Abstract
Variants in the FTO (fat mass and obesity associated) gene are associated with increased body mass index in humans. Here, we show by bioinformatics analysis that FTO shares sequence motifs with Fe(II)- and 2-oxoglutarate–dependent oxygenases. We find that recombinant murine Fto catalyzes the Fe(II)- and 2OG-dependent demethylation of 3-methylthymine in single-stranded DNA, with concomitant production of succinate, formaldehyde, and carbon dioxide. Consistent with a potential role in nucleic acid demethylation, Fto localizes to the nucleus in transfected cells. Studies of wild-type mice indicate that Fto messenger RNA (mRNA) is most abundant in the brain, particularly in hypothalamic nuclei governing energy balance, and that Fto mRNA levels in the arcuate nucleus are regulated by feeding and fasting. Studies can now be directed toward determining the physiologically relevant FTO substrate and how nucleic acid methylation status is linked to increased fat mass.
Recent studies have revealed a strong association between common variants in the first intron of FTO and obesity in both children and adults, with ~16% of studied populations homozygous for the risk alleles (1-4). As adults, these individuals weigh ~3 kg more than those homozygous for the low risk alleles as a result of a specific increase in fat mass (2). FTO mRNA is expressed in a wide range of human tissues (2). The Fto gene was first cloned after identification of a fused-toe mutant mouse whose phenotype results from a 1.6-Mb deletion of six genes, including Fto (5).
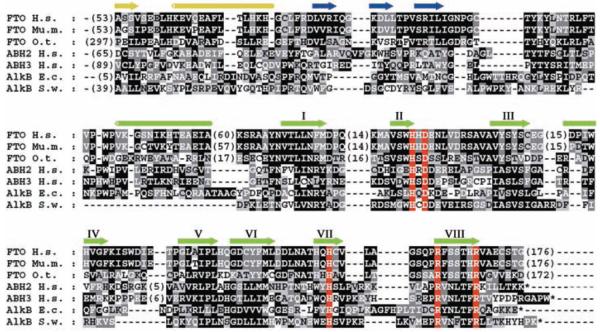
Sequence analysis predicts that FTO protein contains a double-stranded beta-helix (DSBH) fold homologous to those of Fe(II) and 2-oxoglutarate (2OG) oxygenases [for a review of these enzymes, see (6)] (Fig. 1). The predicted DSBH fold of FTO contains four conserved residues characteristic of Fe(II) and 2OG binding sites (7, 8), and its sequence is highly conserved in organisms ranging from mammals to green algae (Fig. 1 and fig. S1). 2OG oxygenases are involved in diverse processes, including DNA repair, fatty acid metabolism, and posttranslational modifications, for example, proline hydroxylation and histone lysine demethylation [reviewed in (6, 9)]. They require nonheme iron [Fe(II)] as a cofactor, use oxygen and, almost always, 2OG as cosubstrates, and produce succinate and carbon dioxide as by-products.
Fig. 1.
Multiple sequence alignment of FTO from human (Homo sapiens; Hs), mouse (Mus musculus; Mm) and green algae (Ostreococcus tauri; Ot) with Escherichia coli (Ec) and Shewanella woodyi (Sw) AlkB and human ABH2 and ABH3. A comparison of Ot FTO with the nonredundant protein sequence database at the National Center for Biotechnology Information using PSI-BLAST (28) revealed significant similarity (E < 0.005) between human FTO, bacterial AlkB, and its human homologs, within two iterations. Conserved residues highlighted in red are histidine and carboxylate (Asp or Glu) Fe(II) binding residues of 2OG oxygenases (His-228/231, Asp-230/233, and His-304/307 in murine Fto/human FTO, respectively), as well as an arginine (Arg-313/316), which binds the 2OG C-5 carboxylate in a 2OG oxygenase subfamily (7, 8). The cylinders and arrows represent α helices and β strands, respectively, assigned as in a crystal structure of ABH3 (29). Secondary structure in yellow represents the N-terminal region, blue strands represent an assigned (29) substrate binding lid for ABH3, green strands labeled with roman numerals identify the eight β strands that form the conserved double-stranded beta-helix of the 2OG oxygenases. GenInfo numbers: human FTO: 122937263; mouse Fto: 6753916; green algae FTO: 116060758; human ABH2: 48717226; human ABH3: 21040275; E. coli AlkB: 113638; S. woodyi AlkB: 118070714.
To determine whether FTO is a 2OG oxygenase, we expressed the murine Fto gene in Escherichia coli and purified N-terminally hexa-His tagged Fto (10). Some 2OG oxygenases catalyze 2OG turnover without a “prime” substrate provided that a reducing agent, typically ascorbate, is present (uncoupled turnover). 2OG uncoupled turnover assays with Fto, monitoring conversion of [1-14C]-2OG into [14C]-carbon dioxide (10), revealed that Fto catalyzed 2OG decarboxylation, a reaction that was stimulated by ascorbate and FeSO4 (fig. S2A). 2OG turnover was inhibited by known 2OG oxygenase inhibitors (fig. S2B) and by the absence of Fe(II) and ascorbate.
We next considered the identity of the prime FTO substrate. Among 2OG oxygenases with known substrates, the FTO sequence is most similar to that of the E. coli enzyme AlkB (11) and its eukaryotic homologs, members of the ABH (AlkB homolog) family (Fig. 1). AlkB is a DNA repair enzyme that repairs cytotoxic 1-methyladenine (1-meA) and 3-methylcytosine (3-meC) lesions by methyl group hydroxylation, followed by a retro-aldol reaction. Among the various human ABHs, only two, ABH2 and ABH3, have been shown to exhibit DNA demethylation activity analogous to that of AlkB (12, 13). ABH 2 and ABH3 are ubiquitously expressed, and their expression is not known to be altered by physiological stimuli.
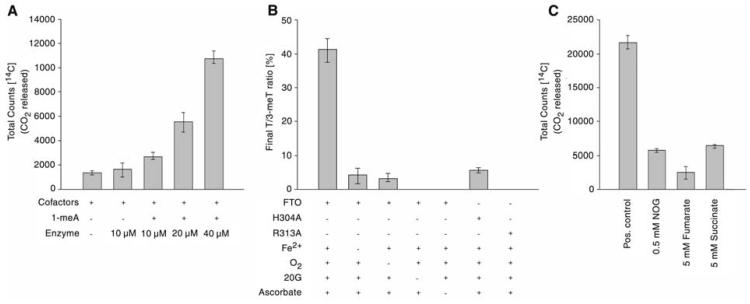
We screened potential Fto substrates, including a synthetic single-stranded 1-methyl adenine (1-meA) methylated oligonucleotide, Lys-9 methylated histone H3, hypoxia-inducible factor-1α (HIF-1α) subunit fragments, IκBα, coenzyme A derivatives, and other known substrates of human 2OG oxygenases (10). Only the 1-meA methylated oligonucleotide stimulated turnover of 2OG above control levels (Fig. 2A). This activity was inhibited by N-oxalylglycine, fumarate, and succinate, which were also inhibitors in the 2OG uncoupled turnover assays (Fig. 2C).
Fig. 2.
Fto is a 2-oxoglutarate–dependent DNA demethylase in vitro. (A) Demethylation of 1-meA is dependent on Fto in 2OG decarboxylation assays; (B) Cofactor/cosubstrate dependence of FTO activity on a 3-meT substrate shown by LC-MS. Data shown represent ratios of thymine to 3-meT in ss-DNA. The oxygen control reaction was carried out in an atmosphere of <1% O2; (C) Inhibition of Fto-catalyzed 1-meA demethylation in 2OG decarboxylation assays. All assays were performed in triplicate, at least, with error bars denoting ±SD.
Using a liquid chromatography–mass spectrometry assay that directly monitors DNA demethylation [without the need for radiolabeled (co-)substrates or coupled assays (10)], we demonstrated that Fto catalyzes Fe(II)- and 2OG-dependent DNA demethylation. This activity was stimulated by ascorbate, as observed for other 2OG oxygenases (6) (Fig. 2B). Significantly reduced turnover was observed when the reaction was performed under reduced oxygen conditions. The production of succinate was verified by 1H nuclear magnetic resonance (400 MHz) analyses and that of formaldehyde was confirmed by derivatization with pentafluorophenylhydrazine.
To test the predicted role of the assigned Fe(II) binding and 2OG 5-carboxylate binding residues, His-304 and Arg-313 alanine substitution mutants were constructed (10) (fig. S3). The His-304 mutant showed reduced 2OG turnover, whereas the Arg-313 mutant ablated activity (Fig. 2B), consistent with observations on other 2OG oxygenases.
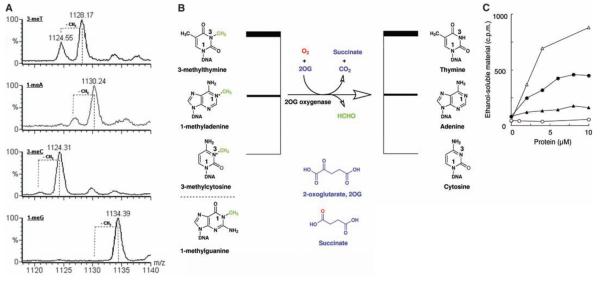
We next investigated Fto activity with single-stranded oligonucleotides (ss-DNA) methylated at a single position: 1-methyladenine (1-meA), 1-methylguanine (1-meG), 3-methylcytosine (3-meC), and 3-methylthymine (3-meT) (Fig. 3B). Under the assay conditions at pH 7.5, Fto exhibited a preference for 3-meTover 1-meA or 3-meC in ss-DNA; 1-meG was not an Fto substrate (Fig. 3A). The preference of Fto for 3-meT substrates was also observed in assays measuring the release of formaldehyde from randomly methylated poly(dA) and poly(dT) (Fig. 3C). Consistent with previous reports (14, 15) we found that recombinant ABH3 demethylated 1-meA, with only very low levels of 3-meT demethylation observed under our conditions (Fig. 3C).
Fig. 3.
In vitro activity of Fto with ss-DNA substrates. (A) Demethylation of ss-DNA substrates by Fto. LC-MS data for the incubation of synthetic 15-nucleotide oligomers of the form 5′-TT(methylated base)TTTTTTTTTTTT-3′ with Fto, cofactors, and cosubstrates; positions of expected peaks for demethylated substrates are indicated by dotted lines. Smaller peaks at higher masses than reactant peaks probably arise from Na+ and K+ adducts of the methylated oligonucleotides. m/z = mass-to-charge ratio, shown in units of Dalton. (B) Stoichiometry of the Fto reaction. (C) Release of formaldehyde from methylated poly(dA) and poly(dT). FTO and ABH3 were assayed for demethylase activity by incubation with [14C]-methylated poly(dA) or [14C]-methylated poly(dT) [total counts per minute (c.p.m.) 1000 or 800, respectively] at 37°C for 15 min. Release of ethanol-soluble [14C]-formaldehyde was monitored. FTO -○-, -●-; ABH3 -∆-, -▲-. [14C]-methylated poly(dA) -○-, -∆-; [14C]-methylated poly(dT) -●-, -▲-. In the absence of 2OG, no significant activity was detected; the ethanol soluble material released by FTO or ABH3 was at background level (less than 100 c.p.m).
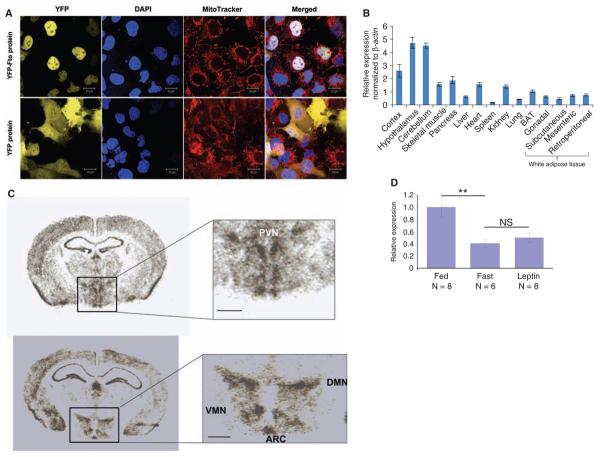
A protein with nucleic acid demethylase activity might be expected to localize to the cell nucleus. Indeed, confocal imaging of COS-7 cells (from simian kidney) transfected with a plasmid encoding for yellow fluorescent protein (YFP)–tagged Fto, or with YFP alone, revealed that YFP-Fto is concentrated in the nucleus, whereas YFP is present only in the cytoplasm (10) (Fig. 4A). YFP-Fto did not colocalize with mitochondria (Fig. 4A).
Fig. 4.
Expression studies of Fto protein and mRNA in mice. (A) Subcellular localization of murine Fto in COS-7 cells. Confocal fluorescence images of COS-7 cells expressing YFP-Fto or YFP show YFP-Fto localizing to the nucleus. Nuclei were visualized with 4′,6-diamidino-2-phenylindole (DAPI) staining and mitochondria with MitoTracker. Colocalization of YFP (yellow) and DAPI (blue) in the merged images produces a white signal. (B) Relative expression of Fto mRNA in different tissues. Bar graphs show the relative expression of Fto mRNA normalized to β actin across a panel of different tissues. Data are represented as the mean (±SE) of six independent mice per tissue. (C) In situ hybridization of murine Fto in brain. PVN, paraventricular nucleus; VMN, ventromedial nucleus; DMN, dorsomedial nucleus; ARC, arcuate nucleus; scale bar = 500 μm. (D) Nutritional regulation of Fto mRNA expression in ARC. Bar graphs show the change in expression of Fto mRNA in the arcuate nucleus of the hypothalamus in the fed, fasted, and leptin-treated-while-fasted state. Response is expressed in terms of fold induction of the fasted and leptin-treated expression over the fed expression. The P value was calculated using a two-tailed distribution unpaired Student's t test. **P < 0.01. Data are represented as the mean (±SE) of at least six independent mice per group.
The FTO SNPs associated with adiposity are intronic and may exert functional effects through altered expression of FTO mRNA. If FTO regulates energy homeostasis, it might be more highly expressed in tissues involved in the control of energy balance and be influenced by nutritional signals. We examined Fto mRNA levels in multiple murine tissues by quantitative reverse transcription polymerase chain reaction (Fig. 4B). Consistent with human data (2), mouse Fto mRNA was detected in all tissues studied, with the highest expression in the brain. Within the brain, high levels were found in the hypothalamus, an area that plays a key role in the control of energy homeostasis. In situ hybridization of hypothalamic slices revealed that Fto mRNA was highly expressed in arcuate (ARC), paraventricular (PVN), dorsomedial (DMN), and ventromedial (VMN) nuclei, all sites of critical importance for the control of energy balance (Fig. 4C).
To determine whether Fto mRNA expression is nutritionally regulated, we examined Fto mRNA levels in laser-dissected hypothalamic nuclei (ARC, VMN, and PVN) from three groups of mice (freely feeding, fasted for 48 hours, and 48 hours fasted treated with daily intraperitoneal injections of leptin, respectively). The leptin-supplemented group was included to examine whether any changes associated with fasting were caused by the reduction of circulating leptin that occurs during starvation (16). In the arcuate nucleus, Fto mRNA levels were reduced by ~60% in fasted mice and were not rescued by leptin supplementation (Fig. 4D). Fto mRNA levels in the PVN and VMN were unchanged (10).
2OG oxygenase–catalyzed posttranslational hydroxylation is central to transcriptional regulation in the hypoxic response (17, 18), and 2OG oxygenases catalyze histone demethylation (19). The catalytic activity of FTO may similarly regulate the transcription of genes involved in metabolism by nucleic acid demethylation. Alternatively, it is possible that FTO, as proposed for ABH2, can act as a nucleic acid repair enzyme (20): There is evidence that breakdown of genomic repair processes leads to obesity and metabolic syndrome (21-23).
Under our assay conditions at physiological pH, the preferred substrate of FTO was 3-methylthymine in DNA, a minor but stable lesion that is generated upon exposure of DNA to methylating agents. Verification of whether this methylated base in DNA, or an as-yet-uninvestigated modification of DNA or possibly RNA, is the physiologically relevant substrate(s) of FTO is a key objective of future work. It is noteworthy that although both E. coli AlkB and human ABH3 demethylate both DNA and RNA (24, 25), it remains unclear whether RNA demethylation is physiologically relevant.
It is now important to determine whether altered FTO demethylase activity underlies the enhanced fat mass associated with the FTO gene variant and whether this alteration in activity is associated with increased food intake, decreased energy expenditure, or both. The majority of human genetic defects associated with obesity have their principal impact on the function of the hypothalamus (26), and our findings with respect to its hypothalamic expression and nutritional regulation suggest that this may also be the case for FTO. Fto is inhibited by Krebs cycle intermediates, in particular fumarate (Fig. 2C), as proposed for the HIF-1α hydroxylases (27). Disease states with elevated fumarate/succinate levels may therefore modulate FTO activity. Further functional studies of Fto may lead to new insights into the pathogenesis of obesity and possibly new avenues for treatment.
Supplementary Material
Acknowledgments
We thank J. M. Claverie, C. Notredame, Z. Kostrouch, and A. Hattersley for useful discussions; R. Read and G. Chavali for SF9 cells, the PMIB vector, and technical advice; and D. Rimmington and D. Lyon for additional technical help. This work was supported by the Biochemical and Biological Research Council, the Medical Research Council, Cancer Research UK, European Community FP6 LSHM-CT-503041/518153/512013, and the Wellcome Trust. F.M.A. is a Royal Society Research Professor. C.J.S. is a cofounder of ReOx, a company that aims to exploit the hypoxic response for therapeutic benefit.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/1151710/DC1
Materials and Methods
Figs. S1 to S4
References
References and Notes
- 1.Dina C, et al. Nat. Genet. 2007;39:724. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 2.Frayling TM, et al. Science. 2007;316:889. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott LJ, et al. Science. 2007;316:1341. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scuteri A, et al. PLoS Genetics. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peters T, Ausmeier K, Dildrop R, Rüther U. Mamm. Genome. 2002;13:186. doi: 10.1007/s00335-001-2142-7. [DOI] [PubMed] [Google Scholar]
- 6.Clifton IJ, et al. J. Inorg. Biochem. 2006;100:644. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 7.Valegard K, et al. Nature. 1998;394:805. doi: 10.1038/29575. [DOI] [PubMed] [Google Scholar]
- 8.Aravind L, Koonin E. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-research0007. research0007.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hausinger RP. Crit. Rev. Biochem. Mol. Biol. 2004;39:21. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 10.Materials and methods and supplementary data are available on Science Online.
- 11.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Nature. 2002;419:174. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 12.Aas PA, et al. Nature. 2003;421:859. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 13.Duncan T, et al. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16660. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koivisto P, Robins P, Lindahl T, Sedgwick B. J. Biol. Chem. 2004;279:40470. doi: 10.1074/jbc.M407960200. [DOI] [PubMed] [Google Scholar]
- 15.Delaney JC, Essigmann JM. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14051. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahima RS, et al. Nature. 1996;382:250. doi: 10.1038/382250a0. [DOI] [PubMed] [Google Scholar]
- 17.Ivan M, et al. Science. 2001;292:464. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 18.Jaakkola P, et al. Science. 2001;292:468. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 19.Klose RJ, et al. Nature. 2006;442:312. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 20.Ringvoll J, et al. EMBO J. 2006;25:2189. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vartanian V, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1864. [Google Scholar]
- 22.van der Pluijm I, et al. PLoS Biol. 2007;5:e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mostoslavsky R, et al. Cell. 2006;124:315. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 24.Ougland R, et al. Mol. Cell. 2004;16:107. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Lee DH, et al. J. Biol. Chem. 2005;280:39448. doi: 10.1074/jbc.M509881200. [DOI] [PubMed] [Google Scholar]
- 26.Farooqi IS, O'Rahilly S. Annu. Rev. Med. 2005;56:443. doi: 10.1146/annurev.med.56.062904.144924. [DOI] [PubMed] [Google Scholar]
- 27.Selak MA, et al. Cancer Cell. 2005;7:77. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 28.Schaffer AA, et al. Nucleic Acids Res. 2001;29:2994. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sundheim O, et al. EMBO J. 2006;25:3389. doi: 10.1038/sj.emboj.7601219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.