CONSPECTUS
Targeting the minor groove of DNA through binding to a small molecule has long been considered an important molecular-recognition strategy in biology. A wide range of synthetic heterocyclic molecules bind non-covalently in the minor groove of the double helix and are also effective against a number of human and animal diseases. A classic structural concept, the isohelicity principle, has guided much of this work: such heterocyclic molecules require a shape that complements the convex surface of the minor groove. Researchers have used this principle to design molecules that can read DNA sequences. This principle also predicts that molecules that lack the complementary shape requirement would only bind weakly to DNA. Recently, however, researchers have unexpectedly found that some essentially linear compounds, which do not have this feature, can have high DNA affinity.
In this Account, we discuss an alternative recognition concept based on these new findings. We demonstrate that highly structured water molecules can play a key role in mediating between the ligand and DNA minor groove without loss of binding affinity. Combined structural and thermodynamic approaches to understanding the behavior of these molecules have shown that there are different categories of bound water in their DNA complexes. For example, application of this water-bridging concept to the phenylamidine platform has resulted in the discovery of molecules with high levels of biological activity and low non-specific toxicity. Some of these molecules are now in advanced clinical trials.
Introduction
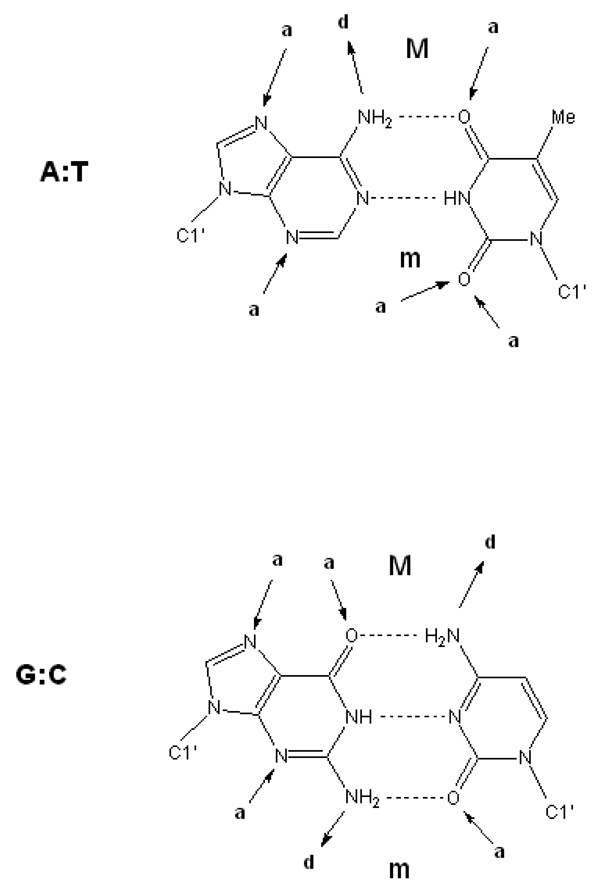
Designed synthetic dicationic compounds that specifically recognize the DNA minor groove are providing clinically useful therapeutic agents against a number of diseases and serving as well-defined model systems giving insight into the structural and thermodynamic features that characterize DNA molecular recognition.1–4 The interactions of dicationic organic molecules with the minor groove are characterized by both general and specific factors. The former include electrostatics, van der Waals contacts, hydrophobicity and groove hydration. Specific factors in particular sequences include H-bonding groups, steric differences and water/ion arrangements. The minor groove contains primarily hydrogen-bond acceptor groups, the purine N3 and pyrimidine O2 at the floor of the groove as well as the deoxyribose O4' at the groove walls (Figure 1).3,5–7 The 2-NH2 of G makes the third H-bond in G•C base pairs and can prevent deep penetration into the minor groove. Structured solvent and increased electrostatic potential in A/T sequences also distinguish A/T from G/C minor groove binding sites.3,5–10 Thus, for compounds that bind in the minor groove, shape that complements the groove curvature (“isohelicity”) and functional group placement, charge and H-bonding groups have been identified as critical components for binding affinity and recognition.2,3,9–16
Figure 1.
Schematic of the Watson-Crick base pairs showing hydrogen-bond donors and acceptors involving the bases.
Recent studies on a range of heterocyclic dications, particularly diamidines and analogs, have significantly extended and modified our understanding of how such compounds can interact with the minor groove, and how ligand curvature, dihedral twist, charge, and in particular interactions with water molecules in the DNA complex can lead to highly specific and very strong interactions with the groove. Crystallographic and thermodynamic studies have provided complementary insights into the details of minor-groove binding. Diamidines are currently the primary class of reversible DNA minor groove binding compounds with therapeutic utility, and we survey here their DNA interactions, comparing and contrasting structural features for classical recognition of the minor groove with new models for non-classical interactions. In particular, the fundamental molecular recognition features among compound shape, water, and base edges will be described and correlated with DNA-ligand binding thermodynamics.
General features of DNA–minor groove complexes at the -GAATTC- site
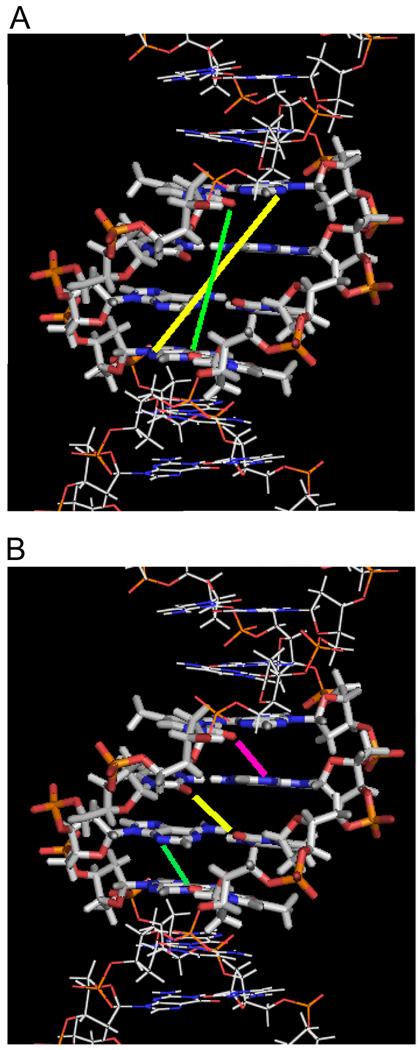
The first B-form DNA crystal structure to be solved was of the DNA duplex, d(CGCGAATTCGCG)2, and has subsequently been studied in detail by both solution and crystallographic studies.3,6,7,17–22 It contains a core of four A–T base pairs, and has been frequently used as a platform for binding of specific DNA minor groove interactions. Figure 2 shows a solution structure of this duplex (“A2T2”) and some critical distances between H-bond groups on base pair edges at the floor of the groove. The right-hand twist of B-DNA creates unequal inter-strand acceptor–acceptor distances for a given base pair separation (Figure 2A). For the AATT site, for example, the O2 of T8 to O2 of T20 is 9.1 Å, while the distance between N3 of the complementary bases A17 and A5 is 12.6 Å (the base numbering scheme on the duplex strands is given in Figure 3). These are linear distances so that in order to form direct H-bonds with the two DNA groups, a bound ligand must have a concave curvature to complement the convex shape subtended by successive base edges along the floor of the groove. The distance difference for H-bond acceptors is a critically important feature for minor groove binding. Shorter ligands that just cover the four base pairs in the AATT sequence will opt for H-bonding with the O2 of T8 and O2 of T20 while longer, crescent-shaped, ligands prefer the N3s of the complementary A17 and A5. These distances change with sequence and this can lead to specificity in recognition of different AT sequences with appropriately designed compounds.
Figure 2.
Interstrand distances of H-bond acceptor groups at the AATT site in d(CGCGAATTCGCG)2. (A) Two sets of H-bond acceptors are within a four-base pair length. The N3 of A5 to N3 of A17 distance (yellow) is longer than the O2 of T8 to O2 of T20 distance (green), and it is a through-base distance so that a bound compound must have some curvature in order to form H-bonds with the two Ns. (B) The shortest inter-strand acceptor-acceptor distances (3.7 Å) within the minor groove of -AATT- are illustrated. The three lines indicate O2 of T8 to N3 of A18 (magenta), O2 of T7 to O2 of T19 (yellow), and N3 of A6 to O2 of T20 (green). Compound -NH- groups (from amidine, amide, benzimidazole, or indole) can form bifurcated H-bonds with these three pairs of H-bond acceptors.
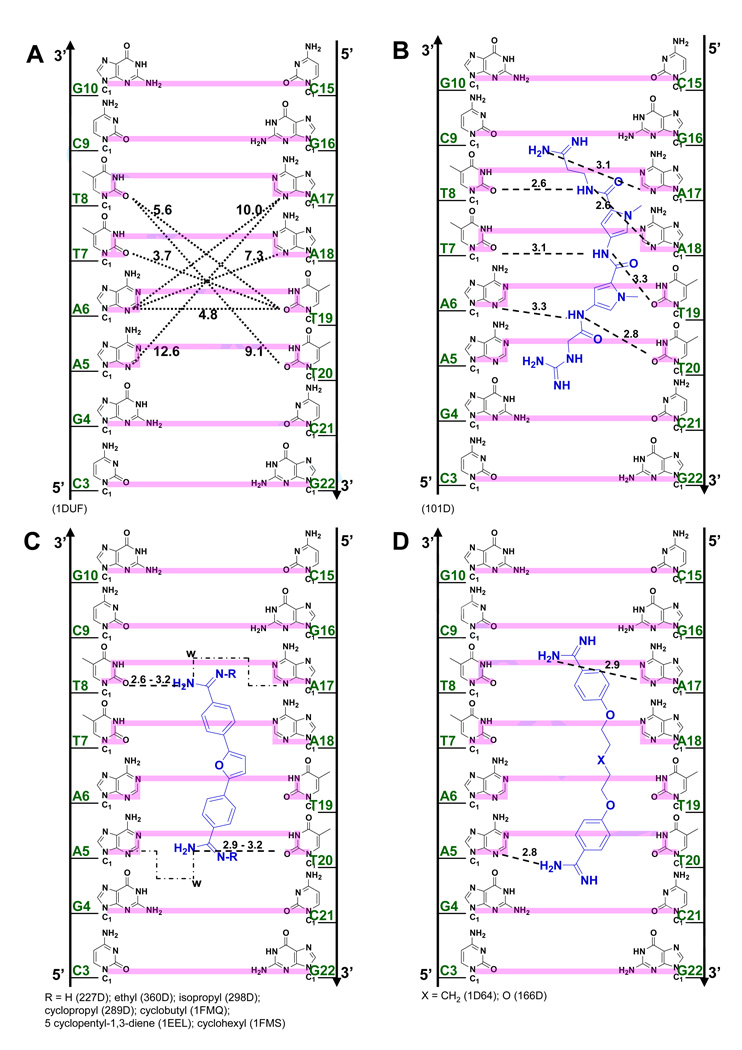
Figure 3.
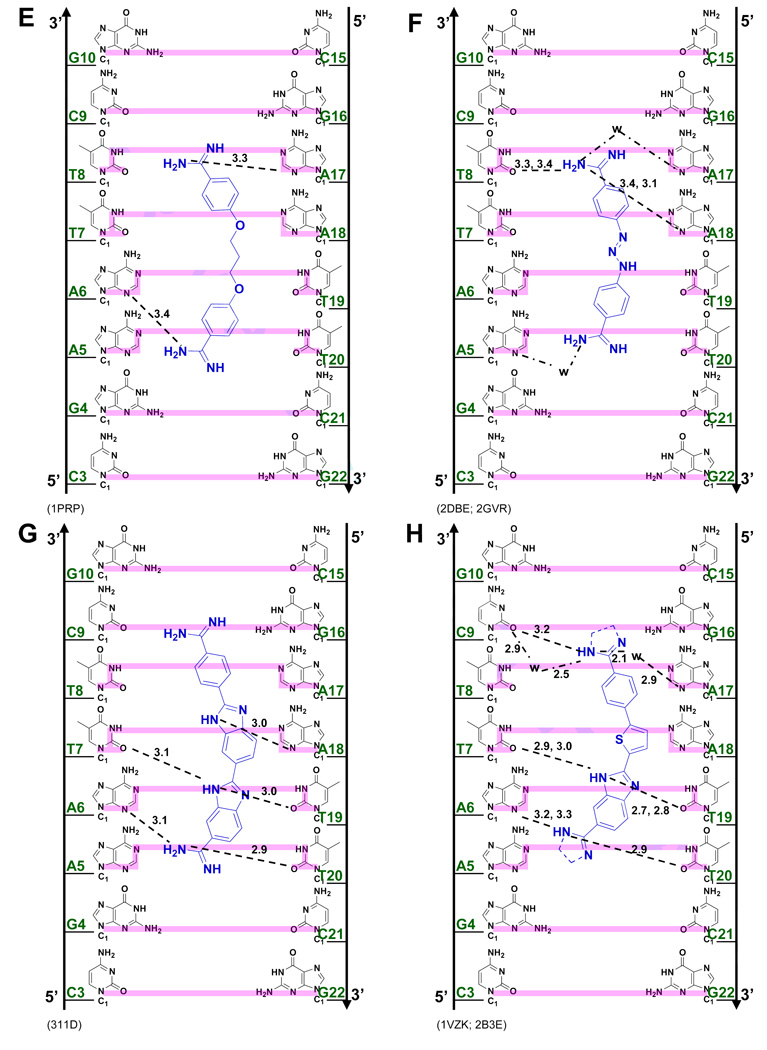
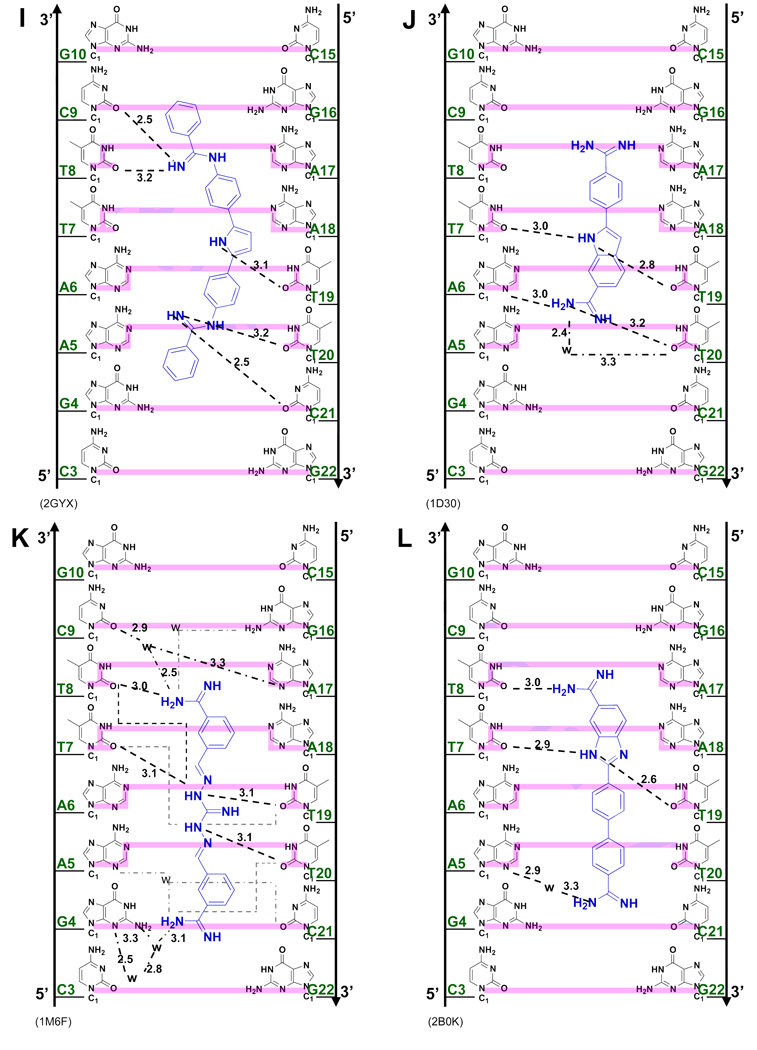
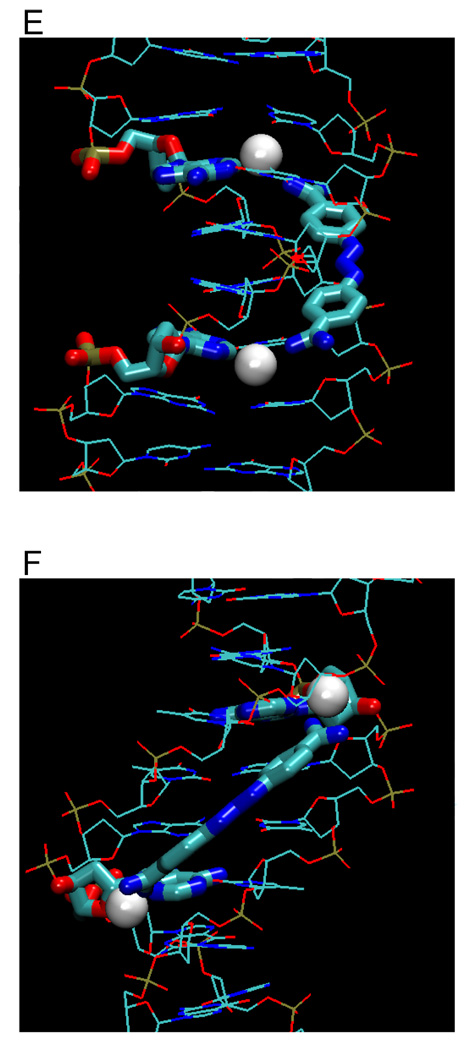
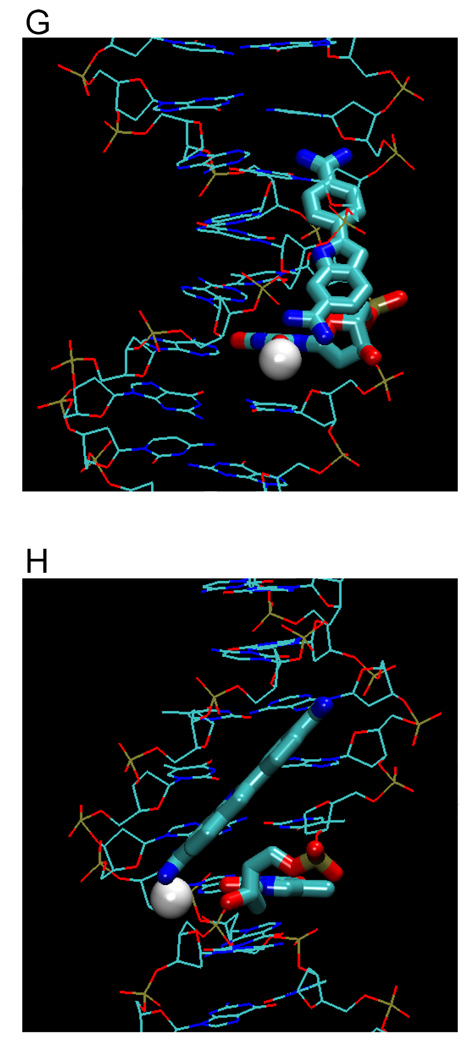
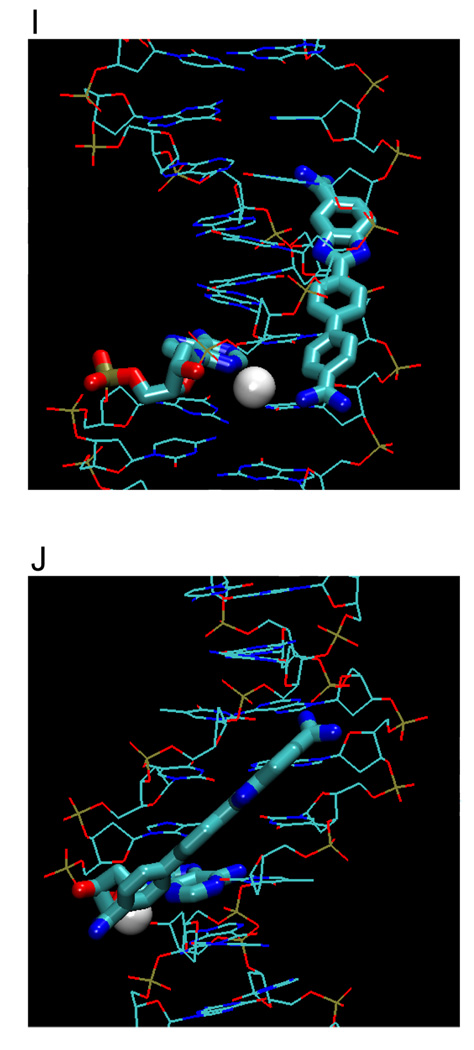
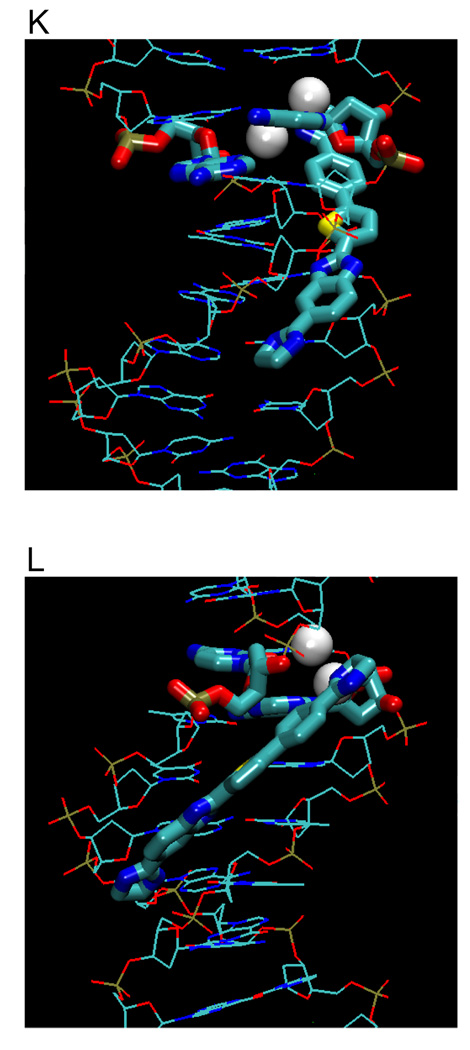
(A) Crystal structures (with PDB IDs) of the dodecamer duplex d(CGCGAATTCGCG)2 with some important inter-strand H-bond acceptor distances and with minor groove binding diamidines and analogs (B - L). The light magenta lines depict the base edges that form the floor of the groove at which specific interactions occur and the dashed lines indicate compound-DNA H-bonds. Water molecules that connect the bound compound to the minor groove floor are indicated with “w”. In (K), lighter gray lines indicate the interaction of another equivalent structure.
Figure 2B illustrates the three shortest (3.7 Å) inter-strand H-acceptor distances on adjacent bases (O2 of T8 to N3 of A18, O2 of T7 to O2 of T19, and N3 of A6 to O2 of T20) at the AATT site. These atoms frequently interact with ligand indole or benzimidazole -NH- groups to form bifurcated H-bonds with the central region of the AATT sequence (Figure 3). The netropsin-AATT structure, (Figure 3B), provided the basis for the classical minor groove recognition model that emphasizes appropriate ligand curvature to match the groove, ligand and DNA complementary H-bonding groups and displacement of water/ions from the -AATT- groove on complex formation.23,24 Based on the shape-complementarity concept, the classical model has been used to predict that ligands with incorrect curvature would bind poorly to DNA. The netropsin amide -NH- groups arranged in a crescent fashion, take advantage of the shortest inter-strand H-bond acceptor distances in the minor groove by forming an array of bifurcated H-bonds with N3 of A6 and O2 of T20, O2 of T7 and O2 of T19, and O2 of T8 and N3 of A18 (Figure 3B). In contrast to many diamidine compounds (discussed below), isothermal titration calorimetry (ITC) studies indicated that the interaction between netropsin and AATT is enthalpically driven with a free energy of −12 kcal/mole, presumably due in part to the large number of H-bonds formed in this complex.25
Figure 3 illustrates specific interactions of all synthetic dicationic amidine analogs that have been crystallized with AATT. For assistance in correlating the compound names, structures, and PDB identifications, a reference list is shown in Table 1. The diamidines and analogs in Figures 3C–I closely match the curvature and H-bonding groups of AT sequences in the DNA minor groove, an isohelical interaction, as well as complement the negative charge of the DNA phosphates. This is the "classical type" of minor groove complex that provides AT base pair sequence recognition. A different and unexpected type of complex, based on the classical model, however, is observed for the compounds bound to AATT in Figures 3J–L. Comparison of these two distinct complex classes shows that the requirements for minor groove interactions to occur can result in the shape of the compounds deviating significantly from an isohelical match to the groove.
Table 1.
| Ligand | Structure shown in Figure | PDB IDs |
|---|---|---|
| Netropsin | 3B | 101D |
| Furamidine & closely related | 3C | 227D, 360D, 298D, 289D, |
| ligands | 1FMQ, 1EEL, 1FMS | |
| Pentamidine & derivative | 3D | 1D64, 166D |
| Propamidine | 3E | 1PRP |
| Berenil | 3F | 2DBE, 2GVR |
| DB185 | 3G | 311D |
| DB818 & DB819 | 3H | 1VZK, 2B3E |
| DB884 | 3I | 2GYX |
| DAPI | 3J | 1D30 |
| CGP 40215A | 3K | 1M6F |
| DB921 | 3L | 2B0K |
Structures and thermodynamics of diamidine complexes with the A/T minor groove
In many of the DNA complexes in Figure 3 the amidine or modified-amidine cationic group is linked to the groove floor-base edge and/or groove wall-O4’ by bridging water molecules that flank the ends of the bound molecule. While interactions with the groove walls and phosphate groups can enhance overall complex stability, they generally have more limited effects on sequence specificity. We will therefore emphasize the interactions among the ligand, the base edges and bridging water molecules at the floor of the minor groove where sequence specificity is primarily defined.
Furamidine, DB75 (R=H in Figure 3C), is a therapeutically useful heterocyclic dication that has been crystallized as an AATT complex (PDB 227D). A neutral, orally available prodrug of the compound is currently in Phase III clinical trials against human African trypanosomiasis.26 The compound selectively targets repeated A/T sequences in the trypanosome mitochondrial kinetoplast DNA and has exhibited no significant toxicity to patients at therapeutic levels.1,2 The primary H-bond contacts from DB75 and derivatives to DNA are consistently between the amidines and O2s of T8 and T20.27–31 In addition, water molecules are observed to bridge the amidine -NH2 and N3 of A5 (PDB 1 EEL and 289D) or both N3 of A5 and N3 of A17 (298D, 360D, and 1FMQ). Under physiological conditions, DB75 and close analogs have a binding constant, Ka, with the AATT sequence in the range of 1 – 5×107 M−1 with a small ΔH and large T•ΔS for binding.31,32 Shape recognition and van der Waals interactions thus appear to play a larger role in specific recognition of -AATT- by these compounds than with netropsin. This set of diamidines (Figure 3C) thus provides a well-defined structural and thermodynamic benchmark for comparison with other minor groove binders. It should be noted that the thermodynamic values reported in this paper are at 25°C and since these complexes have negative heat capacity change values for binding, the ΔH and T•ΔS will become more negative or less positive, depending on the initial values.
Unlike the relatively rigid diphenylfuran core of DB75 and analogs (Figure 3C), pentamidine33 and derivatives34 (Figure 3D) have flexible linkers connecting two benzamidines. Pentamidine also binds to the four base pairs of AATT (PDB 1D64), but it utilizes the N3s of A5 and A17 as amidine H-bond acceptors rather than O2 of the complementary T20 and T8 as with furamidine. Since the separation of the two inner-facing amidine -NH2 groups in bound pentamidine (15.3 Å) is significantly longer than in DB75 (12.8 Å), pentamidine interacts with the longer inter-strand H-bond acceptor pair within AATT (H-bond distances shown in Figure 3D). The shorter diphenylfuran derivatives select the closer O2 of T8 to O2 of T20 for H-bonding (Figure 3C).
Propamidine (Figure 3E) is a shorter version of pentamidine but the amidine N-N distance (14.6 Å) in its AATT complex (PDB 1PRP) is longer than that of furamidine. While one of the amidines of propamidine forms a direct H-bond with N3 of A17 (as with pentamidine), the other amidine cannot reach the N3 of A5 of the opposite strand and it is closer to N3 of A6 (3.4 Å). The propamidine molecule is longer than furamidine but interacts with N3 of A17 and N3 of A6 because it is more acutely concave than furamidine. This curvature helps it wrap around the base edges for the N3 distances noted above. In spite of its relatively long H-bonds (to N3 of A6), propamidine binds more strongly than pentamidine,35 probably because of a smaller conformational entropy loss when the trimethylene linker fits into the minor groove relative to the pentamethylene of pentamidine. Both pentamidine and propamidine bind more weakly than furamidine and ITC studies indicate that their interactions are entropically driven.
Berenil (diminazene) (Figure 3F)36 resembles furamidine and is an important antitrypanosomal drug in animals. In its AATT complex berenil (PDB 2DBE, 2GVR) has an amidine N-N distance (13.3 Å) that is slightly longer than that of furamidine. However, it only binds across the -ATT- site with a weak (3.4 Å) H-bond from one amidine to O2 of T8. The other amidine interacts with N3 of A5 through a bridging water molecule in the groove. The longer propamidine also covers three base pairs (-ATT-) but no water molecule is required to link it to the floor of the groove. A crystal structure with a longer AAATTT site37 shows that berenil binds to the central -AATT- by forming H-bonds with inter-strand O2s of T in much the same way that DB75 binds to DNA. Berenil binds with a slightly lower affinity than furamidine (1×107 and 2×107 M−1, respectively)38 and with a small ΔH and large T•ΔS of binding. The furamidine, pentamidine and berenil group of compounds thus illustrate the flexibility of amidine-DNA H-bonding interactions that are possible for ligands of similar size within an expanded classical model of minor groove complex formation. The compounds, and to a lesser extent DNA, have sufficient conformational flexibility to allow these differences in AATT recognition to optimize the Gibbs energy of binding in each case.
Compounds with additional types of H-Bond interactions
DB185 (Figure 3G) is distinct from furamidine but similar in structure to Hoechst 33258, another well-established minor groove binder. DB185 binds to the minor groove of AATT much more strongly than furamidine or Hoechst 33258.39 It is one of a small number of DNA minor groove binders with a high affinity (Ka > 1011 M−1). The strong binding results in part from the H-bond network between the ligand and DNA, van der Waals interactions and extensive water release from the minor groove and ligand.39–41 A bifurcated H-bond is formed between the terminal benzimidazole -NH- and the inter-strand O2 of T7 and T19 as well as between the amidine attached to this benzimidazole and N3 of A6 and O2 of T20 (Figure 3G). Extensive water bridges are also observed at the amidine attached to benzimidazole with water molecules linked to O2 of C21 and O4’ of G22 on the backbone.
DB818 (amidine) and DB819 (imidazoline) have the furan of DB75 converted to a thiophene and a phenyl replaced by a benzimidazole (Figure 3H).42,43 The larger size of the sulfur atom (compared to oxygen in furan) and resulting bond angle changes become magnified at the molecular extremities in terms of overall ligand shape. The leads to a lower curvature and a superior fit within the minor groove with a binding affinity that is over 30 times greater for DB818 than for furamidine.43 This change is due to the thiophene substitution since the benzimidazole derivative of DB75 has similar binding affinity to DB75.44,45 With DB818 (Figure 3H, PDB 1VZK), there are direct amidine H-bonds to O2 of C9 and bifurcated H-bonds to O2 of T20 and N3 of A6. There are also two water molecules mediating the interactions between one of the imidazoline ends with O2 of C9 and N3 of A17.
DB884 was the first reversed-amidine to be crystallized with DNA (Figure 3I),46 In its AATT complex one rev-Am -NH- forms a bifurcated H-bond with intra-strand O2 of T8 and O2 of C9 while the other reversed-amidine interacts with the opposite strand through a bifurcated H-bond with intra-strand O2 of T20 and O2 of C21 (Figure 3I). An important feature in this complex that differs from furamidine is an H-bond between the pyrrole -NH- and O2 of T19. Although reversed-amidines generally bind more weakly than the corresponding amidines, DB884 is an exception, with a Ka similar to that of furamidine.46 Replacement of the pyrrole NH with a NCH3 or with O (furan) leads to a destabilization of the complex indicating the importance of the pyrrole-DNA H-bond.46 The extra H-bond contributes to a larger ΔH on binding of DB884 relative to DB75.46
The phenyl-indole diamidine, DAPI, has been used for many years as a DNA stain. Although DAPI is small, it binds to AATT with a higher affinity than DB75.25,31 The indole -NH- forms a bifurcated H bond with the inter-strand O2s of T7 and T19 (Figure 3J).47 The amidine attached to this indole forms a bifurcated H-bond with N3 of A6 and O2 of T20 while a flanking water molecule also bridges the amidine with O2 of T20. The other amidine is above the floor of the minor groove but no water molecule is observed in the relatively low resolution DAPI structure. As with many diamidine compounds, ITC shows that the binding enthalpy contributes less than half of the binding free energy.25
"Nonclassical" minor groove complexes
The minor groove ligands described above obey the requirement for isohelicity to complement the DNA minor groove. Recent results, however, have shown that the "classical model", which is described above and which originated when the database of structures was small, is only one way to obtain high affinity and specific DNA minor groove recognition. The AATT complex of CGP 40215A (Figure 3K) differs significantly from the classical model (Figures 3C–I)48 because of the linear shape of the compound. In contrast to pentamidine (of similar size), only one of the amidines of CGP 40215A can form direct H-bond contacts with AATT. While this would be expected to significantly reduce the binding affinity, CGP 40215A actually binds stronger than the curved pentamidine (40 - 50 times stronger) or furamidine (5 times stronger).
In the DNA complex (PDB 1M6F) one amidine of CGP 40215A forms H-bonds with O2 of T8 along with indirect H-bonds, through a bridging water molecule, with N3 of the complementary A17, and O2 of C9. An -NH- on the linker forms a bifurcated H-bond with the inter-strand O2 of T7 and T19, the shortest inter-strand H-bond acceptor distance in the minor groove (Figure 3A). The amidine group at the other end of CGP 40215A cannot form a direct H-bond with the base edges because it protrudes away from the groove floor, but instead forms an H-bond bridge with two water molecules to N2 and N3 of G4. A molecular dynamics simulation suggests that due to the symmetry of the ligand and the AATT DNA duplex, an equivalent structure exists with a similar pattern of H-bonds. This compound thus shows a distinct mode of groove binding in which water molecules can compensate for the lack of curvature of the ligand and can complete the crescent shape requirement for minor groove recognition and strong binding.48 Thermodynamic studies indicate that the binding enthalpy contributes slightly more than entropy to the binding free energy.49
To provide additional insight into and tests of the curvature requirements for minor groove binding, DB921 and other relatively linear diamidines have been designed and synthesized. DB921 is an adaptation of DAPI that lengthens the molecule without introducing additional curvature. In the DB921 complex (PDB 2B0K), the benzimidazole group is in the groove while the biphenyl-amidine projects to the top edge of the minor groove. The benzimidazole -NH- forms a bifurcated H-bond with the inter-strand O2s of T7 and T19 and the amidine on the benzimidazole forms an H-bond with the O2 of T8. This structure creates an optimum pocket for a direct bridging water molecule between the phenyl-amidine and N3 of A5 (Figure 3L),50 which forms a link of flexible orientation and H-bonding ability between the amidine and the N3 H-bond acceptor on the groove floor and is sandwiched between the base and the amidine. This water interaction differs from that of berenil in which a terminal water molecule is linked to an acceptor atom of the next base along the sequence. Surprisingly, the binding constant for DB921 is more than 25 times higher than that of furamidine and ITC results reveal that the interaction with AATT is entropically driven.50 This demonstrates that a concave shape is not a requirement for a ligand to have strong and specific interactions with the minor groove.
Water-mediated H-bonds in diamidine complexes
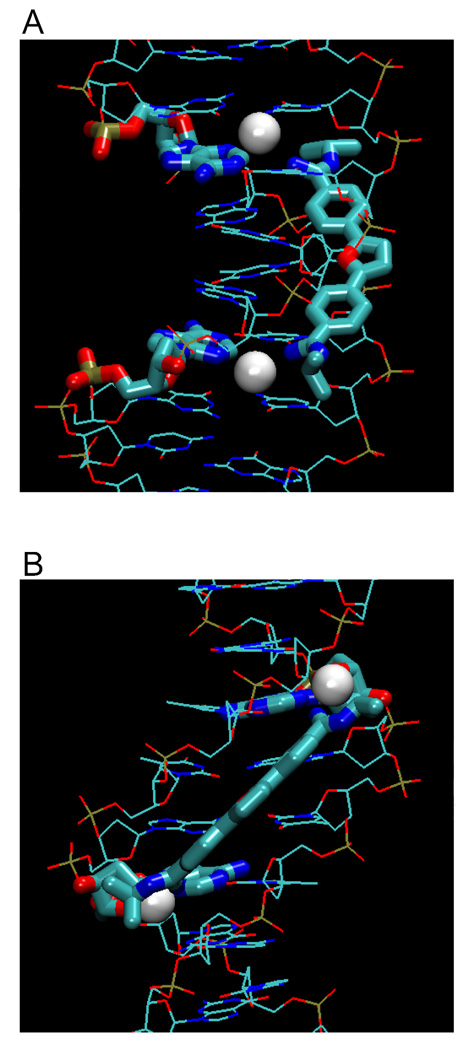
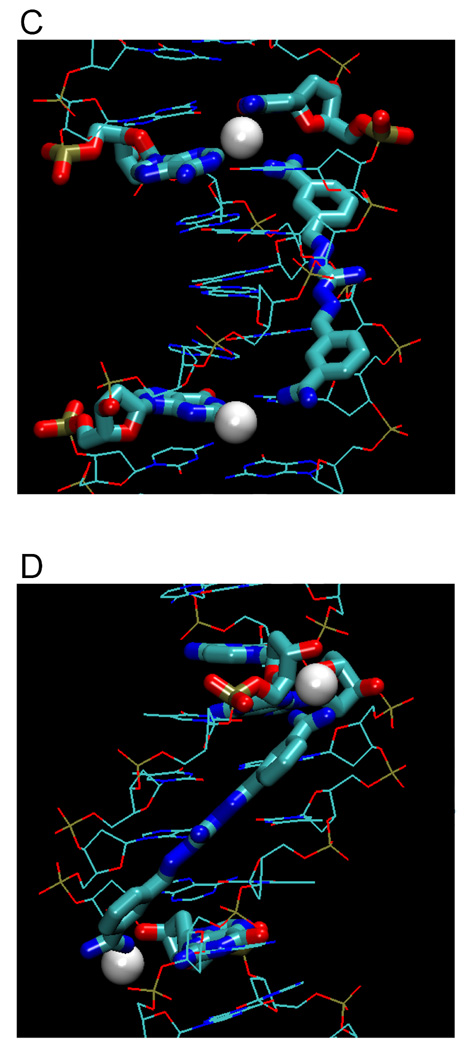
Figure 4 illustrates examples of hydrated amidine groups (from Figure 3) within the minor groove, showing different types of ligand-DNA water bridges. Only water molecules that are within 3.3 Å or less between ligand and base edge(s) are shown. Figures 4A, B show different views of a furamidine derivative (PDB 298D) that forms two water bridges with N3s of A17 and A5. Figures 4C and 4D illustrate water bridges of the linear CGP 40215A with −NH2 of G4 (lower) or with O2 of C9 and N2 of A17. Figures 4E, F show berenil with water bridges to N3 of A5 or A17. DAPI forms a water bridge at its indole end with O2 of T20 (Figures 4G, H). In a water-dependent interaction that is very different from the classical model, the phenylamidine of DB921 and the first A base of AATT sandwich a buried water molecule that is an essential and intrinsic part of the complex (Figures 4I, J). The imidazoline-thiophene derivative DB819 (Figures 4K, L) has two types of water interactions: one water molecule is between the imidazoline and N3 of A17 while the other reaches to the O2 of C9 on the opposite strand. Clearly, water is an important component of most diamidine minor groove complexes and in cases such as DB921, it is an essential part of the complex structure.
Figure 4.
Types of amidinium-water-base interactions. Bridging waters in furamidine (A, B), CGP40215A (C, D), berenil (E, F), DAPI (G, H), sandwiching water of DB921 (I, J), and a mix of both types with DB819 (K, L). Water molecules are shown as white spheres, ligands and interacting nucleotides as tubes, and other nucleotides as stick representations.
New and extended design features for biologically-active diamidines
Binding to sites, such as the A/T-rich kinetoplast of trypanosomes and leishmania or the A/T-rich nuclear DNA of the malaria parasite, provide excellent targets for biological activity with little host toxicity.2 This mirrors the "AT island" concept proposed for minor groove binding anticancer drugs and it may be possible that appropriately designed diamidines can also be developed as anticancer agents.51 For effective cell uptake the compound size should be kept small and synergistic biological effects that can take place at multiple A/T sequences, such as those in kinetoplast DNA, offer attractive biological targets. The inclusion of water interactions and ligand flexibility into the design of new ligands is a challenging problem, and additional compounds and DNA interactions studies will be required to establish the limits of this new approach to achieve satisfactory binding affinity, sequence selectivity, site recognition, and biological activity. Results with DB921 (Figures 4I, J) illustrate the utility of water as a flexible adapter for DNA complexes and provide optimism for this design approach.
More drastic differences in diamidine-DNA interactions are observed with DB293 and DB1242 which form cooperative stacked dimers to recognize more complex sequences with G–C base pairs than is possible for the compounds in Figure 3.44,52 A single molecule of DB1242, for example, can extend over 3–4 base pairs but by stacking of two molecules, a five base-pair sequence including four G–C base pairs can be targeted.52 Positively-charged amidine, reversed-amidine, or guanidine groups (Figure 3) are favorable for hydration, solubility, stacking with the minor groove, H-bonding and electrostatic interactions with the anionic field of the minor groove. Benzamidine motifs are not only important for DNA binding because of charge complementarity to the DNA and as H-bond donors but also for cellular uptake.53–55 Amidine derivatives, thus, continue to offer exciting opportunities for development of drugs that target DNA. Polyamides that target the minor groove have excellent sequence recognition capability9,10,12,14,56 and many useful applications in fundamental molecular recognition. Unlike the amidines, however, the polyamides have poor cell uptake and limited stability with no potential for oral drug administration. They have, therefore, not been used in any clinical application. Ongoing research with the new amidine derivatives, such as DB293 and DB1242, which recognize mixed DNA sequences, may provide a bridge between the therapeutic and sequence design areas.
Acknowledgements
Long term collaborative efforts on diamidine-DNA complexes with David W. Boykin and coworkers (Georgia State University, Atlanta, GA), Christian Bailly (Institut de Recherche Pierre Fabre, Toulouse, France), J. Ed Hall and Richard R. Tidwell (University of North Carolina, Chapel Hill, NC) have been both scientifically productive and personally enjoyable. Almost all of the diamidines in this review were synthesized by Professor Boykin and coworkers and this project would not have been possible without their efforts, suggestions and collaboration. Support from National Institutes of Health, the Bill and Melinda Gates Foundation, the Georgia Research Alliance (to WDW), and CRUK (to SN) is gratefully acknowledged.
Biographies
Binh Nguyen received his Ph.D. in biophysical chemistry (2002) from Georgia State University, Atlanta, GA. He conducted postdoctoral research at Georgia State University and is currently at Washington University in St. Louis.
Stephen Neidle has a Ph.D. in chemical crystallography from Imperial College, London (1970) and a DSc from the University of London. He is currently Professor of Chemical Biology at the School of Pharmacy, University of London.
W. David Wilson obtained his Ph.D. in biophysical chemistry in 1970 at Purdue University. He began work on peptide-DNA interactions as well as anticancer drug design with Dr. Edmond Gabbay at the University of Florida in Gainesville, Florida. He is currently Regents' Professor of Chemistry at Georgia State University.
Footnotes
All three authors have a strong interest in DNA structure and interactions, in the design of compounds to specifically recognize DNA and in the targeting of DNA sequences and structures for therapeutic uses.
REFERENCES
- 1.Tidwell RR, Boykin DW. In: DNA and RNA Binders: From Small Molecules to Drugs. Demeunynck M, Bailly C, Wilson WD, editors. Vol. 2. Weinheim: WILEY-VCH; 2003. pp. 414–460. [Google Scholar]
- 2.Wilson WD, Nguyen B, Tanious FA, Mathis A, Hall JE, Stephens CE, Boykin DW. Dications that target the DNA minor groove: compound design and preparation, DNA interactions, cellular distribution and biological activity. Curr. Med. Chem. Anticancer Agents. 2005;5:389–408. doi: 10.2174/1568011054222319. [DOI] [PubMed] [Google Scholar]
- 3.Neidle S. DNA minor-groove recognition by small molecules. Nat. Prod. Rep. 2001;18:291–309. doi: 10.1039/a705982e. [DOI] [PubMed] [Google Scholar]
- 4.Mathis AM, Bridges AS, Ismail MA, Kumar A, Francesconi I, Anbazhagan M, Hu Q, Tanious FA, Wenzler T, Saulter J, Wilson WD, Brun R, Boykin DW, Tidwell RR, Hall JE. Diphenyl furans and Aza Analogs: Effects of structural modification on in vitro activity, DNA binding, and accumulation and distribution in trypanosomes. Antimicrob. Agents Chemother. 2007;51:2801–2810. doi: 10.1128/AAC.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl. Acad. Sci. U S A. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wing R, Drew H, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980;287:755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- 7.Shui X, McFail-Isom L, Hu GG, Williams LD. The B-DNA dodecamer at high resolution reveals a spine of water on sodium. Biochemistry. 1998;37:8341–8355. doi: 10.1021/bi973073c. [DOI] [PubMed] [Google Scholar]
- 8.Lavery R, Pullman B, Zakrzewska K. Intrinsic electrostatic properties and base sequence effects in the structure of oligonucleotides. Biophys. Chem. 1982;15:343–351. doi: 10.1016/0301-4622(82)80017-3. [DOI] [PubMed] [Google Scholar]
- 9.Dervan PB, Burli RW. Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol. 1999;3:688–693. doi: 10.1016/s1367-5931(99)00027-7. [DOI] [PubMed] [Google Scholar]
- 10.Reddy BS, Sondhi SM, Lown JW. Synthetic DNA minor groove-binding drugs. Pharmacol. Ther. 1999;84:1–111. doi: 10.1016/s0163-7258(99)00021-2. [DOI] [PubMed] [Google Scholar]
- 11.Goodsell D, Dickerson RE. Isohelical analysis of DNA groove-binding drugs. J. Med. Chem. 1986;29:727–733. doi: 10.1021/jm00155a023. [DOI] [PubMed] [Google Scholar]
- 12.Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 13.Zasedatelev AS. Geometrical correlations useful for design of sequence-specific DNA narrow groove binding ligands. FEBS Lett. 1991;281:209–211. doi: 10.1016/0014-5793(91)80395-j. [DOI] [PubMed] [Google Scholar]
- 14.Wemmer DE. Designed sequence-specific minor groove ligands. Annu. Rev. Biophys. Biomol. Struct. 2000;29:439–461. doi: 10.1146/annurev.biophys.29.1.439. [DOI] [PubMed] [Google Scholar]
- 15.Nikitin AM, Gursky GV. New structural motifs isohelical to DNA. Dokl. Biochem. Biophys. 2003;390:139–142. doi: 10.1023/a:1024408206286. [DOI] [PubMed] [Google Scholar]
- 16.Nikitin AM, Rodin SA, Pis'menskii VF, Surovaya AN, Gursky GV. A new pseudopeptide motif for designing specific DNA-binding compounds capable of recognizing long DNA sequences. Dokl. Biochem. Biophys. 2002;384:167–171. doi: 10.1023/a:1016028415768. [DOI] [PubMed] [Google Scholar]
- 17.Drew HR, Dickerson RE. Structure of a B-DNA dodecamer. III. Geometry of hydration. J. Mol. Biol. 1981;151:535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- 18.Drew HR, Wing RM, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl. Acad. Sci. U S A. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tereshko V, Minasov G, Egli M. A "Hydrat-Ion" spine in a B-DNA minor groove. J. Am. Chem. Soc. 1999;121:3590–3595. [Google Scholar]
- 20.Tjandra N, Tate S-i, Ono A, Kainosho M, Bax A. The NMR structure of a DNA dodecamer in an aqueous dilute liquid crystalline phase. J. Am. Chem. Soc. 2000;122:6190–6200. [Google Scholar]
- 21.Woods KK, McFail-Ison L, Sines CC, Howerton SB, Stephens RK, Williams LD. Monovalent cations sequester within the A-tract minor groove of [d(CGCGAATTCGCG)]2. J. Am. Chem. Soc. 2000;122:1546–1547. [Google Scholar]
- 22.Johansson E, Parkinson G, Neidle S. A new crystal form for the dodecamer C-G-C-G-A-A-T-T-C-G-C-G: symmetry effects on sequence-dependent DNA structure. J. Mol. Biol. 2000;300:551–561. doi: 10.1006/jmbi.2000.3907. [DOI] [PubMed] [Google Scholar]
- 23.Goodsell DS, Kopka ML, Dickerson RE. Refinement of netropsin bound to DNA: bias and feedback in electron density map interpretation. Biochemistry. 1995;34:4983–4993. doi: 10.1021/bi00015a009. [DOI] [PubMed] [Google Scholar]
- 24.Patel DJ. Antibiotic-DNA interactions: intermolecular nuclear Overhauser effects in the netropsin-d(C-G-C-G-A-A-T-T-C-G-C-G) complex in solution. Proc. Natl. Acad. Sci. U S A. 1982;79:6424–6428. doi: 10.1073/pnas.79.21.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freyer MW, Buscaglia R, Nguyen B, Wilson WD, Lewis EA. Binding of netropsin and 4,6-diamidino-2-phenylindole to an A2T2 DNA hairpin: a comparison of biophysical techniques. Anal. Biochem. 2006;355:259–266. doi: 10.1016/j.ab.2006.04.049. [DOI] [PubMed] [Google Scholar]
- 26.Midgley I, Fitzpatrick K, Taylor LM, Houchen TL, Henderson SJ, Wright SJ, Cybulski ZR, John BA, McBurney A, Boykin DW, Trendler KL. Pharmacokinetics and metabolism of the prodrug DB289 (2,5-bis[4-(N-methoxyamidino)phenyl]furan monomaleate) in rat and monkey and its conversion to the antiprotozoal/antifungal drug DB75 (2,5-bis(4-guanylphenyl)furan dihydrochloride) Drug Metab. Dispos. 2007;35:955–967. doi: 10.1124/dmd.106.013391. [DOI] [PubMed] [Google Scholar]
- 27.Laughton CA, Tanious F, Nunn CM, Boykin DW, Wilson WD, Neidle S. A crystallographic and spectroscopic study of the complex between d(CGCGAATTCGCG)2 and 2,5-bis(4-guanylphenyl)furan, an analogue of berenil. Structural origins of enhanced DNA-binding affinity. Biochemistry. 1996;35:5655–5661. doi: 10.1021/bi952162r. [DOI] [PubMed] [Google Scholar]
- 28.Guerri A, Simpson IJ, Neidle S. Visualisation of extensive water ribbons and networks in a DNA minor- groove drug complex. Nucleic Acids Res. 1998;26:2873–2878. doi: 10.1093/nar/26.12.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trent JO, Clark GR, Kumar A, Wilson WD, Boykin DW, Hall JE, Tidwell RR, Blagburn BL, Neidle S. Targeting the minor groove of DNA: crystal structures of two complexes between furan derivatives of berenil and the DNA dodecamer d(CGCGAATTCGCG)2. J. Med. Chem. 1996;39:4554–4562. doi: 10.1021/jm9604484. [DOI] [PubMed] [Google Scholar]
- 30.Simpson IJ, Lee M, Kumar A, Boykin DW, Neidle S. DNA minor groove interactions and the biological activity of 2,5-bis. Bioorg. Med. Chem. Lett. 2000;10:2593–2597. doi: 10.1016/s0960-894x(00)00511-4. [DOI] [PubMed] [Google Scholar]
- 31.Mazur S, Tanious FA, Ding D, Kumar A, Boykin DW, Simpson IJ, Neidle S, Wilson WD. A thermodynamic and structural analysis of DNA minor-groove complex formation. J. Mol. Biol. 2000;300:321–337. doi: 10.1006/jmbi.2000.3869. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Kumar A, Boykin DW, Wilson WD. Sequence and length dependent thermodynamic differences in heterocyclic diamidine interactions at AT base pairs in the DNA minor groove. Biophys. Chem. 2007;131:1–14. doi: 10.1016/j.bpc.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards KJ, Jenkins TC, Neidle S. Crystal structure of a pentamidine-oligonucleotide complex: implications for DNA-binding properties. Biochemistry. 1992;31:7104–7109. doi: 10.1021/bi00146a011. [DOI] [PubMed] [Google Scholar]
- 34.Nunn CM, Jenkins TC, Neidle S. Crystal structure of gamma-oxapentamidine complexed with d(CGCGAATTCGCG)2. The effects of drug structural change on DNA minor-groove recognition. Eur. J. Biochem. 1994;226:953–961. doi: 10.1111/j.1432-1033.1994.00953.x. [DOI] [PubMed] [Google Scholar]
- 35.Cory M, Tidwell RR, Fairley TA. Structure and DNA binding activity of analogues of 1,5-bis(4- amidinophenoxy)pentane (pentamidine) J. Med. Chem. 1992;35:431–438. doi: 10.1021/jm00081a003. [DOI] [PubMed] [Google Scholar]
- 36.Brown DG, Sanderson MR, Skelly JV, Jenkins TC, Brown T, Garman E, Stuart DI, Neidle S. Crystal structure of a berenil-dodecanucleotide complex: the role of water in sequence-specific ligand binding. Embo J. 1990;9:1329–1334. doi: 10.1002/j.1460-2075.1990.tb08242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown DG, Sanderson MR, Garman E, Neidle S. Crystal structure of a berenil-d(CGCAAATTTGCG) complex. An example of drug-DNA recognition based on sequence-dependent structural features. J. Mol. Biol. 1992;226:481–490. doi: 10.1016/0022-2836(92)90962-j. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen B, Lee MP, Hamelberg D, Joubert A, Bailly C, Brun R, Neidle S, Wilson WD. Strong binding in the DNA minor groove by an aromatic diamidine with a shape that does not match the curvature of the groove. J. Am. Chem. Soc. 2002;124:13680–13681. doi: 10.1021/ja027953c. [DOI] [PubMed] [Google Scholar]
- 39.Tanious FA, Hamelberg D, Bailly C, Czarny A, Boykin DW, Wilson WD. DNA sequence dependent monomer-dimer binding modulation of asymmetric benzimidazole derivatives. J. Am. Chem. Soc. 2004;126:143–153. doi: 10.1021/ja030403+. [DOI] [PubMed] [Google Scholar]
- 40.Kopka ML, Fratini AV, Drew HR, Dickerson RE. Ordered water structure around a B-DNA dodecamer. A quantitative study. J. Mol. Biol. 1983;163:129–146. doi: 10.1016/0022-2836(83)90033-5. [DOI] [PubMed] [Google Scholar]
- 41.Clark GR, Boykin DW, Czarny A, Neidle S. Structure of a bis-amidinium derivative of hoechst 33258 complexed to dodecanucleotide d(CGCGAATTCGCG)2: the role of hydrogen bonding in minor groove drug-DNA recognition. Nucleic Acids Res. 1997;25:1510–1515. doi: 10.1093/nar/25.8.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell NH, Evans DA, Lee MP, Parkinson GN, Neidle S. Targeting the DNA minor groove with fused ring dicationic compounds: comparison of in silico screening and a high-resolution crystal structure. Bioorg. Med. Chem. Lett. 2006;16:15–19. doi: 10.1016/j.bmcl.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 43.Mallena S, Lee MP, Bailly C, Neidle S, Kumar A, Boykin DW, Wilson WD. Thiophene-based diamidine forms a "super" at binding minor groove agent. J. Am. Chem. Soc. 2004;126:13659–13669. doi: 10.1021/ja048175m. [DOI] [PubMed] [Google Scholar]
- 44.Wang L, Bailly C, Kumar A, Ding D, Bajic M, Boykin DW, Wilson WD. Specific molecular recognition of mixed nucleic acid sequences: an aromatic dication that binds in the DNA minor groove as a dimer. Proc. Natl. Acad. Sci. U S A. 2000;97:12–16. doi: 10.1073/pnas.97.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailly C, Tardy C, Wang L, Armitage B, Hopkins K, Kumar A, Schuster GB, Boykin DW, Wilson WD. Recognition of ATGA sequences by the unfused aromatic dication DB293 forming stacked dimers in the DNA minor groove. Biochemistry. 2001;40:9770–9779. doi: 10.1021/bi0108453. [DOI] [PubMed] [Google Scholar]
- 46.Munde M, Lee M, Neidle S, Arafa R, Boykin DW, Liu Y, Bailly C, Wilson WD. Induced fit conformational changes of a "reversed amidine" heterocycle: optimized interactions in a DNA minor groove complex. J. Am. Chem. Soc. 2007;129:5688–5698. doi: 10.1021/ja069003n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsen TA, Goodsell DS, Cascio D, Grzeskowiak K, Dickerson RE. The structure of DAPI bound to DNA. J. Biomol. Struct. Dyn. 1989;7:477–491. doi: 10.1080/07391102.1989.10508505. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen B, Hamelberg D, Bailly C, Colson P, Stanek J, Brun R, Neidle S, Wilson WD. Characterization of a novel DNA minor-groove complex. Biophys. J. 2004;86:1028–1041. doi: 10.1016/s0006-3495(04)74178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen B, Stanek J, Wilson WD. Binding-linked protonation of a DNA minor-groove agent. Biophys. J. 2006;90:1319–1328. doi: 10.1529/biophysj.105.071381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miao Y, Lee MP, Parkinson GN, Batista-Parra A, Ismail MA, Neidle S, Boykin DW, Wilson WD. Out-of-shape DNA minor groove binders: induced fit interactions of heterocyclic dications with the DNA minor groove. Biochemistry. 2005;44:14701–14708. doi: 10.1021/bi051791q. [DOI] [PubMed] [Google Scholar]
- 51.Woynarowski JM. Targeting critical regions in genomic DNA with AT-specific anticancer drugs. Biochim. Biophys. Acta. 2002;1587:300–308. doi: 10.1016/s0925-4439(02)00093-5. [DOI] [PubMed] [Google Scholar]
- 52.Munde M, Ismail MA, Arafa R, Peixoto P, Collar CJ, Liu Y, Hu L, David-Cordonnier MH, Lansiaux A, Bailly C, Boykin DW, Wilson WD. Design of DNA Minor Groove Binding Diamidines That Recognize GC Base Pair Sequences: A Dimeric-Hinge Interaction Motif. J. Am. Chem. Soc. 2007;129:13732–13743. doi: 10.1021/ja074560a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Koning HP, Jarvis SM. Adenosine transporters in bloodstream forms of Trypanosoma brucei brucei: substrate recognition motifs and affinity for trypanocidal drugs. Mol. Pharmacol. 1999;56:1162–1170. doi: 10.1124/mol.56.6.1162. [DOI] [PubMed] [Google Scholar]
- 54.Barrett MP, Fairlamb AH. The biochemical basis of arsenical-diamidine crossresistance in African trypanosomes. Parasitol. Today. 1999;15:136–140. doi: 10.1016/s0169-4758(99)01414-3. [DOI] [PubMed] [Google Scholar]
- 55.Maser P, Luscher A, Kaminsky R. Drug transport and drug resistance in African trypanosomes. Drug Resist. Updat. 2003;6:281–290. doi: 10.1016/j.drup.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 56.Lacy ER, Madsen EM, Lee M, Wilson WD. In: DNA and RNA Binders: From Small Molecules to Drugs. Demeunynck M, Bailly C, Wilson WD, editors. Vol. 2. Weinheim: WILEY-VCH; 2003. pp. 384–413. [Google Scholar]