Abstract
Photoreactivation, one of the first DNA repair pathways to evolve, is the direct reversal of premutagenic lesions caused by ultraviolet (UV) irradiation, catalyzed by photolyases in a light-dependent, single-enzyme reaction. It has been experimentally shown that photoreactivation prevents UV mutagenesis in a broad range of species. In the absence of photoreactivation, UV-induced photolesions are repaired by the more complex and much less efficient nucleotide excision repair pathway. Despite their obvious beneficial effects, several lineages, including placental mammals, lost photolyase genes during evolution. In this study, we ask why photolyase genes have been lost in those lineages and discuss the significance of these losses in the context of the evolution of the genomic mutation rates. We first perform an extensive phylogenomic analysis of the photolyase/cryptochrome family, to assess what species lack each kind of photolyase gene. Then, we estimate the ratio of nonsynonymous to synonymous substitution rates in several groups of photolyase genes, as a proxy of the strength of purifying natural selection, and we ask whether less evolutionarily constrained photolyase genes are more likely lost. We also review functional data and compare the efficiency of different kinds of photolyases. We find that eukaryotic photolyases are, on average, less evolutionarily constrained than eubacterial ones and that the strength of natural selection is correlated with the affinity of photolyases for their substrates. We propose that the loss of photolyase genes in eukaryotic species may be due to weak natural selection and may result in a deleterious increase of their genomic mutation rates. In contrast, the loss of photolyase genes in prokaryotes may not cause an increase in the mutation rate and be neutral in most cases.
Keywords: mutation rate evolution, photolyase, photoreactivation, DNA repair, mutator gene
Introduction
Ultraviolet (UV) light causes premutagenic lesions in DNA that can become mutations after replication if they are not repaired (Friedberg et al. 2006). These lesions can be repaired in eukaryotes Archaea, and Eubacteria by nucleotide excision repair (NER) systems which are neither specific for this kind of lesions nor the most efficient pathway to repair them. A specific UV-damaged DNA repair pathway also exists called photoreactivation. Photoreactivation is the direct reversal of UV-induced lesions catalyzed by a single enzyme called photolyase and using only visible light as an energy source (Kelner 1949 Sancar 1994 Sancar 2000).
There are three kinds of photolyases according to their substrate specificity. Cyclobutane pyrimidine dimers (CPDs) are repaired by CPD photolyases (Sancar et al. 1984 Sancar 1985) which are present in organisms of all kingdoms (Kanai et al. 1997) including some viruses (van Oers et al. 2004). The 6 4-Pyrimidine-pyrimidones or 6 4 photoproducts which are less frequently induced by UV irradiation are repaired by (6-4) photolyases only known to be present in eukaryotes (Todo et al. 1993). Finally a new class of photolyases has been described in eubacteria plants and animals with specificity for CPDs in single-stranded DNA (Brudler et al. 2003 Selby and Sancar 2006). Sequence comparisons distinguish these three kinds of photolyases and suggest further subdivision of CPD photolyases into two classes namely CPD class I and CPD class II (Kanai et al. 1997). In addition plants and animals possess independently derived photolyase paralogs called cryptochromes which are involved in circadian rhythm entrainment and in plants in the light-dependent regulation of developmental pathways (Cashmore et al. 1999). Despite the striking similarity between cryptochromes and photolyases, no photolyase activity of any kind has been observed in cryptochromes (Hsu et al. 1996, Öztürk et al. 2007).
Photolyases are composed of two structural domains an α/β domain and a helical domain which are joined by a linker region (Park et al. 1995 Tamada et al. 1997). Photolyases bind two cofactors or chromophores. The catalytic chromophore is FADH2 in all known cases and it is bound by the helical domain. As a second chromophore which works as a light-harvesting antenna most photolyases have either 8-hydroxy-5-deazaflavin (Eker1981 Eker1990) or 5 10-methenyltetrahydro-folic acid (Johnson1988 Li1991) although flavin mononucleotide (Ueda et al. 2005) and FAD (Fujihashi et al. 2007) have also been found as second chromophores.
Photoreactivation was one of the first DNA repair pathways to evolve (Eisen and Hanawalt 1999) and is thought to be the most effective one in the repair of UV-induced mutagenic lesions (Schul et al. 2002). Although the contribution of photolyases to replication fidelity in the wild is unknown it has been repeatedly shown that they prevent most of the mutations caused by experimental UV irradiation in cell cultures (Asahina et al. 1999 Otoshi et al. 2000 Tanaka et al. 2001 You et al. 2001). Despite the evident advantage that photolyase genes confer to organisms exposed to sunlight they have been lost several times during evolution. One dramatic consequence of the loss of photolyase genes in the placental mammals lineage (Kato et al. 1994) is the complete dependence on the NER pathway to repair UV-induced DNA damage, which makes people with inactivating mutations in genes of that pathway extremely sensitive to UV light (Friedberg et al. 2006). Stressing the deleterious character of the loss of photoreactivation in placental mammals it has been shown that transgenic mice expressing a photolyase gene from the marsupial Potorus tridactylis successfully avoid acute UV effects like sunburn hyperplasia and apoptosis (Schul et al. 2002). In addition the absence of photolyase orthologous genes in the human genome and their presence in the zebrafish genome (this study) is a very likely explanation of the observed higher capacity of zebrafish embryos to repair UV-damaged DNA relative to human cells (Sussman 2007).
The genomic mutation rates per generation vary widely among species (Drake et al. 1998 Lynch 2006 Baer et al. 2007) although the reasons for the variation are still debated (Baer et al. 2007 Lynch 2008). Variation in an organism's ability to repair DNA damage was proposed as one of the main factors to explain the variation in molecular evolutionary rates (Britten 1986). Later Eisen and Hanawalt (1999) showed that there is indeed extensive variation in the endowment of DNA repair pathways among species. Recently Marcobal et al. (2008) contributed compelling evidence that the hypermutability of the bacteria Oenococcus oeni is due to the lack of the mismatch repair genes MutS and MutL in its genome. Thus there is no doubt that the quality and composition of the DNA repair and replication machineries must affect the characteristic spontaneous mutation rate of a species. The absence of photolyase genes in some lineages may contribute to this variation.
Motivated by the likely contribution of the presence or absence of photolyase genes to the diversity of genomic mutation rates among species we aim to understand what factors affect the evolution of photolyases and why photolyase genes may be lost in some lineages. Taking advantage of the detailed knowledge of the photoreactivation mechanism we ask whether the efficiency of photolyases is correlated with the strength of purifying natural selection on their genes. There are three steps in the photoreactivation mechanisms that might be optimized by natural selection: binding to the damaged DNA transduction of energy from visible light and electron transfer from the chromophore FADH2 to the damaged DNA (Sancar 1994). We compare the available measures of photolyase efficiency with their estimated ratio of nonsynonymous to synonymous substitution rates (dN/dS) which is used as a proxy for the strength of purifying selection.
We also perform an extensive phylogenomic analysis of the photolyase/cryptochrome family to determine what species have lost photolyase genes. We reasoned that phylogenetic groups with less evolutionary constrained photolyases should exhibit a higher incidence of gene loss.
Photolyase genes can justifiably be viewed as potential mutator genes in the sense that they influence the spontaneous mutation rate in a wide range of organisms exposed to sunlight (see supplementary table 3 and references therein, Supplementary Material online). Thus we finally discuss our results in the context of the population-genetics factors affecting the evolution of mutator genes.
Materials and Methods
Phylogenomic Analysis
To assemble the most complete set of photolyase homologs possible BlastP searches (Altschul et al. 1990) were performed against the National Center for Biotechnology Information (NCBI) nonredundant database using representative sequences of the photolyase family as queries selected from the main groups identified by Kanai et al. (1997). To include photolyase homologs from unannotated genomic sequences this same set of queries was used in TblastN searches against all completely sequenced and assembled genomes available in NCBI. Additional TblastN searches were submitted to GeneDB (http://www.genedb.org) and the Broad Institute (http:// www.broad.mit.edu) to expand the taxonomic range. The FGENESH+ gene predictor (http://linux1.softberry.com/berry.phtml) was occasionally used to improve the translation of some unannotated eukaryotic genes. Results from different searches were merged and aligned using ClustalX 1.83 (Thompson et al. 1997) and the alignments manually edited using BioEdit (Hall 1999). The linker region between the two functional domains of the photolyase/cryptochrome family was unalignable and consequently excluded. The final alignment contained 882 sequences with a mean length of 405 amino acids. A neighbor-joining tree was obtained using MEGA 4 (Tamura et al. 2007) with 1000 bootstrap replicates. To generate trees with more time-consuming methods only the most divergent 250 sequences were selected using the HHFilter application of the Max-Planck Institute Bioinformatics Toolkit (Biegert et al. 2006) to test the monophyly of the main paralogous groups. For this representative subset of sequences maximum parsimony trees were obtained using PAUP* (Swofford 2002) with 100 bootstrap replicates and Bayesian inference was performed with MrBayes (Ronquist and Huelsenbeck 2003).
The presence or absence of each orthology group identified by phylogenetic inference is reported for all completely sequenced and assembled genomes used in the homology searches. To determine whether the presence–absence diversity in a genus was due to a recent loss or a recent acquisition by horizontal gene transfer, the phylogeny of the members of that genus with completely sequenced genomes was inferred by the neighbor-joining tree of their 16S ribosomal RNA (rRNA) subunits. Muscle was used for the alignments and MEGA 4 to build the trees. The trees were rooted with the closest relative of a different genus, and the gene-loss and gene-gain events were determined by the most parsimonius scenario. The maximum composite likelihood method was used to compute the distances.
Estimation of the Ratio of Nonsynonymous to Synonymous Substitution Rates
We were interested in comparing the strength of natural selection to preserve photolyase genes among taxonomic groups and among gene families. We reasoned that less evolutionarily constrained genes would be more easily lost by random genetic drift.
We used the ratio of nonsynonymous to synonymous substitutions dN/dS to measure the strength of purifying selection on sets of photolyase genes. Groups of very similar orthologous sequences usually belonging to the same genus were identified from the phylogenetic inference and analyzed separately. Their nucleotide sequences were retrieved and codons were aligned with ClustalW as implemented in BioEdit (Hall 1999). Maximum-likelihood estimates of dN/dS were calculated with PAML 4 (Yang 2007). The pairwise synonymous distances were checked within each set of sequences to make sure that the saturation of synonymous substitutions was not affecting the dN/dS estimates. When necessary sets were redefined to fulfill the condition that pairwise synonymous distances were not larger than 1.2 substitutions per site.
A constant dN/dS was assumed for all branches in order to get a representative estimate for the whole group. One hundred bootstrap replicates were performed to estimate the variance of each dN/dS estimate. To get a dN/dS value representative of a more diverse group of genes, the weighted mean of several dN/dS estimates was calculated, using the inverse of the sample variance as weights. To make comparisons between weighted means, the data were first log transformed, to make it normal. Then, the weighted standard deviations (SDs) were estimated following Bland and Kerry (1998), and the welch correction of the t-test was applied.
Results
Evolutionary History of Photolyase Genes
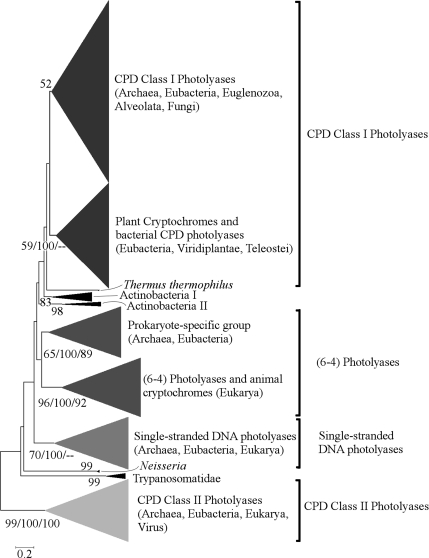
Our extensive phylogenetic analysis of the photolyase/cryptochrome family not only confirms most of the known features of the evolutionary history of photolyases and cryptochromes (Kanai et al. 1997) but also reveals some unexpected findings. Six main groups are modestly to highly supported by bootstrap values in the neighbor-joining and the maximum parsimony trees and by posterior probabilities in the Bayesian tree (from bottom to top in fig. 1): CPD class II photolyases; single-stranded DNA photolyases; (6-4) photolyases and animal cryptochromes; a previously undescribed prokaryotic group of unknown activity; a heterogeneous group including plant cryptochromes and some bacterial CPD photolyases; and a large and weakly supported group including all other bacterial archaeal, and eukaryotic CPD class I photolyases. The remaining sequences which cannot be assigned to any of these groups belong to five small divergent groups with narrow taxonomic ranges. These are shown in black in figure 1 and are labeled according to their species composition: Trypanosomatidae Neisseria Actinobacteria I Actinobacteria II and Thermus thermophilus.
FIG. 1.—
Unrooted neighbor-joining tree of the photolyase/cryptochrome family. Branches are labeled first with the their bootstrap support (percentage) and, when available, with: second, the bootstrap support of an equivalent branch in the maximum parsimony consensus tree; and third, with the posterior probability of an equivalent branch, according to the Bayesian analysis. Groups with the same tone are proposed to be orthologous, based on the their complementary taxonomic distribution and their proximity, despite the lack of bootstrap support in two cases (see Discussion).
The CPD class I photolyases and the heterogeneous group of plant cryptochromes and other bacterial CDP photolyases are relatively close to each other and have complementary taxonomic distributions: no species has genes of both kinds (supplementary table 1, Supplementary Material online) suggesting that they are orthologous. Two fish proteins of unknown function (XP_683212.3 from Danio rerio and CAF92156 from Tetraodon nigroviridis) are grouped together with plant cryptochromes with 96% bootstrap support in the neighbor-joining tree. Despite intense Blast searches no other animal sequences were found with such a close relationship to plant cryptochromes. Table 1 shows the main groups of organisms where each photolyase subfamily has been found. For the sake of simplicity and additional reasons argued in the discussion, plant cryptochromes and related sequences of fish and bacterial origin have been added to the CPD class I subfamily in tables 1 and 2.
Table 1.
Presence of Photolyase Genes in the Main Groups of Organisms
| Taxon | Class Ia | Class II | (6-4) Photolyases | Prokaryote Groupb | Single Strand |
| Bacteria | |||||
| Acidobacteria | + | ||||
| Actinobacteria | + | + | |||
| α-Proteobacteria | + | + | + | + | |
| Bacteroidetes | + | + | + | + | |
| β-Proteobacteria | + | + | + | ||
| Cyanobacteria | + | + | +c | + | + |
| Chlamydiae | + | ||||
| Chlorobi | + | ||||
| Chloroflexi | + | ||||
| δ-Proteobacteria | + | + | |||
| ε-Proteobacteria | + | ||||
| Firmicutes | + | + | |||
| γ-Proteobacteria | + | + | + | ||
| Planctomycetes | + | + | |||
| Spirochaeta | + | ||||
| Thermotogae | + | ||||
| Archaea | |||||
| Crenarcheota | + | ||||
| Euryarchaeota | + | + | + | + | |
| Eukarya | |||||
| Chromalveolata | + | + | + | + | |
| Excavatad | + | ? | ? | ? | |
| Opisthokonta | + | + | + | + | |
| Plantae | + | + | + | + | |
| Virus | |||||
| Poxviridae | + | ||||
| Baculoviridae | + |
Includes plant cryptochromes and related sequences. See text.
Putative orthologs of (6-4) photolyases.
A (6-4) photolyase-like gene is exceptionally found in the genome of the cyanobacteria Gloeobacter violaceous (glr1749), probably as a result of horizontal gene transfer from green algae.
In addition to CPD class I photolyases, some species from Trypanosomatidae have another photolyase homolog of uncertain origin.
Table 2.
Number of recent photoyase Gene Losses (L) and Gains (G) Inferred in Eubacterial Genera, Classified by Photolyase Subfamily
| Class I | Class II | Prokaryote | Single Strand | |||||
| L | G | L | G | L | G | L | G | |
| Azoarcus [0.015] | 1 | |||||||
| Bacilus [0.134] | 1 | |||||||
| Buchnera [0.128] | 1 | |||||||
| Corynebacterium [0.104] | 1 | |||||||
| Flavobacterium [0.056] | 1 | 1 | ||||||
| Francisella [0.002] | 1 | 1 | ||||||
| Frankia [0.026] | 1 | |||||||
| GGeobacter [0.086] | 1 | |||||||
| Mesorhizobium [0.034] | 1 | |||||||
| Mycobacterium [0.076] | 1 | |||||||
| Prochlorococus [0.053] | 1 | 2 | 1 | |||||
| Pseudoalteromonas [0.116] | 1 | |||||||
| Ralstonia [0.047] | 1 | |||||||
| Rhizobium [0.009] | 1 | |||||||
| Rhodobacter [0.008] | 1 | |||||||
| Rickettsia [0.040] | 1 | |||||||
| Shewanella [0.178] | 1 | 1 | ||||||
| Staphylococus [0.048] | 1 | |||||||
| Streptococus [0.030] | 1 | |||||||
| Synechococcus [0.280] | 1 | 1 | ||||||
| Vibrio [0.074] | 2 | 2 | ||||||
CPD class I subfamily includes traditional CPD class I photolyases and CPD photolyases grouped together with plant cryptochromes (see fig. 1 and text). “Prokaryote” refers to the prokaryote-specific group next to the eukaryotic (6-4) photolyases in figure 1. The total length of the 16S RNA tree of each genus is shown in brackets. About 100 more eubacterial genera, with one or more completely sequened and assembled genomes, did not show presence-absence variation of photolyase genes.
Photolyase Gene-Loss Events
Phylogenetic analysis of the photolyase/ cryptochrome family allowed us to identify recent events of gene loss. We identified `recent’ gene-loss and gene-gain events from the rooted 16S rRNA subunit trees of all genera with presence–absence variation of any photolyase gene (data not shown).
Table 2 summarizes the events identified. Most losses involve CPD class I photolyases in eubacterial species. Among the 44 eubacterial genera with more than 1 completely sequenced genomes 21 show presence–absence variation in at least one of the four subfamilies of photolyases.
Among eukaryotes at least five fungal species (Schizosaccharomyces pombe Yarrowia lipolytica Cryptococcus neoformans Candida albicans and Ashbya gossypii) two protozoans (Guillardia theta and Dictyostelium discoideum) and the nematodes Caenorhabditis elegans and C. briggsae seem to have lost all photolyase genes. In addition placental mammals have only cryptochromes with no photolyase activity.
Genus-specific loss is identified within eukaryotes, but there are currently very few eukaryotic genera with more than one completely sequenced genome. The entire list of presence–absence data is available in the Supplementary Material online.
Comparison of dN/dS Estimates
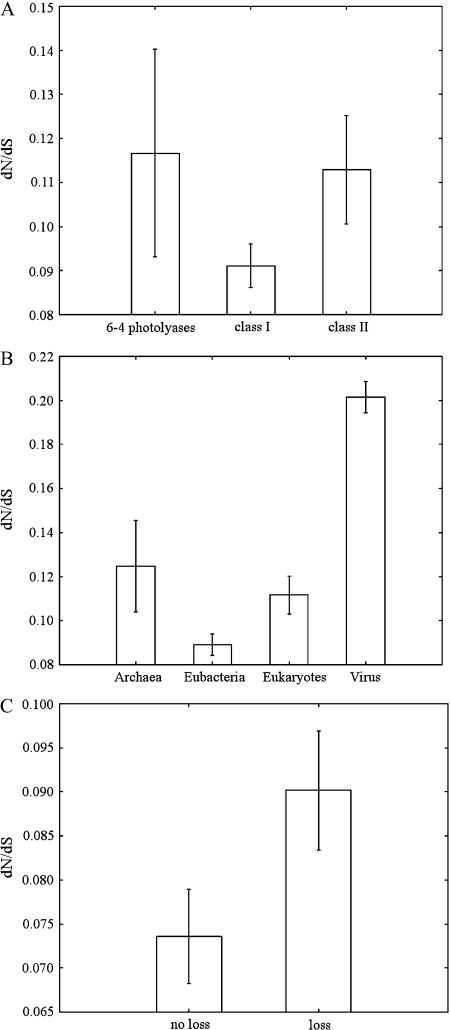
For every set of sequences in the photolyase/cryptochrome family with adequate similarity the maximum likelihood estimate of the ratio of nonsynonymous to synonymous substitution rates was obtained. To ensure the quality of the estimates the sets of sequences were defined in such a way that no pairwise synonymous distance within a set was larger than 1.2 substitutions per site. In all, 33 CPD class I photolyase sets 12 CPD class II photolyase sets, and 2 (6-4) photolyase sets were analyzed (supplementary table 2, Supplementary Material online). Cryptochromes were excluded from the analysis because their functions are not believed to directly affect the genomic mutation rate.
The overall mean dN/dS was 0.0963 ± 0.0039 which as usually interpreted means that about 90% of replacement substitutions are effectively purged by natural selection. CPD class I photolyases seem to have the lowest dN/dS estimates among the three subfamilies (fig. 2) although the difference is not significant at the 0.05 level in any of the three comparisons (Welch test P value 0.06105 for CPD class I/CPD class II comparison). The mean dN/dS estimate for eubacterial photolyases is significantly lower, at the 0.05 level, than that for photolyases from all other kingdoms. Because most eukaryotic photolyases used in this comparison are of class II differences among subfamilies may contribute to the difference between eubacterial and eukaryotic dN/dS values. Comparisons involving the Archaea are impaired by the fact that only 1 dN/dS estimate is available in this kingdom.
FIG. 2.—
Comparison of the mean dN/dS estimates among photolyase paralogs (A), among kingdoms (B), and between genera with and without an observed photolyase gene loss (C). In C, only dN/dS estimates for CPD class I photolyases of eubacterial genera are used, and dN/dS values of “no-loss” genera were weighted by the evolutionary time sampled. Error bars indicate the standard error of the mean, except for Archaea in panel B, where only one dN/dS estimate was used. In that case, the error bar indicates the SD of the estimate, obtained with the bootstrap method.
In all, 29 dN/dS estimates correspond to CPD class I photolyase genes of 22 bacterial genera. Among these genera, five show evidence of a CPD class I photolyase gene-loss event, whereas two other experienced a recent gain of the same gene, according to the most parsimonious mapping of events on the 16S rRNA trees. On average, 3.65 CPD class I photolyase genes are lost per base substitution of the 16S rRNA genes, whereas 1.46 are gained by lateral gene transfer. We expected photolyase genes to be more evolutionary constrained in genera in which they are lost at a lower rate. The mean dN/dS of CPD class I photolyase genes in genera with an observed gene loss is 0.0902 ± 0.0024, which is not significantly different than the mean dN/dS of those genes in genera without any gene loss nor any gene gain, at the 0.05 level (0.08620 ± 0.0014; P value 0.38). However, this comparison is very conservative because the detection of a gene-loss event may have failed in many genera because of insufficient evolutionary time sampled, rather than due to a lower gene-loss rate. To limit this bias, we computed the weighted mean of the dN/dS estimates of genera without an observed gene loss using as weights the sum of branch lengths of their corresponding 16S rRNA trees. This weighted mean becomes 0.07360 ± 0.00260 (fig. 2C), which is significantly different from the mean dN/dS estimate of genera with an observed gene loss.
Comparative Review of the Efficiency of DNA Photolyases
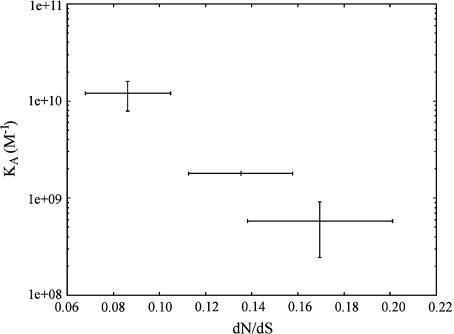
We expect photolyases submitted to stronger natural selection to be more efficient in the repair of DNA damage than less evolutionarily constrained homologs. To test this hypothesis we review published measures of photolyase efficiency for different species and types of photolyases (table 1). Although functional data are limited to photolyases from few species a clear direct relationship is observed between the strength of selection and the affinity of CPD class I photolyases for their substrate (fig. 3). The photolyases with the best documented properties are also the most different ones: the Escherichia coli and the Saccharomyces cerevisiae CPD class I photolyases. The difference in their binding constants, KA suggests that under equal enzyme concentrations about 20 times more pyrimidine dimers would be bound to the yeast photolyase than to the E. coli photolyase. Accordingly and in contrast to the general tendency for eukaryotes Saccharomyces CPD class I photolyases experience a more efficient natural selection (dN/dS = 0.08617 ± 0.01855) than Enterobacteriaceae CPD class I photolyases (0.16948 ± 0.03145).
FIG. 3.—
Relationship between the binding constants of photolyases for their substrates (KA) and the dN/dS estimates obtained with the genes encoding those photolyases and their closest orthologs. Binding constant values correspond to the measures reported in table 1, or to their means and standard errors, when more than one measure is available for the same species. Measures from Escherichia coli and Salmonella typhimurium CPD class I photolyases are pooled in the same mean because the corresponding dN/dS estimate also describes both species. Horizontal error bars are SDs of dN/dS estimates, obtained by bootstrap.
A measure of the specific affinity of photolyases for their substrate is the discrimination ratio or selectivity factor which is the ratio between the enzyme's binding constant for pyrimidine dimers and its binding constant for nondamaged DNA. This ratio is very similar for yeast and E. coli photolyases.
The efficiency of the light-dependent reaction step is characterized by the quantum yield φ which is the probability that an absorbed photon repairs the dimer. In this step the E. coli CPD class I photolyase seems to be the most efficient one. This appears insufficient to offset the disparity in binding affinities.
DISCUSSION
Evolutionary History of the Photolyase/Cryptochrome Family
The overall picture of the early evolution of the photolyase/cryptochrome family emerging from the present analysis depicts four different photolyases already present in the common ancestor of Bacteria Archaea, and Eukarya: CPD class I photolyases CPD class II photolyases (6-4) photolyases and single-stranded DNA photolyases. All 4 subfamilies have been repeatedly lost (see table 1) and at least two of them evolved different functions (plant and animal cryptochromes).
The prokaryote-specific group of photolyase homologs depicted in figure 1 next to the eukaryotic group of (6-4) photolyases and animal cryptochromes is the main addition to the photolyase/cryptochrome family arising from this work. This is the first evidence for the existence of this group of proteins in prokaryotes present in at least five eubacterial and 1 archaeal phylum (table 1). The wide taxonomic range of this gene lineage suggests a very ancient origin and, together with its position in the tree (fig. 1) raises the possibility that the evolution of (6-4) photolyases predates the evolution of eukaryotes.
Up to now photoreactivation of 6 4 photoproducts has only been observed in eukaryotes including Drosophila melanogaster (Todo et al. 1993) Xenopus laevis (Todo et al. 1997) and Arabidopsis thaliana (Nakajima et al. 1998). The (6-4) photolyase orthologs are also present in some fungal species and in the Stramenopiles Phytophthora and Thalassiosira pseudonana (data not shown). In the animal lineage cryptochromes mainly involved in the entrainment to the circadian clock and cell cycle regulation, originated from the gene duplications of (6-4) photolyase genes (reviewed in Öztürk et al. 2007). Remarkably (6-4) photolyases are not known in mammals although mammals do have cryptochromes.
The proteins included in the prokaryote-specific group have not been functionally characterized to the best of our knowledge but their plausible orthology to (6-4) photolyases raises the possibility that they too are (6-4) photolyases which if true would in turn imply that the last common ancestor of Bacteria Archaea, and Eukarya had (6-4) photolyase activity. Because photoreactivation is a direct and efficient DNA repair mechanism present in the last common ancestor of all cellular forms of life (Eisen and Hanawalt 1999) it is plausible that (6-4) photolyases evolved together with CPD photolyases before the formation of the stratospheric ozone shield (Rothschild 1999) and before the appearance of the more sophisticated bacterial and eukaryotic NER pathways (Eisen and Hanawalt 1999) which can also repair 6 4 photoproducts (Friedberg et al. 2006).
Several eubacterial CPD photolyases were grouped together with plant cryptochromes with reasonable bootstrap support (see fig. 1). The eubacterial components of this group have been considered a different class of CPD photolyases namely of class III on the basis of a more limited analysis (Öztürk et al. 2007). We propose that they are CPD class I photolyases for the following reasons. First the low resolution of the topology of the inner branches of the CPD class I cluster and its low bootstrap support do not allow us to definitely distinguish it from the rest of eubacterial CPD photolyases grouped together with the plant cryptochromes. And second our survey of all completely sequenced prokaryotic genomes revealed no species having photolyase genes of both the proposed CPD class III and the CPD class I. Without definitive proof of paralogy the more parsimonious hypothesis of orthology between CPD class I photolyases and the eubacterial CPD photolyases grouped with plant cryptochromes should not be ruled out.
To refute the existence of a third class of CPD photolyases implies that plant cryptochromes are also orthologous to CPD class I photolyases. Given the known existence of CPD class I photolyases in other eukaryotic groups such as fungi (Kanai et al. 1997) the ciliate Tetrahymena thermophila and protozoans of the Trypanosomatidae family (this study; data not shown) the orthology of plant cryptochromes and CPD class I photolyases is not that surprising. However, the finding of two fish proteins as a sister clade of plant cryptochromes with 98% bootstrap support in the neighbor-joining tree was unexpected. This is the first report of animal CPD class I photolyases.
Recent Gene Losses and Evolutionary Constraints
Our observations suggest that photolyase genes are lost from eubacterial genomes at a high rate: more than three times faster than single base substitutions in 16S rRNA among the genomes surveyed for the dN/dS analysis. Such a high rate of gene loss among prokaryotes is accompanied by similar but lower rate of gene gain by horizontal gene transfer. As a result the repertoire of photolyases may be highly variable along the evolution of a bacterial lineage.
The mean dN/dS estimate of photolyase genes (0.0963 ± 0.0039) suggests that they are all under moderate purifying selection far from neutrality but still less constrained than core genes from enteric bacteria (drmN/dS ∼ 0.03 Charlesworth and Eyre-Walker 2006). The comparison of functional data of CPD class I photolyases of yeast and E. coli confirms our expectation that photolyases under stronger selection must be more efficient than less constrained ones because yeast photolyase has a higher affinity for its substrates than E. coli photolyase (table 1 and fig. 3). Despite the fact that yeast CPD class I photolyases are under more efficient purifying selection than their enteric orthologs natural selection is on average more efficient on eubacterial than in eukaryotic photolyase genes (fig. 2B). Therefore eukaryotic photolyases should be on average less efficient than eubacterial ones due to the higher proportion of nonsynonymous substitutions fixed in the former as a consequence of weak selection.
Although the reduced efficiency of natural selection on eukaryotic photolyases may arise from a number of causes ranging from the existence of redundant DNA repair pathways to a lower incidence of UV light in the sampled species’ environments it is plausible and consistent with this large-scale pattern that eukaryotic photolyases are not purged of all deleterious substitutions due to their smaller effective population sizes which make natural selection less efficient (Lynch 2007). Under this scenario the complete loss of photolyase activity in many eukaryotic lineages including placental mammals may not be adaptive. In general if the selective advantage of the repair of UV-induced DNA damage by photolyases is lower than 1/2Ne in a diploid eukaryote with small effective population size (Ne) natural selection will be unable to maintain gene function (Lynch 2008).
One remaining question is why eubacterial photolyase genes, actively purged of deleterious mutations by natural selection, should be lost so often. One plausible explanation is that photolyase genes may not be useful in environments with low UV irradiation. In these situations, photolyase genes would start accumulating mutations at a neutral rate until their inactivation. We reasoned that if a photolyase gene was recently lost in a species for this reason, closely related species of the same genus, with presumably similar ecological constraints, should accumulate a higher proportion of nonsynonymous substitutions. And that is exactly what we observe among eubacterial CPD class I photolyases (fig. 2C). Another line of evidence suggesting that the loss of photolyase genes is not deleterious in eubacterial species comes from a detailed inspection of supplementary table 1, Supplementary Material online. Among the species surveyed for the presence of photolyase genes, there are 46 photosynthetic eubacterial strains, belonging to 23 different genera. Photosynthetic organisms are expected to experience strong selection to keep active photolyase genes, because of their exposure to UV radiation (Rothschild and Cockell 1999, Cockell and Rothschild 1999). The only four photosynthetic taxa without any photolyase genes are the low-light adapted cyanobacterias: Acaryochloris marina, whose main photosynthetic pigment (chlorophyll d) absorbs light in the far-red, and allows it to live out of the reach of UV radiation (Kühl et al. 2005); and the Prochlorococcus marinus strains MIT 9303, MIT 9313, and CCMP1375, which are low-light adapted ecotypes and live in deeper regions of the water column than their high-light adapted relatives.
Therefore, the loss of photolyase genes in eubacterial species that are not exposed to UV radiation may not result in an increase of their genomic mutation rate at all. However, the circumstances under which an allele that increases the genomic mutation rate (a mutator allele) can increase in frequency have been subject to considerable theoretical study (Leigh 1970, Leigh 1973, Gillespie 1981, Holsinger and Feldman 1983, Feldman and Liberman 1986, Holsinger et al. 1986, Liberman and Feldman 1986, Kondrashov 1995, Taddei et al. 1997, Dawson 1998(@, Dawson 1999, Johnson 1999a, Johnson 1999b, Tenaillon et al. 1999, Tanaka et al. 2003, André and Godelle 2006, Lynch 2008), and it is worthy to consider whether photolyase genes can be lost under those circumstances from the genomes of species affected by UV radiation. In asexual populations, where mutator genes remain linked to the excess of mutations that they cause, natural selection on those mutations has a strong indirect effect on the mutator gene (Leigh 1970). During periods of environmental stability, natural selection will promote a lower mutation rate, whereas during episodes of adaptive evolution, mutator alleles may be driven to fixation together with the selected mutations (Tenaillon et al. 1999, Tanaka et al. 2003, André and Godelle 2006). This evolutionary regulation of the mutation rate has been experimentally observed, on a short time scale, in bacterial populations coevolving with phages (Pal et al. 2007). The associated processes are thought to account for the maintenance of mutation-rate polymorphisms in natural eubacterial populations, and they may result in temporal oscillations in the average genomic mutation rate of a bacterial species (Taddei et al. 1997). Mismatch repair genes have been shown to be the mutator genes most frequently lost by mutator strains (Pal et al. 2007). Accordingly, the molecular fingerprints of recurrent losses and reacquisition of mismatch repair genes has been observed in Escherichia coli (Denamur et al. 2000).
Our data suggest that in some cases null photolyase alleles may be positively selected mutator alleles as well. Specifically, the complete absence of photolyase genes in the genome of Ralstonia solanacearum GMI1000 is better explained by the mutator allele model than by the hypothesis of a neutral loss. In contrast to the expectation under the neutral loss hypothesis, the CPD class I photolyase genes of other species of the same genus experience a relatively efficient natural selection (dN/dS = 0.07453 ± 0.01765, supplementary table 2, Supplementary Material online), and R. solanacearum GMI1000, which is a plant pathogenic bacterium, seems to be exposed to UV radiation, at least during some stages of its life cycle. Under the mutator allele model, the loss of a functional photolyase gene, actively purged of deleterious mutations up to that moment, may happen during one of the frequent adaptive episode that a pathogen may experience under the constant adaptation of its host.
Supplementary Material
Supplementary tables 1–3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org).
Table 3.
Biochemical Properties of Photolyases
| Species | Paralog | KA (M−1) | Discrimination Ratio | ϕ |
| Saccharomyces cerevisiae | CPD class I | 0.11−1.93 × 1010a | 0.63−1.40 × 104b | 0.49c |
| Escherichia coli | CPD class I | 0.05−2.20 × 109d | 0.20−7.50 × 104e | 0.59f |
| Salmonella typhimurium | CPD class I | 6.25 × 108g | — | 0.50g |
| Bacillus firmus | CPD class I | 1.8 × 109h | — | 0.75h |
| Xenopus laevis | (6-4) Photolyase | 2.10 × 108i | — | 0.11i |
| Drosophila Melanogaster | (6-4) photolyase | 2.00 × 109j | — | — |
KA is the binding constant and ϕ is the quantum yield.
Harm and Rupert (1968, 1970); Madden and Werbin (1974); Vande Berg Sancar (1998).
Harm (1970); Husain and Sancar (1987); Sancar, Smith, et al. (1987); Li and Sancar (1990).
Husain and Sancar (1987); Sancar, Smith, et al. (1987).
Payne and Sancar (1990). Measures obtained by Sancar, Jons, et al. (1987) and Hitomi et al. (1997) were excluded because they used enzymes lacking the secondary chromophore.
Hitomi et al. (1997). These measures correspond to the enzyme without the secondary chromophore.
Supplementary Material
Acknowledgments
This work was funded by National Institutes of Health grant GM36827 to M.L. and W.K.T. We also thank Tom Doak for his very useful comments and two anonymous reviewers, who helped improve the manuscript substantially.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- André J-B, Godelle B. The evolution of mutation rate in finite asexual populations. Genetics. 2006;172:611–626. doi: 10.1534/genetics.105.046680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina H, Han Z-B, Kawanishi M, Kato T, Jr, Ayaki H, Todo T, Yagi T, Takebe H, Ikenaga M, Kimura SH. Expression of a mammalian DNA photolyase confers light-dependent repair activity and reduces mutations of UV-irradiated shuttle vectors in xeroderma pigmentosum cells. Mutat Res. 1999;435:255–262. doi: 10.1016/s0921-8777(99)00051-8. [DOI] [PubMed] [Google Scholar]
- Baer CF, Miyamoto MM, Denver DR. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat Rev Genet. 2007;8:619–631. doi: 10.1038/nrg2158. [DOI] [PubMed] [Google Scholar]
- Baer ME, Sancar GB. The role of conserved amino acids in substrate binding and discrimination by photolyase. J Biol Chem. 1993;268:16717–16724. [PubMed] [Google Scholar]
- Biegert A, Mayer C, Remmert M, Söding J, Lupas AN. The MPI bioinformatics toolkit for protein sequence analysis. Nucleic Acids Res. 2006;34:W335–W339. doi: 10.1093/nar/gkl217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland JM, Kerry SM. Statistical notes. Weighted comparison of means. Br Med J. 1998;316:129. doi: 10.1136/bmj.316.7125.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ. Rates of DNA sequence evolution differ between taxonomic groups. Science. 1986;231:1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- Brudler R, Hitomi K, Daiyasu H, et al. (11 co-authors). Identification of a new cryptochrome class: structure, function, and evolution. Mol Cell. 2003;11:59–67. doi: 10.1016/s1097-2765(03)00008-x. [DOI] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu Y-J, Liu D. Cryptochromes: blue light receptors for plants and animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- Charlesworth J, Eyre-Walker A. The rate of adaptive evolution in enteric bacteria. Mol Biol Evol. 2006;23:1348–1356. doi: 10.1093/molbev/msk025. [DOI] [PubMed] [Google Scholar]
- Cockell CS, Rothschild LJ. The effects of UV radiation A and B on diurnal variation in photosynthesis in three taxonomically and ecologically diverse microbial mats. Photochem Photobiol. 1999;69:203–210. doi: 10.1562/0031-8655(1999)069<0203:teoura>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- Dawson KJ. Evolutionarily stable mutation rates. J Theor Biol. 1998;194:143–157. doi: 10.1006/jtbi.1998.0752. [DOI] [PubMed] [Google Scholar]
- Dawson J. The dynamics of infinitesimally rare alleles, applied to the evolution of mutation rates and the expression of deleterious mutations. Theor Popul Biol. 1999;55:1–22. doi: 10.1006/tpbi.1998.1375. [DOI] [PubMed] [Google Scholar]
- Denamur E, Lecointre G, Darlu P, et al. (12 co-authors). Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell. 2000;103:711–721. doi: 10.1016/s0092-8674(00)00175-6. [DOI] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Hanawalt PC. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eker APM, Dekker RH, Berends W. Photoreactivating enzyme from Streptomyces griseus—IV. On the nature of the chromophore cofactor in Streptomyces griseus photoreactivating enzyme. Photochem Photobiol. 1981;33:65–72. doi: 10.1111/j.1751-1097.1981.tb04298.x. [DOI] [PubMed] [Google Scholar]
- Eker APM, Kooiman P, Hessels JKC, Yasui A. DNA photoreactivating enzyme from the cyanobacterium Anacystis nidulans. J Biol Chem. 1990;265:8009–8015. [PubMed] [Google Scholar]
- Feldman MW, Liberman U. An evolutionary reduction principle for genetic modifiers. Proc Natl Acad Sci USA. 1986;83:4824–4827. doi: 10.1073/pnas.83.13.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. Washington (DC): ASM Press; 2006. [Google Scholar]
- Fujihashi M, Numoto N, Kobayashi Y, Mizushima A, Tsujimura M, Nakamura A, Kawarabayasi Y, Miki K. Crystal structure of archaeal photolyase form Sulfolobus tokodaii with two FAD molecules: implication of a novel light-harvesting cofactor. J Mol Biol. 2007;365:903–910. doi: 10.1016/j.jmb.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Gillespie JH. Mutation modification in a random environment. Evolution. 1981;35:468–476. doi: 10.1111/j.1558-5646.1981.tb04910.x. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Harm H, Rupert CS. Analysis of photoenzymatic repair of UV lesions in DNA by single light flashes I. In vitro studies with Haemophilus influenza transforming DNA and yeast photoreactivating enzyme. Mutat Res. 1968;6:355–370. doi: 10.1016/0027-5107(68)90053-5. [DOI] [PubMed] [Google Scholar]
- Harm H, Rupert CS. Analysis of photoenzymatic repair of UV lesions in DNA by single light flashes—VI. Rate constants for enzyme-substrate binding in vitro between yeast photoreactivating enzyme and ultraviolet lesions in haemophilus transforming DNA. Mutat Res. 1970;10:291–306. doi: 10.1016/0027-5107(70)90044-8. [DOI] [PubMed] [Google Scholar]
- Harm W. Analysis of photoenzymatic repair of UV lesions in DNA by single light flashes—V. Determination of the reaction-rate constants in E. coli cells. Mutat Res. 1970;10:277–290. doi: 10.1016/0027-5107(70)90043-6. [DOI] [PubMed] [Google Scholar]
- Hitomi K, Kim S-T, Iwai S, Harima N, Otoshi E, Ikenaga M, Todo T. Binding and catalytic properties of Xenopus (6-4) photolyase. J Biol Chem. 1997;272:32591–32598. doi: 10.1074/jbc.272.51.32591. [DOI] [PubMed] [Google Scholar]
- Holsinger KE, Feldman MW. Modifiers of mutation rate: evolutionary optimum with complete selfing. Proc Natl Acad Sci USA. 1983;80:6732–6734. doi: 10.1073/pnas.80.21.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger KE, Feldman MW, Altenberg L. Selection for increased mutation rates with fertility differences between matings. Genetics. 1986;112:909–922. doi: 10.1093/genetics/112.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang R-P, Todo T, Wei Y-F, Sancar A. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry. 1996;35:13871–13877. doi: 10.1021/bi962209o. [DOI] [PubMed] [Google Scholar]
- Husain I, Sancar A. Binding of E. coli DNA photolyase to a defined substrate containing a single T< > T dimer. Nucleic Acids Res. 1987;15:1109–1120. doi: 10.1093/nar/15.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Hamm-Alvarez S, Payne G, Sancar GB, Rajagopalan KV, Sancar A. Identification of the second chromophore of Escherichia coli and yeast DNA photolyases as 5,10-methenyltetrahydrofolate. Proc Natl Acad Sci USA. 1988;85:2046–2050. doi: 10.1073/pnas.85.7.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. The approach to mutation-selection balance in an infinite asexual population, and the evolution of mutation rates. Proc Biol Sci. 1999a;266:2389–2397. doi: 10.1098/rspb.1999.0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. Beneficial mutations, hitchhiking and the evolution of mutation rates in sexual populations. Genetics. 1999b;151:1621–1631. doi: 10.1093/genetics/151.4.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai S, Kikuno R, Toh H, Ryo H, Todo T. Molecular evolution of the photolyase–blue-light photoreceptor family. J Mol Evol. 1997;45:535–548. doi: 10.1007/pl00006258. [DOI] [PubMed] [Google Scholar]
- Kato T, Jr, Todo T, Ayaki H, Ishizaki K, Morita T, Mitra S, Ikenaga M. Cloning of a marsupial DNA photolyase gene and the lack of related nucleotide sequences in placental mammals. Nucleic Acids Res. 1994;22:4119–4124. doi: 10.1093/nar/22.20.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelner A. Effect of visible light on the recovery of Streptomyces griseus conidia from ultra-violet irradiation injury. Proc Natl Acad Sci USA. 1949;35:73–79. doi: 10.1073/pnas.35.2.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov AS. Modifiers of mutation-selection balance: general approach and the evolution of mutation rates. Genet Res. 1995;66:53–69. [Google Scholar]
- Kühl M, Chen M, Ralph PJ, Schreiber U, Larkum AWD. Ecology: A niche for cyanobacteria containing chlorophyll d. Nature. 2005;433:820. doi: 10.1038/433820a. [DOI] [PubMed] [Google Scholar]
- Leigh EG., Jr Natural selection and mutability. Am Nat. 1970;104:301–305. [Google Scholar]
- Leigh EG., Jr The evolution of mutation rates. Genetics. 1973;73:1–18. [PubMed] [Google Scholar]
- Li YF, Sancar A. Active site of Escherichia coli DNA photolyase: mutations at Trp277 alter the selectivity of the enzyme without affecting the quantum yield of photorepair. Biochemistry. 1990;29:5698–5706. doi: 10.1021/bi00476a009. [DOI] [PubMed] [Google Scholar]
- Li YF, Sancar A. Cloning, sequencing, expression and characterization of DNA photolyase from Salmonella typhimurium. Nucleic Acids Res. 1991;19:4885–4890. doi: 10.1093/nar/19.18.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman U, Feldman MW. Modifiers of mutation rate: a general reduction principle. Theor Popul Biol. 1986;30:125–142. doi: 10.1016/0040-5809(86)90028-6. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of eukaryotic gene structure. Mol Biol Evol. 2006;23:450–468. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. Sunderland: Sinauer Associates, Inc. Publishers; 2007. [Google Scholar]
- Lynch M. The cellular, developmental and population-genetic determinants of mutation-rate evolution. Genetics. 2008;180:933–943. doi: 10.1534/genetics.108.090456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden JJ, Werbin H. Use of the membrane binding technique to study the kinetics of yeast deoxyribonucleic acid photolyase reactions. Formation of enzyme-substrate complexes in the dark and their photolysis. Biochemistry. 1974;13:2149–2154. doi: 10.1021/bi00707a024. [DOI] [PubMed] [Google Scholar]
- Malhotra K, Kim ST, Sancar A. Characterization of a medium wavelength type DNA photolyase: purification and properties of photolyase from Bacillus firmus. Biochemistry. 1994;33:8712–8718. doi: 10.1021/bi00195a012. [DOI] [PubMed] [Google Scholar]
- Marcobal AM, Sela DA, Wolf YI, Makarova KS, Mills DA. The role of hypermutability in the evolution of the genus Oenococcus. J Bacteriol. 2008;190:564–570. doi: 10.1128/JB.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima S, Sugiyama M, Iwai S, Hitomi K, Otoshi E, Kim S-T, Jiang C-Z, Todo T, Britt AB, Yamamoto K. Cloning and characterization of a gene (UVR3) required for photorepair of 6-4 photoproducts in Arabidopsis thaliana. Nucleic Acids Res. 1998;26:638–644. doi: 10.1093/nar/26.2.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otoshi E, Yagi T, Mori T, Matsunaga T, Nikaido O, Kim S-T, Hitomi K, Ikenaga M, Todo T. Respective roles of cyclobutane pyrimidine dimers, (6-4)photoproducts, and minor photoproducts in ultraviolet mutagenesis of repair-deficient xeroderma pigmentosum A cells. Cancer Res. 2000;60:1729–1735. [PubMed] [Google Scholar]
- Öztürk N, Song SH, Özgür S, Selby CP, Morrison L, Partch C, Zhong C, Sancar A. Structure and function of animal cryptochromes. Cold Spring Harb Symp Quant Biol. 2007;72:119–131. doi: 10.1101/sqb.2007.72.015. [DOI] [PubMed] [Google Scholar]
- Pal C, Maciá MD, Oliver A, Schachar I, Buckling A. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature. 2007;450:1079–1081. doi: 10.1038/nature06350. [DOI] [PubMed] [Google Scholar]
- Park HW, Kim ST, Sancar A, Deisenhofer J. Crystal structure of DNA photolyase from Escherichia coli. Science. 1995;268:1866–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- Payne G, Sancar A. Absolute action spectrum of E-FADH2 and E-FADH2-MTHF forms of Escherichia coli DNA photolyase. Biochemistry. 1990;29:7715–7727. doi: 10.1021/bi00485a021. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rothschild LJ. The influence of UV radiation on protistan evolution. J Eukaryot Microbiol. 1999;46:548–555. doi: 10.1111/j.1550-7408.1999.tb06074.x. [DOI] [PubMed] [Google Scholar]
- Rothschild LJ, Cockell CS. Radiation: microbial evolution, ecology, and relevance to Mars missions. Mutat Res. 1999;430:281–291. doi: 10.1016/s0027-5107(99)00140-2. [DOI] [PubMed] [Google Scholar]
- Sancar A. Structure and function of DNA photolyase. Biochemistry. 1994;33:2–9. doi: 10.1021/bi00167a001. [DOI] [PubMed] [Google Scholar]
- Sancar GB. Sequence of the Saccharomyces cerevisiae PHR1 gene and homology of the PHR1 photolyase to E. coli photolyase. Nucleic Acids Res. 1985;13:8231–8246. doi: 10.1093/nar/13.22.8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar G. Enzymatic photoreactivation: 50 years and counting. Mutat Res. 2000;451:25–37. doi: 10.1016/s0027-5107(00)00038-5. [DOI] [PubMed] [Google Scholar]
- Sancar GB, Jorns MS, Paynet G, Fluke DJ, Rupert CS, Sancar A. Action mechanism of Escherichia coli DNA photolyase III. Photolysis of the enzyme-substrate complex and the absolute action spectrum. J Biol Chem. 1987;262:492–498. [PubMed] [Google Scholar]
- Sancar GB, Smith FW, Lorence MC, Rupert CS, Sancar A. Sequence of the Escherichia coli photolyase gene and protein. J Biol Chem. 1984;259:6033–6038. [PubMed] [Google Scholar]
- Sancar GB, Smith FW, RReid, Payne G, Levy M, Sancar A. Action mechanism of Escherichia coli DNA photolyase I. Formation of the enzyme-substrate complex. J Biol Chem. 1987;262:478–485. [PubMed] [Google Scholar]
- Schul W, Jans J, Rijksen YMA, et al. (11 co-authors) Enhanced repair of cyclobutane pyrimidine dimers and improved UV resistance in photolyase transgenic mice. EMBO J. 2002;21:4719–4729. doi: 10.1093/emboj/cdf456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc Natl Acad Sci USA. 2006;103:17696–17700. doi: 10.1073/pnas.0607993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman R. DNA repair capacity of zebrafish. Proc Natl Acad Sci USA. 2007;104:13379–13383. doi: 10.1073/pnas.0706157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. Version 4. Sunderland: Sinauer Associates; 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods) [Google Scholar]
- Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon PH, Godelle B. Role of mutator alleles in adaptive evolution. Nature. 1997;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- Tamada T, Kitadokoro K, Higuchi Y, Inaka K, Yasui A, de Ruiter PE, Eker APM, Miki K. Crystal structure of DNA photolyase from Anacystis nidulans. Nat Struct Biol. 1997;4:887–891. doi: 10.1038/nsb1197-887. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Nakajima S, Ihara M, Matsunaga T, Nikaido O, Yamamoto K. Effects of photoreactivation of cyclobutane pyrimidine dimers and pyrimidine (6-4) pyrimidone photoproducts on ultraviolet mutagenesis in SOS-induced repair-deficient Escherichia coli. Mutagenesis. 2001;16:1–6. doi: 10.1093/mutage/16.1.1. [DOI] [PubMed] [Google Scholar]
- Tanaka MM, Bergstrom CT, Levin BR. The evolution of mutator genes in bacterial populations: the roles of environmental change and timing. Genetics. 2003;164:843–854. doi: 10.1093/genetics/164.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Toupance B, Le Nagard H, Taddei F, Godelle B. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics. 1999;152:485–493. doi: 10.1093/genetics/152.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gbson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todo T, Kim S-T, Hitomi K, Otoshi E, Inui T, Morioka H, Kobayashi H, Ohtsuka E, Toh H, Ikenaga M. Flavin adenine dinucleotide as a chromophore of the Xenopus (6-4)photolyase. Nucleic Acids Res. 1997;25:764–768. doi: 10.1093/nar/25.4.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todo T, Takemori H, Ryo H, Ihara M, Matsunaga T, Nikaido O, Sato K, Nomura T. A new photoreactivating enzyme that specifically repairs ultraviolet light-induced (6-4)photoproducts. Nature. 1993;361:371–374. doi: 10.1038/361371a0. [DOI] [PubMed] [Google Scholar]
- Ueda T, Kato A, Kuramitsu S, Terasawa H, Shimada I. Identification and characterization of a second chromophore of DNA photolyase from Thermus thermophilus HB27. J Biol Chem. 2005;280:36237–36243. doi: 10.1074/jbc.M507972200. [DOI] [PubMed] [Google Scholar]
- van Oers MM, Herniou EA, Usmany M, Messelink GJ, Vlak JM. Identification and characterization of a DNA photolyase-containing baculovirus from Chrysodeixis chalcites. Virology. 2004;330:460–470. doi: 10.1016/j.virol.2004.09.032. [DOI] [PubMed] [Google Scholar]
- Vande Berg BJ, Sancar GB. Evidence for dinucleotide flipping by DNA photolyase. J Biol Chem. 1998;273:20276–20284. doi: 10.1074/jbc.273.32.20276. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- You Y-H, Lee D-H, Yoon J-H, Nakajima S, Yasui A, Pfeifer GP. Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J Biol Chem. 2001;276:44688–44694. doi: 10.1074/jbc.M107696200. [DOI] [PubMed] [Google Scholar]
- Zhao X, Liu J, Hsu DS, Zhao S, Taylor J-S, Sancar A. Reaction mechanism of (6-4) photolyase. J Biol Chem. 1997;272:32580–32590. doi: 10.1074/jbc.272.51.32580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.