28S rRNA confirmed the integrity of the RNA extracted. RNA representative of cells constituting the whole liver was extracted from an entire (scraped) tissue section.
RNA amplification and cRNA labeling
Purified total RNA obtained from the microdissected cells was subjected to 1.5 rounds of linear amplification using T7 bacteriophage RNA-polymerase-driven in vitro transcription, with materials and protocol provided in the RiboAmp™ HS RNA Amplification Kit (KIT0205; Arcturus). The final amplification and labeling of dsDNA product was performed using the Enzo BioArray HighYield RNA Transcript Labeling Kit (Enzo Life Sciences, Framingdale, NY, USA).
Microarray hybridization
Microarray analysis was performed using the GeneChip Mouse Genome® 430 2.0 array and the recommended instruments (GeneChip Scanner 3000), with reagents and protocols provided by the supplier (Affymetrix, Santa Clara, CA, USA). This GeneChip covers the transcribed mouse genome on a single array with 45 000 probe sets that analyze the expression level of over 39 000 transcripts.
Bioinformatics and data mining
Each group consisted of three samples; each sample was pooled from three sets of captured material derived from individual, sham-operated or bile-duct-ligated mice. The expression signals were normalized using the standardization and normalization of microarray data (SNOMAD) program[14]. Concordantly absent expression signals were removed from analysis. A t test was used to perform paired comparisons of the gene expression levels between sham-operated and bile-duct-ligated animals. A 5% false discovery rate (FDR) correction controlled for multiple comparisons. The q value of a test measured the minimum FDR rate incurred when calling that test significant. q values were computed from the unadjusted P values, using the Q-VALUE program written by Storey and Tibshirani[15]. Significant, differentially expressed genes were grouped into functional categories using the GenMAPP 2 (http://www.GenMAPP.org) and MAPPFinder programs by integrating the annotations of the Gene Ontology Project (ftp://ftp.geneontology.org/go/gene-associations)[16,17].
Quantitative real-time reverse-transcriptase polymerase chain reaction (RT-PCR)
Purified RNA extracted from microdissected Kupffer cells was treated with DNase (10 U DNase I/μg total RNA) and reverse transcribed with Sensiscript Reverse Transcriptase according to the protocols provided by the supplier (Qiagen). Quantitative gene expression analysis was performed using the MX4000 Multiplex Quantitation QPLR System (Stratagene, La Jolla, CA, USA) and SYBR green technology. QuantiTect SYBR Green PCR Master Mix (12.5 μL; Qiagen) was mixed in 96-well optical plates with an equal volume of RNase-free water that contained 0.6 μmol/L of the forward and reverse primers, and cDNA corresponding to 70 ng of total RNA input. The plates were heated for 2 min at 50°C, and then 15 min at 95°C to activate the HotStart Taq DNA polymerase. Subsequently, 45 cycles consisting of 30 s at 95°C followed by 1 min at 55°C and 30 s at 72°C were run. To verify the generation of single PCR products, melting curves were constructed by heating the samples to 95°C with a ramp time of 20 min at the end of the run. 18S rRNA was used as the housekeeping standard. The threshold cycle (i.e. the number of PCR cycles required in order for the fluorescent signal to reach a fixed intensity) was determined and the number of RNA copies was calculated from standard curves. The mean mRNA copies/105 18S rRNA copies ± SD for samples derived from three mice treated comparably was reported. The PCR primers listed in Table 1 were designed using Primer3 software (Whitehead Institute, Cambridge, MA, USA) and purchased from Qiagen.
Table 1.
PCR primers
| Gene | GenBank accession size | Orientation | Primer sequence | Amplicon size |
| KCR | D88577 | Forward | AAATGACCTCAGCTCCCAGA | 103 |
| Reverse | TTCACCAGCCCTTTCCATAC | |||
| CSF-1 receptor (CD115) | X06368 | Forward | TGGCCTTCCTTGCTTCTAAA | 114 |
| Reverse | ATGTCCCTAGCCAGTCCAA | |||
| HMGCS1 | NM_145942 | Forward | GCGGCTAGAAGTTGGAACAG | 196 |
| Reverse | AGCATATCGTCCATCCCAAG | |||
| Lipocalin 2 | NM_008491 | Forward | CCAGTTCGCCATGGTATTTT | 169 |
| Reverse | GGTGGGGACAGAGAAGATGA | |||
| Hck | BC010478 | Forward | GCCTCAAAAACAGAGCCAAG | 150 |
| Reverse | GTACAGTGCGACCACAATGG | |||
| 18S rRNA | XR_000144 | Forward | AATGGTGCTACCGGTCATTCC | 192 |
| Reverse | ACCTCTCTTACCCGCTCTC |
Statistical analysis
The results were analyzed using the SigmaStat statistics program (Jandel Scientific, San Rafael, CA, USA). Individual means were compared using a non-paired Student’s t test or a Mann-Whitney rank sum test. Data derived from three or more groups were compared by one-way analysis of variance (ANOVA) followed by a Tukey test, to identify the groups that differed significantly (P < 0.05).
RESULTS
Carbon particles co-localize with fluorescent latex beads and anti-F4/80 staining
LCM and analysis of the genes expressed by a specific cell type in a heterogeneous population (i.e. Kupffer cells among other hepatic cells in the study reported here) depends upon the rapid identification of those cells. The intravenous inoculation of India ink and subsequent ingestion of carbon particles by Kupffer cells provides a well-documented means of rapidly labeling Kupffer cells in vivo for laser capture. At 18 h or more post-inoculation of India ink, the livers were perfused in situ with a balanced salt solution, dissected, and frozen at -80°C, and 6-μm liver sections were prepared from representative tissue wedges. The carbon particles were visualized microscopically by overlaying the sections with xylene (Figure 1). To ensure the particles were phagocytosed by Kupffer cells, mice were injected simultaneously with India ink and fluorescent latex beads (Sigma-Aldrich Chemical Co., St. Louis, MO, USA), which are often used to track and label Kupffer cells in vivo. Using conventional light and UV light to visualize fluorescent emissions, two images were made with a confocal microscope, and both images were merged using Adobe Photoshop. As shown in Figure 2A-C, the carbon particles and fluorescent latex beads co-localized. The liver sections were also stained with a streptavidin-conjugated antibody specific for F4/80, a surface marker expressed by macrophages including Kupffer cells, and biotinylated Cy3 was used to visualize antibody binding. Confocal images of the carbon particles and bound antibody were merged and found to co-localize as well (Figure 2D-F). Notably, areas with an accumulation of carbon particles stained intensely with F4/80 (white arrows), whereas single carbon spots (black arrows) were not stained with F4/80. This observation was considered when selecting areas for LCM.
Figure 2.
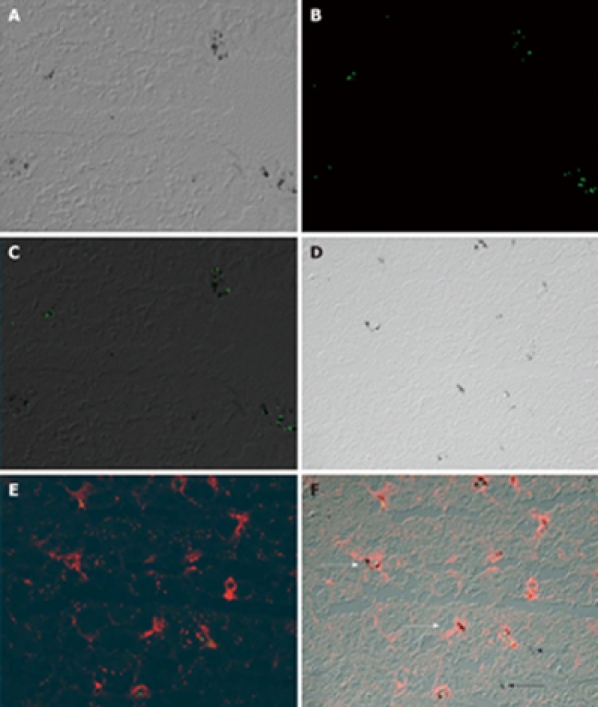
Carbon particles co-localize with ingested fluorescent latex beads and macrophages stained immunohistochemically. A-C: Kupffer cells lining the liver sinusoids were identified by the presence of carbon particles in mice inoculated iv with India ink. Fluorescent latex beads mixed with India ink co-localized with the carbon particles. D-F: A carbon-particle-containing liver section was stained sequentially with streptavidin-conjugated anti-pan macrophage marker F4/80 and biotin-Cy3, and visualized by fluorescence microscopy. An accumulation of carbon particles stained intensely with F4/80 (white arrows), whereas single carbon spots (black arrows) were not stained with F4/80.
LCM of carbon-positive areas enriches for Kupffer cell receptor (KCR) and colony-stimulating factor-1 receptor (CSF-1R) mRNA
Labeling Kupffer cells with India ink in vivo prior to liver dissection negates in vitro staining prior to LCM. This drastically reduced the time between tissue sectioning and actually capturing the cells (approximately 15 min elapsed time) (Figure 3). To ascertain that material was captured, tissue sections and caps were examined by conventional microscopy. A liver section with carbon particles overlaid with xylene is shown in Figure 4A; the red-crossed-through circles show the location of carbon-positive areas on the static image obtained with the LCM system. After microdissection, the same section displayed gaps where the captured areas were removed (Figure 4B). Captured material on caps before RNA extraction with lysis buffer is shown in Figure 4C. Overlaying the cap with water revealed carbon particles associated with this material (Figure 4D).
Figure 3.
Experimental timeline. Schematic outline of the experimental approach used to isolate Kupffer cells labeled in vivo with carbon. Notably, the elapsed time between liver dissection, sectioning and LCM of carbon-labeled Kupffer cells was only about 11 min.
Figure 4.
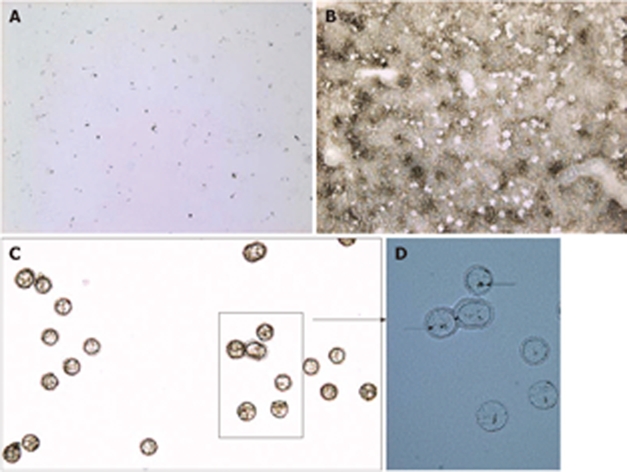
Images documenting LCM of carbon-labeled Kupffer cells. A: Static image of carbon-labeled liver section overlaid with xylene (10 × magnification); B: Same tissue section after the evaporation of xylene and capture of carbon-labeled cells (10 ×); C: Captured cells visualized on the CapSure LCM cap (20 ×); D: Same cap mounted with water and a coverslip showing carbon within microdissected cells (arrows, 40 ×).
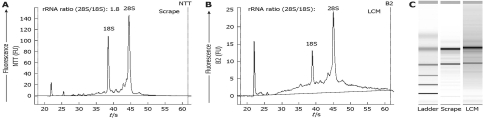
The RNA was extracted from the captured material on caps, and the quality and quantity were assessed using a bioanalyzer and Lab-on-Chip technology (Agilent Technologies). Degradation appeared only slight relative to the RNA extracted from an entire tissue section scraped from a slide (Figure 5). Indeed, the quality of RNA obtained by LCM proved to be high and the quantity sufficient for subsequent analyses by real-time RT-PCR and microarray analysis. The amount of RNA isolated ranged from 100 to 1000 pg per 1000 Kupffer cells captured.
Figure 5.
High quality RNA is extracted from material obtained by LCM. RNA size and quality were analyzed with the Agilent 2100 Bioanalyzer. A, B: Profiles of total RNA extracted from scrapes of the entire liver section (A) and from laser-captured cells (B); C: Electrophoresis gel of the same RNA.
The carbon-labeled material obtained by LCM was significantly enriched in KCRs and CSF-1R RNA transcripts relative to the transcripts determined in the tissue scrape (Figure 6). This further substantiates the association of carbon with and preferential dissection of Kupffer cells by LCM in accordance with our previous findings[6].
Figure 6.
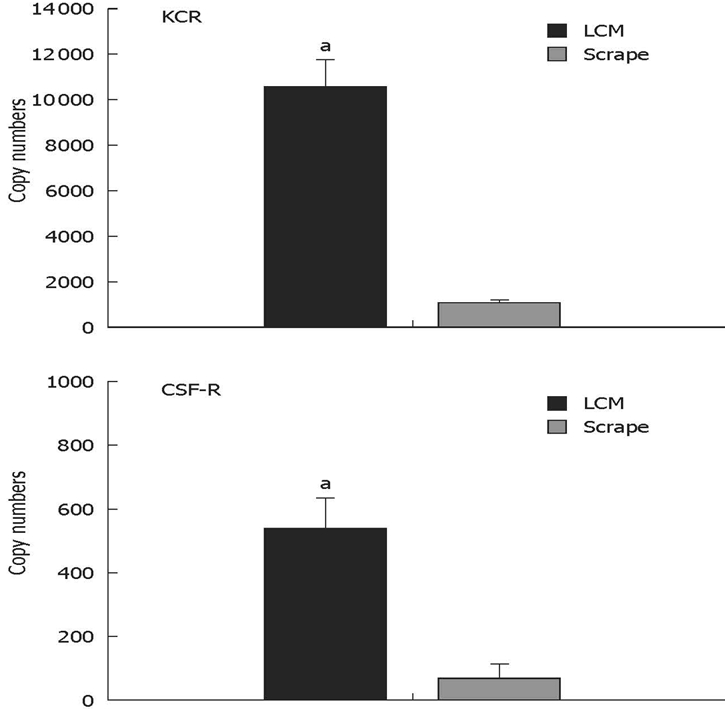
Kupffer-cell-specific mRNA transcripts are enriched in the laser-captured material. Carbon-labelled cells or total liver section scrapes were obtained from three mice; Kupffer cell receptor and CSF-1 receptor mRNA transcripts were quantified by real-time RT-PCR. aSignificantly more than extracted from total tissue scrapes (P < 0.005; non-paired Student t test).
Comparison of microarray data generated from Kupffer cells dissected from sham-operated and from bile-duct-ligated mice
Kupffer cells play a vital role in suppressing tissue damage that occurs in a mouse model of cholestatic liver injury. To identify the genes involved, groups of mice inoculated iv with India ink 18 h previously underwent bile-duct ligation or sham operation. The livers were dissected at 6 h post-surgery and sectioned; the carbon-labeled cells were obtained by LCM; the RNA was extracted, purified and amplified; and DNA microarray analysis was performed using the GeneChip Mouse Genome® 430 2.0 array. A total of 493 genes were found to be more than 2.5-fold up- or down-regulated (q value < 10) when the microarray data from bile-duct-ligated and sham-operated animals were compared. Interestingly, most of the genes that exhibited significant change were involved in cell growth and maintenance (Table 2: Gene ontology). Sixteen of the genes identified were highly up-regulated or down-regulated (> 5-fold change with a q value < 0.5), and the focus of further analyses. Our primary interest in evaluating these genes was to ascertain their role in maintaining the integrity of the liver during periods of cholestasis. Therefore, the 16 highly up- or down-regulated genes were subjected to a standardized PubMed search (http://www.ncbi.nlm.nih.gov/sites/entrez) with terms related to our model (Table 3). Abstracts dating back 15 years were reviewed and the publications relevant to our model were selected (articles of interest). Based upon this search algorithm, the number of genes of interest was further reduced to 10 (Table 4). Three of these genes, i.e. 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1 (HM GCS1), lipocalin 2, and hemopoietic cell kinase (Hck), were of particular interest in light of the role of Kupffer cells in abrogating cholestatic liver injury.
Table 2.
Significantly up- or down-regulated genes ontology
| Number | Percentage (%) | |
| Cytoplasm | 105 | 16.7 |
| Cell growth and/or maintenance | 87 | 13.8 |
| Integral to membrane | 85 | 13.5 |
| Nucleus | 57 | 9.0 |
| Nucleoside and nucleic acid metabolism | 54 | 8.6 |
| Protein metabolism | 54 | 8.6 |
| Biosynthesis | 34 | 5.4 |
| Purine nucleotide binding | 33 | 5.2 |
| DNA binding | 30 | 4.8 |
| Signal transduction | 26 | 4.1 |
| Response to external stimulus | 24 | 3.8 |
| Transferase activity, transferring phosphorus containing groups | 24 | 3.8 |
| Lipid metabolism | 23 | 3.7 |
| Plasma membrane | 19 | 3.0 |
| Catabolism | 18 | 2.9 |
| Immune response | 18 | 2.9 |
| Phosphorus metabolism | 18 | 2.9 |
| Peptidase activity | 16 | 2.5 |
Table 3.
Standardized Pubmed search with terms relevant to Kupffer cells abrogating liver injury during cholestasis: Each gene up- or down-regulated > 5-fold
| Gene | Fold change |
Search terms |
Articles of interest | ||||||||
| *Gene* | Kupffer cells | Cholestasis | Apoptosis | Cell death | Survival | Inflammation | Liver | Liver repair | |||
| Acetyl-coenzyme A synthetase 2 | 9.61 | 179 | - | - | - | 1 | 2 | - | 19 | - | 2 |
| Hydroxysteroid (17-β) dehydrogenase 2 | 8.26 | 668 | - | - | 4 | 2 | 10 | 3 | 70 | - | 1 |
| RIKEN cDNA 1110025G12 | 7.61 | - | - | - | - | - | - | - | - | - | - |
| HM GCS1 | 5.81 | 181 | - | - | 6 | 6 | 2 | - | 67 | - | 3 |
| RBP 4 | 5.76 | 1044 | 2 | 2 | 10 | 10 | 25 | 42 | 195 | 2 | 3 |
| RIKEN cDNA 1200011D03 gene | 5.75 | - | - | - | - | - | - | - | - | - | - |
| Inter-alpha trypsin inhibitor, heavy chain 2 | 5.46 | 113 | 2 | - | 1 | - | 3 | 19 | 63 | - | 5 |
| Lipocalin 2 | 5.35 | 375 | - | 1 | 24 | 25 | 15 | 48 | 17 | 1 | 15 |
| Hck | -5.67 | 4 | - | - | - | - | - | - | - | - | 3 |
| Hypothetical protein 5930431H10 | -6.15 | - | - | - | - | - | - | - | - | - | - |
| Leucine-rich repeat-containing 5 | -6.26 | 22 | - | - | 1 | 1 | - | - | 1 | - | 2 |
| SAM domain and HD domain, 1 (SAMHD1) | -6.81 | 5 | - | - | - | - | - | - | - | - | - |
| Myristoylated alanine rich protein kinase C substrate | -7.14 | 458 | - | - | 5 | 7 | 10 | 5 | 11 | - | 3 |
| Heterogeneous nuclear ribonucleoprotein A/B | -7.94 | 50 | - | - | 2 | 2 | 2 | - | 5 | - | 3 |
| (SCAN-KRAB-) zinc finger gene 1 | -9.05 | 6 | - | - | 1 | - | 1 | - | - | - | 1 |
| Inactive X specific transcripts | -12.21 | 3 | - | - | - | - | - | - | - | - | - |
Table 4.
Overview of function, source and involvement of the 10 genes identified by Pubmed-search: Degree of up- or down-regulation
| Gene | Fold change | Biological function | Primary cell source | Involvement | Reference |
| Acetyl-Coenzyme A synthetase 2 | 9.61 | Mitochondrial matrix enzyme involved in CoA ligation | Mitochondrial matrix | Activation during fasting | [50,51] |
| HM GCS1 | 5.81 | Cholesterol biosynthesis | Inhibition induces apoptosis, up-regulation involved in liver regeneration | [25,26] | |
| RBP4 | 5.76 | An a2-globulin that transports vitamin A from the liver | Parenchymal liver cells, little in Kupffer cells | Known marker for hepatocellular necrosis | [22,23] |
| Inter-alpha trypsin inhibitor, heavy chain 2 | 5.46 | Plasma protease inhibitor | Kupffer cells | Endothelial growth factor | [27–29] |
| Lipocalin 2 | 5.35 | Iron-siderophore-binding protein | Macrophages, neutrophils | Suppression of inflammation, tissue involution, apoptosis, differentiation of myeloid cells | [31–40] |
| Hck | -5.67 | Protein-tyrosine kinase | Granulocytes, monocytes | Mitogenesis, differentiation, survival, migration | [41–44] |
| Leucine-rich repeat-containing 5 | -6.26 | Interaction with cellular G-proteins | Enzyme inhibition, cell adhesion, cellular trafficking, proliferation and activation of lymphocytes and monocytes | [45,52] | |
| Myristoylated alanine rich protein kinase C substrate | -7.14 | Substrate of protein kinase C | Intracellular signaling, brain development, cellular migration and adhesion, phagocyte activation, phagocytosis | [46,47,53–55] | |
| Heterogeneous nuclear ribonucleoprotein A/B | -7.94 | Chromatin-associated RNA-binding protein | Ubiquitious | RNA handling, proliferation arrest | [56,57] |
| (SCAN-KRAB-) zinc finger gene 1 | -9.05 | Protein interaction domain | Ubiquitious | Cell survival, differentiation | [48,49] |
DISCUSSION
Non-specific phagocytosis of particles taken up in the liver is mediated primarily by Kupffer cells. Kupffer cells constitute 20%-30% of the non-parenchymal cells of the liver, where they reside within the sinusoidal vascular space, predominantly in the periportal area. Here, they are perfectly situated to clear endotoxins from the blood, and to phagocytose microorganisms and debris. For example, latex particles inoculated iv distinguish a latex-labeled Kupffer cell population that does not change over a 3-mo period, substantiating the relatively long life span of these resident tissue macrophages[18]. While endothelial cells lining the blood vessels internalize soluble materials via pinocytosis, Kupffer cells generally ingest particulate matter, e.g. colloidal carbon, via phagocytosis. The restricted ability to internalize carbon particles was used herein to label and subsequently dissect Kupffer cells by LCM. The specificity of this approach is demonstrated by co-localization of the particles with fluorescent latex beads inoculated simultaneously iv, and with anti-F4/80-stained hepatic macrophages (Figure 2). The specificity is further documented by the elevated levels of macrophage-associated RNA transcripts (i.e. CSF-1R and KCR messages) extracted from the carbon-labeled material obtained by LCM, relative to those levels extracted from whole liver sections.
Unlike the majority of cells that constitute the mononuclear phagocyte system, Kupffer cells synthesize interleukin (IL)-10 (an anti-inflammatory cytokine) when exposed to lipopolysaccharide (LPS), rather than proinflammatory cytokines such as IL-12 and IL-18[19]. This finding suggests that Kupffer cells, which are continuously exposed to LPS derived from the intestines via the hepatic portal vein, generally impose an immunosuppressive effect on the hepatic environment. In this regard, it is relevant to note that the ingestion of endotoxin and debris occurs coincidentally with the ability of Kupffer cells to induce tolerance[20]. Moreover, in addition to tolerance, our studies involving a mouse model of cholestasis indicate that Kupffer cells play a general role in suppressing inflammation and liver injury[6].
Differentiating Kupffer cells from non-resident macrophages, which infiltrate the liver during periods of chronic inflammation, and which may or may not display similar effector functions, represents a major challenge in characterizing the activity of Kupffer cells. The relatively short time period between bile-duct ligation and liver dissection enabled the specific identification and isolation of carbon-labeled Kupffer cells in the study reported here. Whether this approach permits the differentiation of Kupffer cells from inflammatory macrophages recruited to cholestatic livers 2-3 d after biliary obstruction is a matter of ongoing investigations in our laboratory.
LCM allows the isolation of tissue sections as small as 8-10 μm[8] and avoids procedures such as collagenase digestion that, conceivably, can alter the gene expression profile of Kupffer cells. Since no staining is necessary, sections derived from the livers of sham-operated and bile-duct-ligated mice could be processed and subsequently dissected within an extremely short period of time. Moreover, while the effects of histological staining on the integrity of RNA are well documented[21], the quality of RNA obtained using our approach proved to be high and the quantity sufficient enough for subsequent analyses (Figure 5).
Microarray analysis comparing Kupffer cells derived from bile-duct-ligated versus sham-operated mice revealed a total of 493 genes that were differentially expressed (2.5-fold, q value < 10). In an effort to restrict further analysis to genes specifically relevant to Kupffer cells and their response to cholestasis, those genes that were up- or down-regulated more than five-fold were subjected to a standardized PubMed search (Table 3), and 10 genes of interest were selected (Table 4). Notably, the expression of four of these genes [retinol binding protein 4 (RBP4), inter-alpha trypsin inhibitor heavy chain 2, lipocalin 2, and Hck] was previously described in Kupffer cells or cells of the mononuclear phagocyte lineage. Of these, only RBP4 appears more common in hepatocytes than in Kupffer cells[22]. RBP4 is often up-regulated in response to hepatocellular necrosis[23] and thus, its expression in Kupffer cells may derive readily from the ingestion of apoptotic hepatocytes[24]. Two of the other up-regulated genes are involved in tissue regeneration: HMGCS1 is critical for hepatocyte regeneration after partial hepatectomy[25,26]; and inter-alpha trypsin inhibitor, a plasma protease inhibitor, promotes endothelial cell growth[27–29].
In addition to promoting tissue regeneration and cell growth, Kupffer cells may induce apoptosis by infiltrating inflammatory cells (e.g. neutrophils) and thus, suppress their contribution to cholestatic liver injury[30]. In this regard, lipocalin 2, the product of one of the up-regulated genes expressed by Kupffer cells present in cholestatic livers induces apoptosis by leukocytes, but not other cell types[31–34]. Moreover, lipocalin has been implicated in myeloid cell differentiation and innate host defenses to bacterial infections[35–40].
In contrast to the genes discussed immediately above, five genes expressed by Kupffer cells were significantly down-regulated in response to cholestasis. Hck, a member of the highly conserved sarcoma family of protein-tyrosine kinases that is preferentially expressed by cells of the myeloid lineage, influences cell migration, adhesion, differentiation and survival[41–44]. Secondly, leucine-rich repeat containing protein 5 promotes the proliferation and/or activation of lymphocytes and monocytes[45]. Thirdly, myristoylated alanine-rich C kinase substrate (a substrate of protein kinase C contained in large amounts in macrophages) has been implicated in phagocyte activation, endocytosis, exocytosis, phagocytosis, and mobility[46,47]. Finally, the SCAN domain is a highly conserved motif found near the N terminus of C(2)H(2) zinc finger transcription factors. Some family members play a role in the transcriptional regulation of genes involved in cell differentiation and survival[48,49].
In summary, LCM offers an effective approach to analyzing gene expression by Kupffer cells without altering liver integrity. Pre-labeling the cells with carbon in vivo renders further staining procedures unnecessary and allows the isolation of sufficient amounts of intact RNA. Thus, the contributions of Kupffer cells to various models of liver injury can be delineated. In this regard, using DNA microarray analysis, we identified 16 genes expressed by Kupffer cells that were highly up- or down-regulated following bile-duct ligation in a mouse model of cholestatic liver injury. A standard PubMed search conducted using terms relevant to the model determined 10 of these genes to be of specific interest and germane to the role of Kupffer cells in suppressing tissue damage. These genes are the subject of ongoing investigation in our laboratory.
COMMENTS
Background
Kupffer cells, resident tissue macrophages that line the liver sinusoids, play a key role in modulating intrahepatic inflammation. Since Kupffer cells represent only a small portion of the entire liver cell population, greatly outnumbered by the parenchymal cells, Kupffer cell isolation faces major technical obstacles. Laser capture microdissection (LCM) offers a method of isolating a single cell type from specific regions of a tissue section. LCM is an essential approach used in conjunction with molecular analysis to study the functional interaction of cells in their native tissue environment.
Research frontiers
LCM circumvents many of the limitations inherent in conventional isolation methods. LCM was created at the National Institutes of Health (Bethesda, MD, USA) and further developed by Arcturus Engineering (Mountain View, CA USA). The principles of LCM entail overlaying the tissue section with a transparent ethylene vinylacetate thermoplastic film. At the point of interest, the film is melted onto the tissue surface with a laser integrated with the microscope optics and then removed, capturing sample areas as small as 3-5 μm from intact tissue sections.
Innovations and breakthroughs
Described herein is a method of labeling and microdissecting mouse Kupffer cells within an extraordinarily short period of time. Kupffer cell labeling is achieved by injecting India ink intravenously, thus circumventing the need for in vitro staining. This method provides an approach to isolating high quality RNA from Kupffer cells without altering the tissue integrity.
Applications
The ability to isolate carbon-labeled Kupffer cells from intact tissue sections using LCM offers an attractive approach to studying gene expression under diverse conditions. Thus, the specific contributions of Kupffer cells to various models of liver injury can be delineated.
Terminology
LCM permits under direct microscopic visualization rapid isolation of selected cell populations from a tissue section. A transparent thermoplastic film is applied to the surface of the tissue section, and a laser pulse then specifically activates the film above the cells of interest. The film is melted onto the tissue surface and then removed, capturing the pre-selected cells. Kupffer cells are resident tissue macrophages that line the liver sinusoids. Here, they are perfectly situated to clear endotoxins from the blood, and to phagocytose microorganisms and debris. Kupffer cells play a key role in modulating inflammation in the liver.
Peer review
In the current study, a new method of isolating Kupffer cells is presented based upon labeling the cells with India ink inoculated iv and microdissection of the carbon-labeled cells by laser capture. The study is well-controlled with regard to the specificity of staining of Kupffer cells with India ink. The method was further validated in a model of cholestasis.
Acknowledgments
The authors thank Dr. Patricia Meitner (Pathology Research, Rhode Island Hospital, directed by Dr. Murray Resnick) for her aid and technical expertise with LCM and Karl Simkevitch for his support with the microarray hybridization.
Footnotes
Supported by NIH Grant DK068097 and funds provided by Rhode Island Hospital, the Deutsche Forschungsgemeinschaft grant (DFG) grant GE1193/1-1 and NIH COBRE Award (RR-P20 RR17695)
Peer reviewers: Dr. Bart Rik De Geest, Center for Molecular and Vascular Biology, Katholieke Universiteit Leuven, Campus Gasthuisberg, Herestraat 49, Leuven 3000, Belgium; Michael Kremer, MD, Skipper Bowles Center for Alcohol Studies, CB# 7178, 3011 Thurston-Bowles Building, University of North Carolina, Chapel Hill, NC 27599, United States
S- Editor Cheng JX L- Editor Kerr C E- Editor Zheng XM
References
- 1.Jaeschke H, Gores GJ, Cederbaum AI, Hinson JA, Pessayre D, Lemasters JJ. Mechanisms of hepatotoxicity. Toxicol Sci. 2002;65:166–176. doi: 10.1093/toxsci/65.2.166. [DOI] [PubMed] [Google Scholar]
- 2.Munthe-Kaas AC, Berg T, Seglen PO, Seljelid R. Mass isolation and culture of rat kupffer cells. J Exp Med. 1975;141:1–10. doi: 10.1084/jem.141.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knook DL, Sleyster EC. Separation of Kupffer and endothelial cells of the rat liver by centrifugal elutriation. Exp Cell Res. 1976;99:444–449. doi: 10.1016/0014-4827(76)90605-4. [DOI] [PubMed] [Google Scholar]
- 4.Valatas V, Xidakis C, Roumpaki H, Kolios G, Kouroumalis EA. Isolation of rat Kupffer cells: a combined methodology for highly purified primary cultures. Cell Biol Int. 2003;27:67–73. doi: 10.1016/s1065-6995(02)00249-4. [DOI] [PubMed] [Google Scholar]
- 5.Smedsrød B, Pertoft H, Eggertsen G, Sundström C. Functional and morphological characterization of cultures of Kupffer cells and liver endothelial cells prepared by means of density separation in Percoll, and selective substrate adherence. Cell Tissue Res. 1985;241:639–649. doi: 10.1007/BF00214586. [DOI] [PubMed] [Google Scholar]
- 6.Gehring S, Dickson EM, San Martin ME, van Rooijen N, Papa EF, Harty MW, Tracy TF Jr, Gregory SH. Kupffer cells abrogate cholestatic liver injury in mice. Gastroenterology. 2006;130:810–822. doi: 10.1053/j.gastro.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 7.Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta LA. Laser capture microdissection: molecular analysis of tissue. Science. 1997;278:1481,1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 8.Simone NL, Bonner RF, Gillespie JW, Emmert-Buck MR, Liotta LA. Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet. 1998;14:272–276. doi: 10.1016/s0168-9525(98)01489-9. [DOI] [PubMed] [Google Scholar]
- 9.Salvidio E, Crosby WH. Thrombocytopenia after intravenous injection of India ink. J Lab Clin Med. 1960;56:711–716. [PubMed] [Google Scholar]
- 10.Gregory SH, Wing EJ. Neutrophil-Kupffer cell interaction: a critical component of host defenses to systemic bacterial infections. J Leukoc Biol. 2002;72:239–248. [PubMed] [Google Scholar]
- 11.Knolle PA, Gerken G. Local control of the immune response in the liver. Immunol Rev. 2000;174:21–34. doi: 10.1034/j.1600-0528.2002.017408.x. [DOI] [PubMed] [Google Scholar]
- 12.Parker GA, Picut CA. Liver immunobiology. Toxicol Pathol. 2005;33:52–62. doi: 10.1080/01926230590522365. [DOI] [PubMed] [Google Scholar]
- 13.Khandoga A, Stampfl A, Takenaka S, Schulz H, Radykewicz R, Kreyling W, Krombach F. Ultrafine particles exert prothrombotic but not inflammatory effects on the hepatic microcirculation in healthy mice in vivo. Circulation. 2004;109:1320–1325. doi: 10.1161/01.CIR.0000118524.62298.E8. [DOI] [PubMed] [Google Scholar]
- 14.Colantuoni C, Henry G, Zeger S, Pevsner J. SNOMAD (Standardization and NOrmalization of MicroArray Data): web-accessible gene expression data analysis. Bioinformatics. 2002;18:1540–1541. doi: 10.1093/bioinformatics/18.11.1540. [DOI] [PubMed] [Google Scholar]
- 15.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 17.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Widmann JJ, Cotran RS, Fahimi HD. Mononuclear phagocytes (Kupffer cells) and endothelial cells. Identification of two functional cell types in rat liver sinusoids by endogenous peroxidase activity. J Cell Biol. 1972;52:159–170. doi: 10.1083/jcb.52.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knolle P, Schlaak J, Uhrig A, Kempf P, Meyer zum Büschenfelde KH, Gerken G. Human Kupffer cells secrete IL-10 in response to lipopolysaccharide (LPS) challenge. J Hepatol. 1995;22:226–229. doi: 10.1016/0168-8278(95)80433-1. [DOI] [PubMed] [Google Scholar]
- 20.Crispe IN, Giannandrea M, Klein I, John B, Sampson B, Wuensch S. Cellular and molecular mechanisms of liver tolerance. Immunol Rev. 2006;213:101–118. doi: 10.1111/j.1600-065X.2006.00435.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Owens JD, Shih JH, Li MC, Bonner RF, Mushinski JF. Histological staining methods preparatory to laser capture microdissection significantly affect the integrity of the cellular RNA. BMC Genomics. 2006;7:97. doi: 10.1186/1471-2164-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaner WS, Hendriks HF, Brouwer A, de Leeuw AM, Knook DL, Goodman DS. Retinoids, retinoid-binding proteins, and retinyl palmitate hydrolase distributions in different types of rat liver cells. J Lipid Res. 1985;26:1241–1251. [PubMed] [Google Scholar]
- 23.Amacher DE, Adler R, Herath A, Townsend RR. Use of proteomic methods to identify serum biomarkers associated with rat liver toxicity or hypertrophy. Clin Chem. 2005;51:1796–1803. doi: 10.1373/clinchem.2005.049908. [DOI] [PubMed] [Google Scholar]
- 24.Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology. 2006;43:S54–S62. doi: 10.1002/hep.21060. [DOI] [PubMed] [Google Scholar]
- 25.Wheeler MD, Smutney OM, Check JF, Rusyn I, Schulte-Hermann R, Thurman RG. Impaired Ras membrane association and activation in PPARalpha knockout mice after partial hepatectomy. Am J Physiol Gastrointest Liver Physiol. 2003;284:G302–G312. doi: 10.1152/ajpgi.00175.2002. [DOI] [PubMed] [Google Scholar]
- 26.Dimitroulakos J, Marhin WH, Tokunaga J, Irish J, Gullane P, Penn LZ, Kamel-Reid S. Microarray and biochemical analysis of lovastatin-induced apoptosis of squamous cell carcinomas. Neoplasia. 2002;4:337–346. doi: 10.1038/sj.neo.7900247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daveau M, Jean L, Soury E, Olivier E, Masson S, Lyoumi S, Chan P, Hiron M, Lebreton JP, Husson A, et al. Hepatic and extra-hepatic transcription of inter-alpha-inhibitor family genes under normal or acute inflammatory conditions in rat. Arch Biochem Biophys. 1998;350:315–323. doi: 10.1006/abbi.1997.0515. [DOI] [PubMed] [Google Scholar]
- 28.Chan P, Risler JL, Raguenez G, Salier JP. The three heavy-chain precursors for the inter-alpha-inhibitor family in mouse: new members of the multicopper oxidase protein group with differential transcription in liver and brain. Biochem J. 1995;306(Pt 2):505–512. doi: 10.1042/bj3060505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida E, Sumi H, Tsushima H, Maruyama M, Mihara H. Distribution and localization of inter-alpha-trypsin inhibitor and its active component acid-stable proteinase inhibitor: comparative immunohistochemical study. Inflammation. 1991;15:71–79. doi: 10.1007/BF00917911. [DOI] [PubMed] [Google Scholar]
- 30.Gujral JS, Farhood A, Bajt ML, Jaeschke H. Neutrophils aggravate acute liver injury during obstructive cholestasis in bile duct-ligated mice. Hepatology. 2003;38:355–363. doi: 10.1053/jhep.2003.50341. [DOI] [PubMed] [Google Scholar]
- 31.Devireddy LR, Teodoro JG, Richard FA, Green MR. Induction of apoptosis by a secreted lipocalin that is transcriptionally regulated by IL-3 deprivation. Science. 2001;293:829–834. doi: 10.1126/science.1061075. [DOI] [PubMed] [Google Scholar]
- 32.Tong Z, Wu X, Kehrer JP. Increased expression of the lipocalin 24p3 as an apoptotic mechanism for MK886. Biochem J. 2003;372:203–210. doi: 10.1042/BJ20021696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miharada K, Hiroyama T, Sudo K, Nagasawa T, Nakamura Y. Lipocalin 2 functions as a negative regulator of red blood cell production in an autocrine fashion. FASEB J. 2005;19:1881–1883. doi: 10.1096/fj.05-3809fje. [DOI] [PubMed] [Google Scholar]
- 34.Miharada K, Hiroyama T, Sudo K, Nagasawa T, Nakamura Y. Efficient enucleation of erythroblasts differentiated in vitro from hematopoietic stem and progenitor cells. Nat Biotechnol. 2006;24:1255–1256. doi: 10.1038/nbt1245. [DOI] [PubMed] [Google Scholar]
- 35.Liu M, Prisco M, Drakas R, Searles D, Baserga R. 24p3 in differentiation of myeloid cells. J Cell Physiol. 2005;205:302–309. doi: 10.1002/jcp.20400. [DOI] [PubMed] [Google Scholar]
- 36.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 37.Sunil VR, Patel KJ, Nilsen-Hamilton M, Heck DE, Laskin JD, Laskin DL. Acute endotoxemia is associated with upregulation of lipocalin 24p3/Lcn2 in lung and liver. Exp Mol Pathol. 2007;83:177–187. doi: 10.1016/j.yexmp.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Lam KS, Kraegen EW, Sweeney G, Zhang J, Tso AW, Chow WS, Wat NM, Xu JY, Hoo RL, et al. Lipocalin-2 is an inflammatory marker closely associated with obesity, insulin resistance, and hyperglycemia in humans. Clin Chem. 2007;53:34–41. doi: 10.1373/clinchem.2006.075614. [DOI] [PubMed] [Google Scholar]
- 39.Meheus LA, Fransen LM, Raymackers JG, Blockx HA, Van Beeumen JJ, Van Bun SM, Van de Voorde A. Identification by microsequencing of lipopolysaccharide-induced proteins secreted by mouse macrophages. J Immunol. 1993;151:1535–1547. [PubMed] [Google Scholar]
- 40.Chakravarti S, Wu F, Vij N, Roberts L, Joyce S. Microarray studies reveal macrophage-like function of stromal keratocytes in the cornea. Invest Ophthalmol Vis Sci. 2004;45:3475–3484. doi: 10.1167/iovs.04-0343. [DOI] [PubMed] [Google Scholar]
- 41.Hauses M, Tönjes RR, Grez M. The transcription factor Sp1 regulates the myeloid-specific expression of the human hematopoietic cell kinase (HCK) gene through binding to two adjacent GC boxes within the HCK promoter-proximal region. J Biol Chem. 1998;273:31844–31852. doi: 10.1074/jbc.273.48.31844. [DOI] [PubMed] [Google Scholar]
- 42.Podar K, Mostoslavsky G, Sattler M, Tai YT, Hayashi T, Catley LP, Hideshima T, Mulligan RC, Chauhan D, Anderson KC. Critical role for hematopoietic cell kinase (Hck)-mediated phosphorylation of Gab1 and Gab2 docking proteins in interleukin 6-induced proliferation and survival of multiple myeloma cells. J Biol Chem. 2004;279:21658–21665. doi: 10.1074/jbc.M305783200. [DOI] [PubMed] [Google Scholar]
- 43.Schaeffer M, Schneiderbauer M, Weidler S, Tavares R, Warmuth M, de Vos G, Hallek M. Signaling through a novel domain of gp130 mediates cell proliferation and activation of Hck and Erk kinases. Mol Cell Biol. 2001;21:8068–8081. doi: 10.1128/MCB.21.23.8068-8081.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown MT, Cooper JA. Regulation, substrates and functions of src. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 45.Kubota K, Kim JY, Sawada A, Tokimasa S, Fujisaki H, Matsuda-Hashii Y, Ozono K, Hara J. LRRC8 involved in B cell development belongs to a novel family of leucine-rich repeat proteins. FEBS Lett. 2004;564:147–152. doi: 10.1016/S0014-5793(04)00332-1. [DOI] [PubMed] [Google Scholar]
- 46.Carballo E, Pitterle DM, Stumpo DJ, Sperling RT, Blackshear PJ. Phagocytic and macropinocytic activity in MARCKS-deficient macrophages and fibroblasts. Am J Physiol. 1999;277:C163–C173. doi: 10.1152/ajpcell.1999.277.1.C163. [DOI] [PubMed] [Google Scholar]
- 47.Hartwig JH, Thelen M, Rosen A, Janmey PA, Nairn AC, Aderem A. MARCKS is an actin filament crosslinking protein regulated by protein kinase C and calcium-calmodulin. Nature. 1992;356:618–622. doi: 10.1038/356618a0. [DOI] [PubMed] [Google Scholar]
- 48.Edelstein LC, Collins T. The SCAN domain family of zinc finger transcription factors. Gene. 2005;359:1–17. doi: 10.1016/j.gene.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 49.Sander TL, Stringer KF, Maki JL, Szauter P, Stone JR, Collins T. The SCAN domain defines a large family of zinc finger transcription factors. Gene. 2003;310:29–38. doi: 10.1016/s0378-1119(03)00509-2. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto J, Ikeda Y, Iguchi H, Fujino T, Tanaka T, Asaba H, Iwasaki S, Ioka RX, Kaneko IW, Magoori K, et al. A Kruppel-like factor KLF15 contributes fasting-induced transcriptional activation of mitochondrial acetyl-CoA synthetase gene AceCS2. J Biol Chem. 2004;279:16954–16962. doi: 10.1074/jbc.M312079200. [DOI] [PubMed] [Google Scholar]
- 51.Fujino T, Kondo J, Ishikawa M, Morikawa K, Yamamoto TT. Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J Biol Chem. 2001;276:11420–11426. doi: 10.1074/jbc.M008782200. [DOI] [PubMed] [Google Scholar]
- 52.Krusche CA, Kroll T, Beier HM, Classen-Linke I. Expression of leucine-rich repeat-containing G-protein-coupled receptors in the human cyclic endometrium. Fertil Steril. 2007;87:1428–1437. doi: 10.1016/j.fertnstert.2006.11.061. [DOI] [PubMed] [Google Scholar]
- 53.Kopitar-Jerala N, Turk B. Cleavage of the myristoylated alanine-rich C kinase substrate (MARCKS) by cysteine cathepsins in cells and tissues of stefin B-deficient mice. Biol Chem. 2007;388:847–852. doi: 10.1515/BC.2007.092. [DOI] [PubMed] [Google Scholar]
- 54.Stumpo DJ, Bock CB, Tuttle JS, Blackshear PJ. MARCKS deficiency in mice leads to abnormal brain development and perinatal death. Proc Natl Acad Sci USA. 1995;92:944–948. doi: 10.1073/pnas.92.4.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arbuzova A, Schmitz AA, Vergères G. Cross-talk unfolded: MARCKS proteins. Biochem J. 2002;362:1–12. doi: 10.1042/0264-6021:3620001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao C, Guo H, Mi Z, Wai PY, Kuo PC. Transcriptional regulatory functions of heterogeneous nuclear ribonucleoprotein-U and -A/B in endotoxin-mediated macrophage expression of osteopontin. J Immunol. 2005;175:523–530. doi: 10.4049/jimmunol.175.1.523. [DOI] [PubMed] [Google Scholar]
- 57.Vávrová J, Janovská S, Rezácová M, Hernychová L, Tichá Z, Vokurková D, Záskodová D, Lukásová E. Proteomic analysis of MOLT-4 cells treated by valproic acid. Mol Cell Biochem. 2007;303:53–61. doi: 10.1007/s11010-007-9455-0. [DOI] [PubMed] [Google Scholar]