Abstract
Aims/hypothesis
The sterol regulatory element-binding factor (SREBF)-1c is a transcription factor involved in the regulation of lipid and glucose metabolism. We have previously found evidence that a common SREBF1c single-nucleotide polymorphism (SNP), located between exons 18c and 19c, is associated with an increased risk of type 2 diabetes. The present study aimed to replicate our previously reported association in a larger case–control study and to examine an additional five SREBF1c SNPs for their association with diabetes risk and plasma glucose concentrations.
Methods
We genotyped six SREBF1c SNPs in two case–control studies (n=1,938) and in a large cohort study (n=1,721) and tested for association with type 2 diabetes and with plasma glucose concentrations (fasting and 120-min post-glucose load), respectively.
Results
In the case–control studies, carriers of the minor allele of the previously reported SNP (rs11868035) had a significantly increased diabetes risk (odds ratio [OR]=1.20 [95% CI 1.04–1.38], p=0.015). Also, three other SNPs (rs2236513, rs6502618 and rs1889018), located in the 5′ region, were significantly associated with diabetes risk (OR ≥1.21, p≤0.006). Furthermore, two SNPs (rs2236513 and rs1889018) in the 5′ region were weakly (p<0.09) associated with plasma glucose concentrations in the cohort study. Rare homozygotes had increased (p≤0.05) 120-min post-load glucose concentrations compared with carriers of the wild-type allele. Haplotype analyses showed significant (p=0.04) association with diabetes risk and confirmed the single SNP analyses.
Conclusions/interpretation
In summary, we replicated our previous finding and found evidence for SNPs in the 5′ region of the SREBF1c gene to be associated with the risk of type 2 diabetes and plasma glucose concentration.
Keywords: Case–control study, Cohort study, Plasma glucose concentration, Polymorphism, Population-based, Sterol regulatory element-binding factor, Type 2 diabetes
Introduction
Dysregulation of fatty acid metabolism may be an underlying cause of type 2 diabetes. Lipotoxicity in peripheral tissues, which is promoted by defective adipose tissue, could be the link between obesity, insulin resistance and type 2 diabetes [1].
Sterol regulatory element-binding factors (SREBFs) are transcription factors that regulate lipid homeostasis by activation of genes involved in fatty acid and cholesterol metabolism. Two genes, SREBF1 located on chromosome 17p11.2 and SREBF2 on 22q13, encode three isoforms of the protein [2]. Through alternative transcription start sites SREBF1 encodes SREBF1a and SREBF1c. Due to its longer acidic transactivation domain, SREBF1a is a more potent transcriptional activator than SREBF1c and can activate all SREBF-responsive genes, including those that mediate the synthesis of cholesterol, fatty acids and triacylglycerols [2]. SREBF1c preferentially activates transcription of genes involved in fatty acid and triacylglycerol synthesis, whereas SREBF2 mainly activates genes that control cholesterol synthesis [2-4]. SREBF1c is the main SREBF1 isoform, being highly expressed in most tissues of mice and humans, including liver, adipose tissue and skeletal muscle [5-7]. SREBF processing is controlled by the cellular sterol content; high sterol levels block the translocation of SREBF to the nucleus and SREBF-mediated gene transcription does not occur [8, 9]. SREBF1c activity is additionally regulated by fatty acids [3]. Animal feeding studies have demonstrated that polyunsaturated fatty acids suppress SREBF1 mRNA expression and protein processing [10, 11]. Furthermore, insulin increases SREBF1c mRNA in skeletal muscle and SREBF1c levels in adipose tissue [7, 12-14]. Given the mediating role of SREBF1c in the nutritionally and insulin-regulated transcription of genes involved in tricacylglycerol and fatty acid synthesis, SREBF1c may underlie the mechanisms that lead to lipotoxicity [1]. The SREBF1c gene can therefore be considered as a good candidate for type 2 diabetes.
There have been only two reports in humans that have examined the relationship between SREBF1 gene variants and diabetes phenotypes to date [15, 16]. We have recently reported significant associations between an intronic C>T single-nucleotide polymorphism (SNP) (rs11868035), located between exons 18c and 19c, and the risk of type 2 diabetes, and plasma concentrations of total and LDL cholesterol [15]. A nearby located synonymous SNP in exon 18 (G952G, rs2297508) was found to be associated with morbid obesity and diabetes in a French population [16]. This G952G variant also influenced cholesterol synthesis in a Finnish population [17] and was associated with hypertriacylglycerolaemia in the men of the French population [16].
The present study aimed to replicate our previously reported association between the intronic C>T SNP (rs11868035) and the risk of type 2 diabetes. To identify whether this SNP is the true variant or whether other polymorphisms in the SREBF1c gene underlie the association with diabetes risk, we selected an additional five SNPs, such that coverage of the genetic variation in the SREBF1c gene and its promoter region was ensured. In total, six SNPs were genotyped in two case–control cohorts (the Cambridgeshire Case–Control Study and the European Prospective Investigation on Cancer [EPIC] Case–Control Study) to examine the contribution of each SNP and their haplotypes to the risk of type 2 diabetes. Subsequently, we genotyped the six SNPs in a large cohort study (the Ely Study) and tested for association with plasma glucose concentrations, fasting and at 120-min post-glucose load.
Subjects and methods
Study populations
Cambridgeshire Case–Control Study
The Cambridgeshire Case–Control Study consists of 552 patients with type 2 diabetes aged 45–76 years, randomly sampled from a population-based diabetes register and 552 controls, recruited from the same population and individually matched for age, sex and geographical location. The study design and methods have been described in detail elsewhere [18]. Briefly, cases were defined by onset of diabetes after the age of 30 years without insulin treatment in the first year following diagnosis. Diabetes was excluded in controls by medical record search, and by an HbA1c measurement of <6%.
The cases and controls were of the same age (controls: 63.8 years [SE 0.33] vs cases: 63.5 years [SE 0.33], p=0.54), but cases had a higher BMI compared with controls (controls: 27.3 kg/m2 [SE 0.18] vs cases: 29.7 [SE 0.22], p<0.001). The average age at onset of type 2 diabetes was 55.7 years (SD 9.0); 9% were treated with insulin, 64% with tablets and 27% followed a diet regimen. Of those with known family history, 32% (171 out of 529) of the cases had a family history for type 2 diabetes compared with 13% (71 out of 539) of the controls.
EPIC Case–Control Study
The EPIC Case–Control Study is nested within the EPIC-Norfolk Study, a population-based cohort study of men and women aged 40–78 years. Both the case–control and full cohort [19, 20] study have been previously described in detail. Briefly, the case–control study consists of 417 incident type 2 diabetes cases and two sets of 417 controls, each matched in terms of age, sex, general practice and recruitment date, with one set additionally matched for BMI. A case was defined by a physician's diagnosis of type 2 diabetes, with no insulin prescribed within the first year following diagnosis, and/or HbA1c >7% at the health check. Controls were randomly selected from the EPIC-Norfolk cohort from among those without diabetes, cancer, stroke or myocardial infarction at baseline and who had not developed diabetes by the time of selection. Potential controls with measured HbA1c levels >6% were excluded. The case and controls of the EPIC Case–Control study were of similar age (controls: 62.2 years [SE 0.3] vs cases: 62.2 years [SE 0.44], p=0.9). The BMI of cases was compared with controls (controls: 27.7 kg/m2 [SE 0.1] vs cases: 29.8 [SE 0.3], p<0.0001). Of those with known family history, 21% (73 out of 354) of the cases had a family history of type 2 diabetes compared with 9.5% (70 out of 741) of the controls.
Ely Study
The Ely Study is a population-based cohort study of the aetiology and pathogenesis of type 2 diabetes and related metabolic disorders. The study design and methods have been described in detail elsewhere [21]. This analysis included 1,721 men and women without diagnosed diabetes in Phase III of the Ely Study. The average age and BMI in the Ely cohort was 61.4 years (SD 9.0) and 27.3 kg/m2 (SD 3.9) for men and 60.8 years (SD 9.1) and 27.2 kg/m2 (SD 5.5) for women. Participants underwent standard anthropo-metric measurements and a 75-g OGTT having fasted since 22.00 h the previous night. Plasma samples were obtained at fasting and 120-min post-glucose load. Glucose concentrations were measured immediately in the routine laboratory using the hexokinase method.
Ethical permission
Ethical permission for the three studies was granted by their respective Local Research Ethics Committee, and study participants had provided informed consent.
SNP selection and genotyping
SNP selection
First, the C>T SNP located between exon 18c and 19c (rs11868035, SNP6) was selected to replicate our previous findings [15]. Subsequently, four additional SNPs (rs2236513 [SNP1], rs6502618 [SNP2], rs4925118 [SNP3] and rs1889018 [SNP4]) were selected to ensure coverage of the genetic variation in the SREBF1c gene and its promoter region (up to 10 kb before the start codon of isoform 1a). Finally, we added a sixth, non-synonymous, SNP (V556M [SNP5]) that was identified by re-sequencing the SREBF1 gene in the Severe Insulin Resistance cohort as described previously [15]. We used the data available in the International HapMap Project-Phase II (release no. 20) to estimate how much of the genetic variation in the SREBF1c gene and its promoter are captured by the selected SNPs [22]. The HapMap reports 13 SNPs that are polymorphic (minor allele frequency [MAF] >1%) in the CEPH population (Utah residents with northern and western European ancestry) [22]. The SNPs selected in the present study capture the genetic variation of 11 of the 13 HapMap SNPs, whereas two rare intronic HapMap SNPs (rs1653007, with MAF=0.033, and rs1108511, with MAF=0.026) remained uncovered.
Genotyping and quality control DNA was extracted from whole blood/EDTA samples (Whatman Biosciences, Ely, UK). Samples were arrayed on 96-well plates with three replicates and one water control per plate. For the case–control populations, cases and control samples were randomly distributed across each 96-well plate, with approximately the same number of cases and controls per plate. Genotyping of samples was performed in 384-well plates at the Wellcome Trust Sanger Institute, Cambridge, using an adaptation of the homogeneous MassExtend protocol supplied by Sequenom, Inc. for the MassArray system (Sequenom, Inc.) [23]. Call rates were on average ≥95% for SNP1, SNP2, SNP3, SNP5 and SNP6 and 87% for SNP4. Concordance rates between duplicate samples were ≥98%.
Statistical methods
A χ2 ‘goodness-of-fit’ test was performed to assess whether the observed genotype frequencies in the controls of the case–control studies, separately and combined, and in the cohort study, were in Hardy–Weinberg equilibrium (HWE). Linkage disequilibrium (LD) between SNPs was estimated in the controls of the combined case–control studies using Haploview V3.2 (http://www.broad.mit.edu/mpg/haploview).
In the case–control studies, separately and combined, each SNP was tested for association with type 2 diabetes in a logistic regression analysis, adjusted for age, sex and BMI. There was no evidence of heterogeneity between the two case–control studies (p>0.3) and the association between the SNPs and the risk of type 2 diabetes was the same in both studies (SNP-by-cohort interaction: p>0.33). Therefore, we present the results of the combined case–control studies only. In the cohort study, association between fasting plasma glucose and 120-min post-load plasma glucose concentrations and the six SNPs was tested in a multiple regression model, adjusting for age and sex. We applied two genetic models: (1) an additive model, which assumes an additive effect for the presence of none (wild-type homozygotes), one (heterozygotes) or two (rare homozygotes) rare alleles, and (2) a general model, which compares the three genotypes irrespectively of their carrier status. Likelihood ratio tests indicated that the general model was not significantly better than the additive model (p=0.35–0.99) such that we report the results of the additive genetic model only.
Haplotype frequencies were estimated using the HAPLOTYPE procedure in SAS/Genetics 9.1 (SAS Institute Inc., Cary, NC, USA). Haplotypes were arranged in frequency order, such that haplotype 1.1.1.1 is the most frequent. Haplotypes prevalent at >5% were retained for further haplotype analysis. In the case–control studies, we tested for association between the haplotypes and type 2 diabetes using a logistic regression analysis, adjusting for age, sex and BMI. In the cohort study, we used a general linear regression model, adjusted for age and sex, and weighted by the probability for haplotype and clustered by individual ID.
Statistical analyses were conducted using Stata SE 8.2 for Windows (StataCorp, College Station, TX, USA). Power calculations were performed using Quanto v1.1.1 (http://hydra.usc.edu/gxe).
Results and discussion
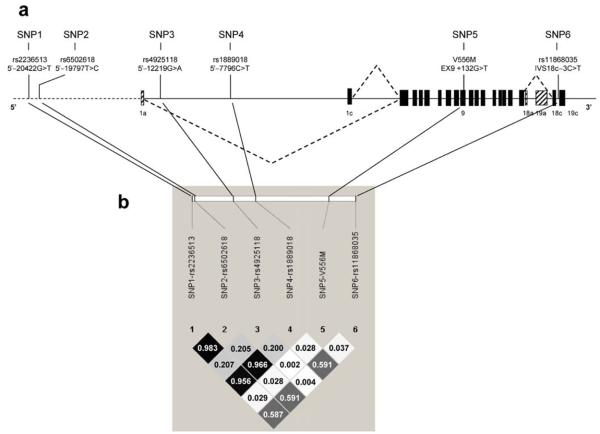
The genotype frequencies of the six SNPs were in HWE in the controls of the case–control studies (Table 1) and in the cohort study (Table 2). Three SNPs, SNP1 (rs2236513), SNP2 (rs6502618) and SNP4 (rs1889018), were in high (r2>0.95) LD with each other (Fig. 1). These three SNPs were in moderate LD (r2=0.59) with the previously reported C>T SNP (SNP6, rs11868035). LD with the two remaining SNPs, SNP3 (rs4925118) and SNP5 (V556M), was low (r2<0.21).
Table 1.
Association between the six SREBF1c SNPs and type 2 diabetes in the combined EPIC Case–Control Study and Cambridgeshire Case–Control Study
| SNP | Location | rs number | Status | Genotype counts (n) |
MAF | HWE p value | OR per minor allele (95% CI) | p value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 1/1 | 1/2 | 2/2 | ||||||||
| SNP1 | 5′-20422G>T | rs2236513 | Case | 318 | 378 | 128 | 0.38 | – | 1.23 (1.07–1.41) | 0.003 |
| Control | 522 | 560 | 140 | 0.34 | 0.58 | – | ||||
| SNP2 | 5′-19797T>C | rs6502618 | Case | 319 | 384 | 129 | 0.39 | – | 1.21 (1.05–1.38) | 0.006 |
| Control | 505 | 554 | 142 | 0.35 | 0.60 | – | ||||
| SNP3 | 5′-12219G>A | rs4925118 | Case | 668 | 135 | 16 | 0.10 | – | 1.03 (0.83–1.28) | 0.77 |
| Control | 983 | 220 | 13 | 0.10 | 0.86 | – | ||||
| SNP4 | 5′-7796T>C | rs1889018 | Case | 300 | 359 | 129 | 0.39 | – | 1.26 (1.10–1.44) | 0.001 |
| Control | 479 | 463 | 137 | 0.34 | 0.13 | – | ||||
| SNP5 | EX9+132G>T | – | Case | 801 | 29 | 1 | 0.02 | – | 1.25 (0.76–2.07) | 0.38 |
| V556M | Control | 1192 | 36 | 0 | 0.01 | 0.60 | – | |||
| SNP6 | IVS18c-3C>T | rs11868035 | Case | 379 | 371 | 89 | 0.33 | – | 1.20 (1.04–1.38) | 0.014 |
| Control | 614 | 515 | 98 | 0.29 | 0.49 | – | ||||
The ORs and p values were obtained from logistic regression analysis using the additive genetic model adjusted for age, sex and BMI. The OR reflects the increased risk with each extra minor allele
Table 2.
Association between the six SREBF1c SNPs and plasma glucose concentrations (fasting and at 120-min post-OGTT) in the Ely Study
| SNP | Location | rs number | Time of sample | HWE p value |
Plasma glucose concentration (mmol/l) |
p valuec | ||
|---|---|---|---|---|---|---|---|---|
| Genotype (95% CI)a |
||||||||
| 1/1 | 1/2 | 2/2 | ||||||
| SNP1 | 5′-20422G>T | rs2236513 | Fasting (n=714/716/196)b |
0.42 | 5.0 (5.0–5.1) | 5.0 (5.0–5.1) | 5.1 (5.0–5.2) | 0.084 |
| At 120 min (n=666/660/184)b | 0.30 | 5.0 (5.8–6.1) | 6.0 (5.8–6.1) | 6.4 (6.1–6.7) | 0.085 | |||
| SNP2 | 5′-19797T>C | rs6502618 | Fasting (n=682/719/191)b |
0.94 | 5.0 (5.0–5.1) | 5.1 (5.0–5.1) | 5.1 (5.0–5.2) | 0.43 |
| At 120 min (n=632/665/178)b | 0.88 | 6.0 (5.8–6.1) | 6.0 (5.9–6.2) | 6.4 (6.1–6.7) | 0.27 | |||
| SNP3 | 5′-12219G>A | rs4925118 | Fasting (n=1,289/321/26)b |
0.25 | 5.0 (5.0–5.1) | 5.1 (5.0–5.2) | 5.0 (4.8–5.3) | 1.00 |
| At 120 min (n=1,199/293/24)b | 0.21 | 6.0 (5.9–6.1) | 6.1 (5.9–6.4) | 6.1 (5.4–7.0) | 0.92 | |||
| SNP4 | 5′-7796T>C | rs1889018 | Fasting (n=630/654/188)b |
0.37 | 5.0 (5.0–5.1) | 5.0 (5.0–5.1) | 5.1 (5.0–5.2) | 0.084 |
| At 120 min (n=585/601/177)b | 0.25 | 5.9 (5.8–6.1) | 5.9 (5.8–6.1) | 6.3 (5.9–6.6) | 0.085 | |||
| SNP5 | EX9+132G>T | – | Fasting (n=1,594/50/1)b |
0.35 | 5.0 (5.0–5.1) | 5.0 (4.8–5.2) | 5.8 (4.4–7.6) | 0.43 |
| V556M | At 120 min (n=1,476/49/1)b | 0.37 | 6.0 (5.9–6.2) | 5.6 (5.1–6.2) | 6.4 (3.3–12.5) | 0.27 | ||
| SNP6 | IVS18c- | rs11868035 | Fasting (n=368/274/48)b |
0.76 | 5.0 (4.9–5.1) | 5.0 (4.9–5.1) | 5.0 (5.0–5.2) | 1.00 |
| 3C>T | At 120 min (n=338/248/43)b |
0.78 | 5.8 (5.8–6.0) | 6.1 (5.8–6.3) | 5.8 (5.2–6.4) | 0.92 | ||
Data are geometric means adjusted for age and sex
Numbers represent genotype count of homozygotes for the common allele, heterozygotes and homozygotes for the minor allele, respectively
The p values were obtained from regression analysis using the additive genetic model adjusted for age and sex
Fig. 1.
a Location of the six SREBF1c SNPs. The SNP location of SNP1, SNP2, SNP3 and SNP4 reflects their relative position (in bp) to the first nucleotide of exon 1c. SNP5 is located 132 bp after first nucleotide of exon 9, and SNP6 is located three nucleotides before the start of exon 19c. b LD (r2) between the six SREBF1c SNPs in the controls of the combined EPIC Study and Cambridgeshire Case–Control Study
In the case–control studies, we tested for association between the six SNPs and type 2 diabetes risk using the additive logistic regression model (Table 1). The minor allele (T-allele) of SNP6 (rs11868035), for which we previously reported significant associations [15], was also in the present study significantly associated with an increased risk of type 2 diabetes. Corroborating evidence comes from a French study that reported a significant association between the minor allele of the G952G variant (rs2297508), located 216 bp downstream of SNP6, and an increased risk of type 2 diabetes [16]. According to data from the International HapMap, LD between SNP6 and the G952G variant is high (D′=1 and r2=0.87) [22], suggesting that the associations found in the present study, our previous study [15] and the French study [16] may be caused by SNP6, the G952G variant or a currently unidentified SREBF1c gene variant that is in LD with both of these SNPs. In addition to SNP6, we also found that the minor alleles of SNP1, SNP2 and SNP4 were significantly associated with an increased risk of type 2 diabetes (Table 1). It should be noted, however, that LD between these three SNPs is high and, therefore, the three association results cannot be considered as independent findings.
In the cohort study, we tested for association between the six SNPs and plasma glucose levels (fasting and at 120 min after loading) using the additive linear regression model. None of the associations reached statistical significance. However, the minor alleles of SNP1 and SNP4 tended to be associated with increased plasma glucose concentrations, supporting the observation from the case–control studies that also showed an increased diabetes risk with the minor alleles of these SNPs. When a recessive genetic model was applied to SNP1 and SNP4, i.e. comparing rare homozygotes with carriers of the wild-type allele, borderline significant associations were obtained for plasma glucose concentrations at 120 min (SNP1: p=0.05 and SNP4: p=0.03).
The case–control study is sufficiently powered (80% at α=0.05) for detecting a genetic effect of 0.05 of odds ratios (ORs) >1.2 for SNPs with a MAF >33% (SNP1, SNP2, SNP4 and SNP6), OR >1.35 for SNP3 (MAF=10%) and OR >1.75 for SNP5 (MAF=2%). The cohort study has sufficient power (80% at α=0.05) for detecting difference between genotypes in fasting plasma glucose concentration of >0.15 mmol/l for SNP1, SNP2, SNP4 and SNP6, of >0.25 mmol/l for SNP3 and of >0.55 mmol/l for SNP5. These power analyses show that the cohort study was not sufficiently powered, as the observed differences were below the detectable level for the given samples size and MAF. Therefore, replication in other cohort studies is required to confirm our findings.
The case–control studies and the cohort study both provide evidence that SNP1, SNP2 and SNP4 may influence plasma glucose concentrations and diabetes risk, respectively. These three SNPs are located in the 5′ region, 7.8–20.4 kb before the start of exon 1c and therefore most probably too distant to be considered as promoter SNPs. However, the HapMap reports a SNP (rs8066560), possibly located in the putative promoter region at −1,099 bp of exon 1c, that is in high LD with the SNPs in the 5′ region (r2>0.95). The promoter region has been shown to be of importance in the transcriptional activation of the gene [24-26]. Analyses of the mouse SREBF1c gene have identified several functional regions in the promoter region, such as a sterol regulatory element complex, a sterol inducible region and a liver X receptor-retinoid X receptor region [24, 25]. The human SREBF1c promoter contains additional binding sites for pyruvate dehydrogenase complex-1 and hepatocyte nuclear factor-4 that upon binding upregulate the promoter activity [26]. Functional studies will be required to identify whether SNP1, SNP2, SNP4 or another SNP in LD affect any of these promoter binding sites and/or the transcriptional activity of SREBF1c.
Haplotype analyses will be more powerful than single SNP analyses when none of the genotyped SNPs are associated with disease, as haplotypes may represent genetic variation that is not captured by any of the independently analysed SNPs. Therefore, we performed haplotype analyses with four haplotype-tagging SNPs (SNP2, SNP3, SNP5 and SNP6). Eight haplotypes were present in the studied populations; however, only three were common (haplotype frequency >5%) and used for further analyses (Table 3). Relative to the most common haplotype (1.1.1.1), haplotype 2.1.1.2 significantly increased the risk of type 2 diabetes in the case–control studies. These analyses show that the simultaneous presence of the rare alleles of SNP2 and SNP6 (haplotype 2.1.1.2) significantly increased the risk of type 2 diabetes, which confirms the results of the single SNP analyses. The effect size for the 2.1.1.2 haplotype is similar to that of the single SNPs, suggesting that the haplotype does not provide additional information beyond the single SNPs. In the cohort study, there was no evidence for association between the haplotypes and plasma glucose concentrations.
Table 3.
Association between SREBF1c haplotypes and risk of type 2 diabetes in the case–control studies and plasma glucose concentrations (fasting and at 120 min) in the Ely Study
| Haplotypea |
p value | |||
|---|---|---|---|---|
| 1.1.1.1 | 2.2.1.1 | 2.1.1.2 | ||
| Case–control studies | ||||
| Haplotype frequency, % (n) | ||||
| Cases | 59.7 (919) | 5.2 (80) | 24.3 (374) | |
| Controls | 62.9 (1,444) | 5.9 (135) | 21.1 (485) | |
| OR (95% CI) | 1 (Reference) | 0.93 (0.70–1.24) | 1.22 | 0.041 |
| Ely Study | ||||
| Haplotype frequency, % | 63.4 | 7.5 | 18.8 | |
| Fasting glucose (mmol/l) (95% CI) | 5.0 (5.0–5.1) | 5.0 (4.9–5.1) | 5.0 (5.0–5.1) | 0.79 |
| 120-min glucose (mmol/l) (95% CI) | 5.9 (5.7–6.0) | 6.2 (5.8–6.6) | 6.1 (5.8–6.4) | 0.10 |
Haplotypes are based on SNP2, SNP3, SNP5 and SNP6
Data are adjusted for age and sex
In summary, our findings support the hypothesis that the SREBF1c gene is a candidate gene for the risk of type 2 diabetes and variation in plasma glucose concentrations. We replicated our previous finding suggesting that the SREBF1c SNP located between exon 18c and 19c (rs11868035) is associated with an increased risk of type 2 diabetes in a new case–control study. In addition, we found convincing evidence for three other SNPs, located in the 5′ region of the SREBF1c gene, to be associated with the risk of type 2 diabetes. Two SNPs (SNP1 and SNP4) in the 5′ region, were also weakly related to plasma glucose concentration in a large cohort, supporting the observed association with diabetes risk in the case–control studies.
Acknowledgements
We are grateful to the volunteers who participated in the Cambridgeshire Case–Control Study, the EPIC-Norfolk Study and the Ely Study. We also thank S. Bumpstead for technical assistance. The Cambridgeshire Case–Control Study was funded by the Wellcome Trust, the Ely Study was funded by the Medical Research Council (MRC) and the Wellcome Trust, and the EPIC-Norfolk Study is supported by programme grants from the UK MRC and Cancer Research UK. S. O'Rahilly and I. Barosso acknowledge support from EU FP6 funding (contract no. LSHM-CT-2003-503041). Additional support was provided by the Stroke Association, British Heart Foundation, Department of Health, Food Standards Agency and the Wellcome Trust.
Abbreviations
- EPIC
European Prospective Investigation on Cancer
- HWE
Hardy–Weinberg equilibrium
- LD
linkage disequilibrium
- MAF
minor allele frequency
- OR
odds ratio
- SNP
single-nucleotide polymorphism
- SREBF1c
sterol regulatory element-binding factor-1c
Footnotes
Duality of interest The authors declare that they have no duality of interest.
References
- 1.Lelliott C, Vidal-Puig A. Lipotoxicity, an imbalance between lipogenesis de novo and fatty acid oxidation. Int J Obes Relat Metab Disord. 2004;28:S22–S28. doi: 10.1038/sj.ijo.0802854. [DOI] [PubMed] [Google Scholar]
- 2.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osborne TF. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J Biol Chem. 2000;275:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- 4.Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimomura I, Bashmakov Y, Shimano H, Horton JD, Goldstein JL, Brown MS. Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver. Proc Natl Acad Sci USA. 1997;94:12354–12359. doi: 10.1073/pnas.94.23.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ducluzeau PH, Perretti N, Laville M, et al. Regulation by insulin of gene expression in human skeletal muscle and adipose tissue: evidence for specific defects in type 2 diabetes. Diabetes. 2001;50:1134–1142. doi: 10.2337/diabetes.50.5.1134. [DOI] [PubMed] [Google Scholar]
- 7.Sewter C, Berger D, Considine RV, et al. Human obesity and type 2 diabetes are associated with alterations in SREBP1 isoform expression that are reproduced ex vivo by tumor necrosis factor-alpha. Diabetes. 2002;51:1035–1041. doi: 10.2337/diabetes.51.4.1035. [DOI] [PubMed] [Google Scholar]
- 8.Espenshade PJ, Li WP, Yabe D. Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proc Natl Acad Sci USA. 2002;99:11694–11699. doi: 10.1073/pnas.182412799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell. 2002;10:237–245. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Nakamura MT, Cho HP, Clarke SD. Sterol regulatory element binding protein-1 expression is suppressed by dietary polyunsaturated fatty acids: a mechanism for the coordinate suppression of lipogenic genes by polyunsaturated fats. J Biol Chem. 1999;274:23577–23583. doi: 10.1074/jbc.274.33.23577. [DOI] [PubMed] [Google Scholar]
- 11.Kim HJ, Takahashi M, Ezaki O. Fish oil feeding decreases mature sterol regulatory element-binding protein 1 (SREBP-1) by down-regulation of SREBP-1c mRNA in mouse liver: a possible mechanism for down-regulation of lipogenic enzyme mRNAs. J Biol Chem. 1999;274:25892–25898. doi: 10.1074/jbc.274.36.25892. [DOI] [PubMed] [Google Scholar]
- 12.Foretz M, Pacot C, Dugail I, et al. ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose. Mol Cell Biol. 1999;19:3760–3768. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JB, Sarraf P, Wright M, et al. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lay SL, Lefrere I, Trautwein C, Dugail I, Krief S. Insulin and sterol-regulatory element-binding protein-1c (SREBP-1C) regulation of gene expression in 3T3-L1 adipocytes. Identification of CCAAT/enhancer-binding protein beta as an SREBP-1c target. J Biol Chem. 2002;277:35625–35634. doi: 10.1074/jbc.M203913200. [DOI] [PubMed] [Google Scholar]
- 15.Laudes M, Barroso I, Luan J, et al. Genetic variants in human sterol regulatory element binding protein-1c in syndromes of severe insulin resistance and type 2 diabetes. Diabetes. 2004;53:842–846. doi: 10.2337/diabetes.53.3.842. [DOI] [PubMed] [Google Scholar]
- 16.Eberle D, Clement K, Meyre D, et al. SREBF-1 gene polymorphisms are associated with obesity and type 2 diabetes in French obese and diabetic cohorts. Diabetes. 2004;53:2153–2157. doi: 10.2337/diabetes.53.8.2153. [DOI] [PubMed] [Google Scholar]
- 17.Laaksonen R, Thelen KM, Paiva H, et al. Genetic variant of the SREBF-1 gene is significantly related to cholesterol synthesis in man. Atherosclerosis. 2006;185:206–209. doi: 10.1016/j.atherosclerosis.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Rathmann W, Haastert B, Icks A, et al. Low faecal elastase 1 concentrations in type 2 diabetes mellitus. Scand J Gastroenterol. 2001;36:1056–1061. doi: 10.1080/003655201750422657. [DOI] [PubMed] [Google Scholar]
- 19.Harding AH, Day NE, Khaw KT, et al. Dietary fat and the risk of clinical type 2 diabetes: The European Prospective Investigation of Cancer-Norfolk Study. Am J Epidemiol. 2004;159:73–82. doi: 10.1093/aje/kwh004. [DOI] [PubMed] [Google Scholar]
- 20.Day NE, Oakes S, Luben R, et al. EPIC-Norfolk: study design and characteristics of the cohort. European prospective investigation of cancer. Br J Cancer. 1999;80:95–103. [PubMed] [Google Scholar]
- 21.Williams DR, Wareham NJ, Brown DC, et al. Undiagnosed glucose intolerance in the community: the Isle of Ely diabetes project. Diabet Med. 1995;12:30–35. doi: 10.1111/j.1464-5491.1995.tb02058.x. [DOI] [PubMed] [Google Scholar]
- 22.The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whittaker P, Bumpstead S, Downes K, Ghori J, Deloukas P. SNP analysis by MALDI-TOF mass spectrometry. In: Celis J, Carter N, Simons K, Small JV, Hunter T, editors. Cell biology: a laboratory handbook. 3rd edn. Academic Press; San Diego: 2006. in press. [Google Scholar]
- 24.Yoshikawa T, Shimano H, Memiya-Kudo M, et al. Identification of liver X receptor–retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol Cell Biol. 2001;21:2991–3000. doi: 10.1128/MCB.21.9.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Memya-Kudo M, Shimano H, Yoshikawa T, et al. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J Biol Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- 26.Tarling E, Salter A, Bennett A. Transcriptional regulation of human SREBP-1c (sterol-regulatory-element-binding protein-1c): a key regulator of lipogenesis. Biochem Soc Trans. 2004;32:107–109. doi: 10.1042/bst0320107. [DOI] [PubMed] [Google Scholar]