Abstract
SRp38 is an atypical SR protein that functions as a general splicing repressor when dephosphorylated. We now show that phosphorylated SRp38 functions as a sequence-specific splicing activator. Unlike characterized splicing activators, SRp38 functions in the absence of other SR proteins but requires a cofactor for activity. SRp38 was able to induce formation of splicing complex A in the absence of the cofactor, but this factor was necessary for progression to complexes B and C. Mechanistically, SRp38 strengthens the ability of the U1 and U2 small nuclear ribonucleoproteins to stably recognize the pre-mRNA. Extending these findings, analysis of alternative splicing of pre-mRNA encoding the glutamate receptor B revealed that SRp38 alters its splicing pattern in a sequence-specific manner. Together, our data demonstrate that SRp38, in addition to its role as a splicing repressor, can function as an unusual sequence-specific splicing activator.
Alternative splicing is a common mechanism for regulating gene expression and increasing protein diversity in metazoan organisms1. In humans, more than 70% of primary transcripts are estimated to undergo alternative splicing2. Splicing is carried out in the spliceosome, in which five small nuclear ribonucleoprotein particles (snRNPs; U1, U2, U4/U6 and U5) and many auxiliary proteins cooperate to accurately recognize the splice sites and catalyze the two steps of the splicing reaction3.
The inclusion of a specific exon in the mature mRNA is largely dependent on the recognition and usage of the flanking splice sites by the splicing machinery4. This seems to be governed by the dynamic formation of protein complexes on the pre-mRNA. Specific sets of splicing regulatory proteins assemble on different pre-mRNAs, generating a ‘splicing’ or ‘mRNP’ code that determines exon recognition5,6. Cis-elements known as exonic splicing enhancers (ESEs) modulate the assembly of regulatory proteins on pre-mRNA and therefore contribute to splice-site choice. Most characterized ESEs have been found adjacent to introns containing (a) weak splice site(s), despite the finding that constitutive exons are also frequently enriched in potential binding sites for splicing regulatory proteins7. ESEs were initially identified as purine-rich sequences8,9, but different motifs have been identified by different strategies10,11.
ESEs are often recognized and bound by SR proteins, a family of highly conserved splicing factors that have key roles in spliceosome activation and in regulation of splice-site selection12–14. SR proteins contain one or two N-terminal RNP-type RNA binding domains (RBD) and a C-terminal arginine- and serine-rich (RS) domain. The RBDs of SR proteins are necessary and sufficient for RNA binding, whereas the RS domains are involved in protein-protein15,16 and perhaps protein-RNA17 interactions. SR proteins function as essential general splicing factors, necessary for spliceosome assembly in in vitro assays12,14,18.
Many studies have identified ESEs that activate 3′ or 5′ splice sites by binding SR proteins19. The ESE-bound SR proteins can stimulate splicing by recruiting the general splicing factor U2AF to weak polypyrimidine tracts via a direct interaction between the RS domains of the SR proteins and U2AF20,21. They are also involved in bridging interactions between ESEs and spliceosomal components, probably mediated by the SRm160 and SRm300 splicing coactivators22,23. These coactivators contain RS domains but without RBD domains, and can form multiple interactions with snRNPs and enhancer-bound SR proteins. In addition, a series of sequential RS domain–RNA contacts at the branch point and the 5′ splice site during splicing complex assembly have been documented, suggesting that RS domain–RNA interactions might also contribute to splicing activation by ESE-bound SR proteins17. ESEs also activate 5′ splice sites, as exemplified by fruitless pre-mRNA splicing in Drosophila melanogaster24.
Previously, we described an unusual member of the SR protein family, SRp38 (also known as FUSIP1), which functions differently from standard SR proteins. Although the domain organization of SRp38 is typical of SR proteins, SRp38 is unable to activate splicing in standard in vitro assays. In contrast to other SR proteins, SRp38 functions as a general splicing repressor25,26. Its repression activity is turned on by tightly regulated dephosphorylation27, and it is required for global inhibition of splicing both in M phase of the cell cycle and following heat shock28. Although present at high levels in numerous cell types and tissues, no function has as yet been assigned to phosphorylated SRp38.
In this report, we address this question by showing that phosphorylated SRp38 functions as a sequence-specific splicing activator in in vitro assays. We show that, unlike other characterized sequence-specific regulators, such as Transformer-2 (TRA2, also known as SFRS10; ref. 9), SRp38 does not require other SR proteins to function but, unlike typical SR proteins, requires a nuclear cofactor for activity. Notably, SRp38 is sufficient to induce formation of spliceosomal complex A in a cell extract lacking SR proteins (S100), but the complexes are stalled and require the cofactor to progress to active splicing complexes. By analysis of alternative splicing of a specific pre-mRNA in vitro and in vivo, we demonstrate that SRp38 can affect the selection of mutually exclusive exons in a sequence-specific manner, reflecting affinity for SRp38. We therefore conclude that SRp38 is a previously uncharacterized type of splicing factor capable of switching from a general repressor to a sequence-specific activator and regulator of alternative splicing.
RESULTS
SRp38 functions as a sequence-specific activator of splicing
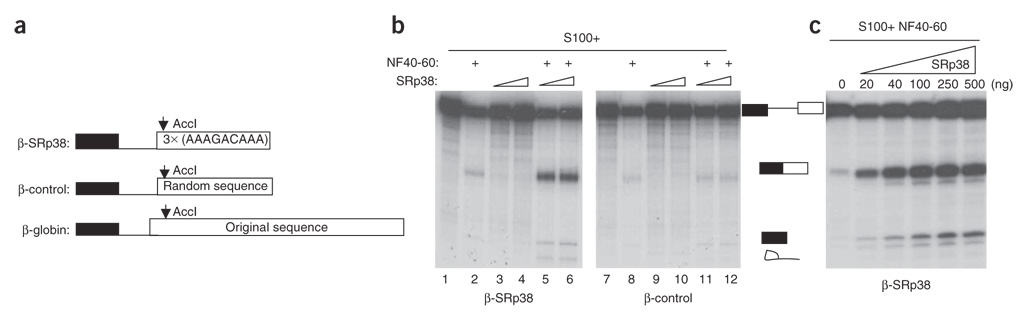
Previous work from our laboratory has shown that SRp38 functions as a general splicing repressor when dephosphorylated. Although that work showed that phosphorylated SRp38 cannot function like other SR proteins, as a general splicing activator26,28, we decided to test whether it might be able to function as a sequence-specific activator. To this end, we took advantage of the high-affinity binding of SRp38 to its consensus recognition sequence (AAAGACAAA), previously determined by SELEX26, and constructed a modified β-globin pre-mRNA substrate in which three copies of this sequence were used to replace sequences in the downstream exon (designated β-SRp38; Fig. 1a). In parallel, we also constructed a control substrate in which random sequences of the same size were inserted into the same position as the SRp38 consensus sequence (Fig. 1a). We then tested whether phosphorylated His-tagged SRp38, purified from recombinant baculovirus-infected insect cells26, could activate splicing of the β-SRp38 RNA in HeLa S100 extract. However, we detected no splicing (Fig. 1b, lanes 3 and 4), even when high concentrations of SRp38 were used (see below).
Figure 1.
SRp38 functions as a sequence-specific activator of splicing. (a) Schematic representation of β-globin and its derivatives, containing either three copies of the SRp38 consensus sequence or a control sequence. (b) Activation of β-SRp38 pre-mRNA splicing by SRp38. Splicing was performed in S100 supplemented with 50 ng (lanes 3, 5, 9 and 11) or 100 ng (lanes 4, 6, 10 and 12) of baculovirus-produced His-SRp38, with or without a nuclear fraction (NF40–60), as indicated above. Products of splicing were analyzed by denaturing PAGE and autoradiography. Splicing products are indicated schematically. (c) Dose-dependent splicing activation of SRp38. The indicated amounts of His-SRp38 were added to splicing reactions performed in S100 supplemented with NF40–60.
One explanation for the inactivity of SRp38 in the above assay is that a cofactor not present in S100 is required for activity. To test this possibility, we prepared ammonium sulfate fractions of HeLa nuclear extract and tested these together with SRp38 and S100. In the presence of a 40–60% saturation cut of nuclear extract (NF40–60), we observed strong activation by SRp38 with the β-SRp38 but not the β-control RNA (Fig. 1b, compare lanes 5 and 6 with lanes 11 and 12). Notably, activation of splicing was dose dependent, with considerable splicing observed with as little as 20 ng of purified His-tagged SRp38 (Fig. 1c). These results indicate that three copies of the SRp38 binding sequence can function as an SRp38-specific ESE, and that SRp38 can indeed function as a splicing activator.
Characteristics of SRp38-dependent splicing activation
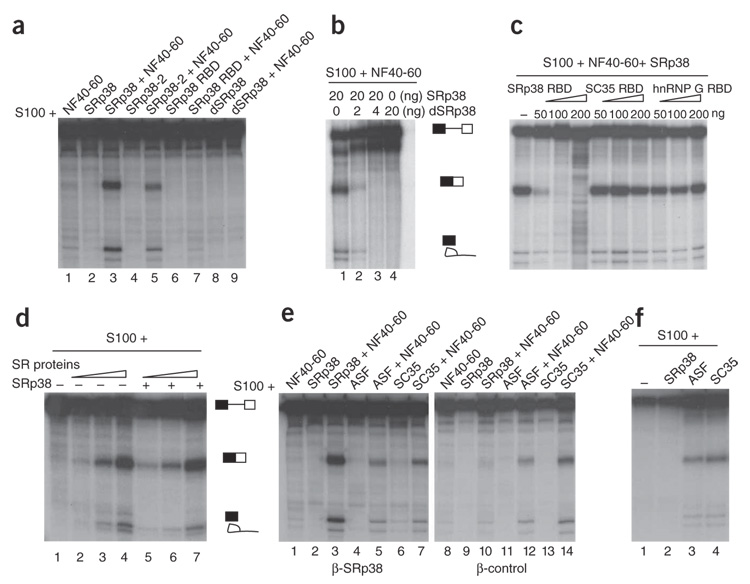
We next tested what properties of the SRp38 RS region affect the protein’s activity in splicing. Purified glutathione S-transferase (GST) derivatives of SRp38, a splice variant, SRp38-2, which contains a short version of the RS domain26, SRp38 RBD, which lacks the whole RS domain, and dephosphorylated SRp38 (dSRp38) were each added to splicing reactions with S100 or S100 plus NF40–60 and the β-SRp38 RNA (Fig. 2a). Whereas SRp38 RBD was completely inactive (lane 7), we did observe splicing activation with SRp38-2, again in a manner dependent on the presence of NF40–60, but splicing efficiency was greatly reduced compared to full-length SRp38 (lane 5, compare to lane 3). Notably, dSRp38 was unable to activate splicing of the β-SRp38 RNA (lane 9). In fact, splicing activated by 20 ng phosphorylated SRp38 was completely repressed by low amounts of dSRp38 (2–4 ng; Fig. 2b, lanes 1–4), consistent with its previously described properties as a general splicing repressor26. These results indicate that the phosphorylated RS domain of SRp38 is required for splicing activation. (The phosphorylation status of both SRp38 and dSRp38 did not change during the splicing reaction; Supplementary Fig. 1 online.) Thus, although the phosphorylation and dephosphorylation state might serve as an on-off switch for standard SR proteins29, its function in the case of SRp38 is to switch from splicing activation to repression.
Figure 2.
Characteristics of SRp38-dependent splicing activation. (a) RS domain requirements for activation. β-SRp38 pre-mRNA was incubated in S100 activated by 50 ng of GST-SRp38, GST–SRp38-2 or GST–SRp38 RBD, or 8 ng of dephosphorylated His-SRp38 (dSRp38), respectively, in the absence (lanes 2, 4, 6 and 8) or presence (lanes 3, 5, 7 and 9) of NF40–60. (b) Repression of splicing by dSRp38. His-dSRp38 (0 ng, 2 ng, 4 ng and 20 ng) was incubated with S100 plus phosphorylated His-SRp38 (20 ng) and NF 40–60 (lanes 10–12). (c) Role of the RBD in SRp38-dependent activation. The indicated amounts of purified GST–SRp38 RBD, His–SC35 RBD or GST–hnRNP G RBD were added to SRp38-dependent splicing assays as indicated. (d) SR proteins do not cooperate with SRp38 for splicing activation. Increasing amounts of purified SR proteins were added to reactions performed in S100 alone (lanes 2–4) or in the presence of 50 ng of GST-SRp38 (lanes 5–7). (e) NF40–60 coactivator activity is specific for SRp38. His-SRp38, His-ASF or His-SC35 (50 ng) was incubated with β-SRp38 or β-SRp38 RNAs in S100 alone or supplemented with NF40–60. (f) Splicing stimulation by ASF/SF2 or SC35 at high concentrations. His-tagged SRp38, ASF/SF2 and SC35 (300 ng) was incubated with β-SRp38 RNA in S100.
We next investigated the specificity that the SRp38 RBD provides in recognizing the β-SRp38 RNA. To this end, we added purified GST–SRp38 RBD, His–SC35 RBD and GST–heterogeneous ribonucleoprotein G (hnRNP G) RBD to β-SRp38 RNA–containing splicing reactions in S100 plus SRp38 and NF40–60. The SC35 RBD shares the highest identity with the SRp38 RBD (46%), whereas the hnRNP G RBD shows only limited identity. The results show that the SRp38 RBD, but not the SC35 or hnRNP G RBDs, inhibited splicing brought about by SRp38 plus NF40–60 (Fig. 2c). Notably, the amount of SRp38 RBD sufficient to block splicing was roughly equivalent to the amount of SRp38 added. The inhibition of splicing thus reflects a specific, competitive interaction between full-length SRp38 and the SRp38 RBD, providing evidence that the SRp38 RBD–RNA interaction is indeed highly specific.
Next, we examined the possible role of standard SR proteins in SRp38 activity. We tested this by determining whether splicing of the β-SRp38 RNA in S100 activated by increasing amounts of purified SR proteins could be enhanced by SRp38 (Fig. 2d). The results show that SRp38 was not able to function with the SR proteins to increase splicing beyond the levels provided by the SR proteins alone (compare lanes 2–4 with 5–7), indicating that SR proteins cannot provide the coactivator activity present in NF40–60.
SRp38 is distinct from other SR proteins in requiring a nuclear fraction to activate splicing in S100. To investigate this requirement further, we examined how NF40–60 influences the activity of standard SR proteins. When ASF/SF2 and SC35 (50 ng) were added to splicing reactions containing the β-SRp38 RNA, splicing was also enhanced by NF40–60 but relatively weakly (Fig. 2e, lanes 1–7). Highlighting the difference between SRp38 and other SR proteins, ASF/SF2 and SC35 plus NF40–60 gave rise to comparable levels of splicing with the β-control RNA, whereas SRp38 was completely inactive (Fig. 2e, lanes 8–14). Given that classical SR proteins typically function in S100 in the absence of a nuclear fraction, we next asked whether higher levels of all three proteins (300 ng) could activate splicing of β-SRp38 (Fig. 2f) or β-control (data not shown) RNA in S100 without NF40–60. Notably, whereas ASF/SF2 and SC35 activated splicing of both substrates, SRp38 was entirely inactive. These results indicate that activation by SRp38, unlike that by other SR proteins, is entirely dependent on a coactivator.
SRp38 facilitates the formation of spliceosomal complex A
We next investigated the mechanism by which SRp38 stimulates splicing. To this end, we first performed spliceosome-assembly assays using the β-SRp38 RNA and S100 extract alone or S100 containing SRp38 in the presence or absence of NF40–60 (Fig. 3a). Unexpectedly, the results of a time course showed that SRp38 in the absence of NF40–60 was able to bring about efficient assembly of what seems to be spliceosomal complex A (Fig. 3a, lanes 10–12). It was especially notable that nearly the entire nonspecific H complex was converted to the A-like complex by SRp38 alone, which is atypical and considerably more efficient than when splicing was in fact activated by SRp38 plus NF40–60 (lanes 14–16). No complex was detected in reactions containing β-SRp38 RNA with S100 (lanes 2–4), with NF40–60 (lanes 6–8) alone or with dSRp38 (Supplementary Fig. 2a online). These results suggest that phosphorylated SRp38 promotes an early step in the splicing pathway by forming early spliceosomal complexes.
Figure 3.
SRp38 promotes formation of spliceosomal complex A. (a) Spliceosome-assembly assays were carried out in S100 complemented with the indicated components and the β-SRp38 pre-mRNA. Splicing complexes were resolved on a 1.5% low-melting agarose gel. (b) SRp38 activates AdML-SRp38 in in vitro splicing assays. Splicing was performed in S100 supplemented with 50 ng of GST-tagged SRp38 with (lane 1) or without (lane 3) NF40–60. Products of splicing were analyzed by denaturing PAGE and autoradiography. (c) Spliceosome assembly was performed the same as in a, except with the AdML-SRp38 pre-mRNA. NE, nuclear extract. (d) Spliceosome-assembly assays were carried out as in c, except with the additions indicated at the top. Anti-U1, anti-U2, anti-U5 or anti-U6 snRNA oligonucleotides (5 µM) were added to reaction mixtures. Endogenous ATP was depleted by preincubating S100 at 30 °C for 40 min.
We next wished to confirm the identities of the complexes described above and extend our findings to a second RNA substrate. To this end, we constructed a modified adenovirus major late substrate RNA (AdML-SRp38) containing three copies of the SRp38 binding sites in its second exon. SRp38 gave rise to the same sequence-dependent splicing activation with this RNA as observed with β-SRp38 RNA and again required NF40–60 for activity (Fig. 3b). We then examined the effect of SRp38 on spliceosome assembly with the AdML-SRp38 RNA. We observed efficient formation of a spliceosomal complex in S100 supplemented with SRp38 alone (lanes 2–3) but not in S100 (lanes 8–9) or with SRp38 alone (lanes 11–12) (Fig. 3c). Notably, the mobility of the complex was identical to the A complex observed in nuclear extract (compare lanes 2 and 3 with 5 and 6). Furthermore, B and C complexes were again not detected (lanes 1–3; longer exposure in lanes 13–15). To provide evidence that the apparent stalled A complex could give rise to B and C complexes in the presence of NF40–60, we performed spliceosome assays in which NF40–60 was added to reaction mixtures containing SRp38 plus S100. B and C complexes were formed in addition to A complex (lanes 16–18). These results suggest that SRp38 promotes formation of spliceosomal A complexes, but these are stalled in the absence of NF40–60.
The above experiments provide evidence that SRp38 can facilitate A complex formation in S100. To provide additional evidence that these complexes indeed correspond to A complex, we first analyzed whether snRNPs were involved in their formation. For this, we used antisense RNA oligonucleotides complementary to U1, U2, U5 and U6 snRNAs to test which, if any, of these could inhibit complex formation in S100 plus SRp38. Complex formation with the AdML-SRp38 RNA was efficiently blocked by antisense oligonucleotides against U1 (lane 2) and U2 (lane 3) snRNAs but not against U5 (lane 4) or U6 (lane 5) snRNAs (Fig. 3d), indicating that U1 and U2 snRNPs were involved in complex formation, consistent with the properties of A complex. In addition, depletion of ATP (lane 6), lack of Mg2+ (lane 7) or incubation of splicing reactions on ice (lane 8) completely inhibited complex formation, ruling out the possibility that the observed complex might be related to the ATP-independent E complex. We used the same strategy to characterize the complex formed on the β-SRp38 substrate (Fig. 3a). Notably, the anti-U1 and anti-U2 oligonucleotides blocked complex formation, whereas the anti-U5 oligonucleotide did not, indicating that A complex but not B complex was formed on this substrate (Supplementary Fig. 2b). Together, these results demonstrate that SRp38 facilitates the formation of A complex in splicing-competent S100 extracts, but the complex is stalled in the absence of a cofactor.
SRp38 strengthens pre-mRNA recognition by U1 and U2 snRNPs
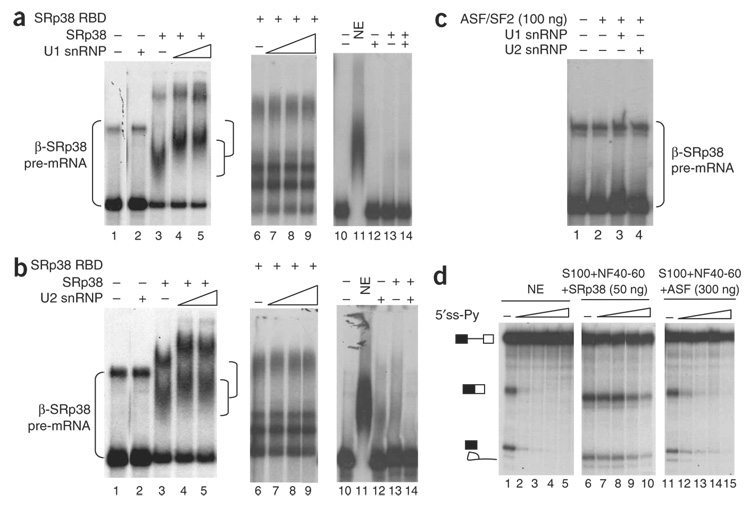
We next investigated the mechanism by which SRp38 facilitates complex A formation. One possibility is that it enhances interactions of U1 and/or U2 snRNPs with the pre-mRNA. To investigate this, we carried out gel-shift assays with purified U1 and U2 snRNPs and SRp38 with the β-SRp38 RNA used in splicing (Fig. 4). As expected, SRp38 bound to the RNA tightly (Fig. 4a, lane 3), whereas, under the conditions used16, U1 snRNP alone did not interact with β-SRp38 mRNA (lane 2). However, the presence of SRp38 stimulated formation of a stable (heparin-resistant) complex of U1 snRNP, SRp38 and RNA (lanes 4 and 5). The SRp38 RBD, although it was able to form an RNA-protein complex by itself, was unable to facilitate formation of a U1 snRNP ternary complex (lanes 6–9), indicating that, as with splicing activation, the RS domain is necessary for complex formation. Notably, the SRp38–U1 snRNP–RNA complex formed only with the β-SRp38 pre-mRNA and not the β-control pre-mRNA (lanes 10–14). Ternary complex formation was also dependent on an intact 5′ splice site in the β-SRp38 pre-mRNA, as a 5′-splice-site mutation that abolished splicing also blocked SRp38-dependent U1 snRNP binding (Supplementary Fig. 3 online). A similar cooperative interaction was observed between SRp38 and U2 snRNP (Fig. 4b). We also performed gel shift assays using the β-SRp38 RNA with ASF/SF2. No complex was observed when 100 ng of ASF/SF2 was added to reaction mixtures, in the presence or absence of snRNPs (Fig. 4c). These data indicate that SRp38, in a binding site–dependent manner, can facilitate recruitment of U1 and U2 snRNPs to a pre-mRNA to stabilize 5′-splice-site and branch-site recognition, respectively.
Figure 4.
SRp38 interacts with both U1 and U2 snRNP complexes on the SRp38 substrate. (a,b) Purified U1 snRNP (a) or U2 snRNP (b), GST-tagged SRp38 (150 ng) or GST-tagged SRp38 RBD (250 ng) were incubated with β-SRp38 pre-mRNA (lanes 1–9) or β-control pre-mRNA (lanes 10–14). Following addition of heparin (0.8 mg ml−1), complexes were resolved by 5% nondenaturing PAGE. Increasing amounts (300 ng and 600 ng) of U1 snRNP (a) or U2 snRNP (b) were incubated with SRp38 and β-SRp38 RNA (lanes 4, 5 and 6). The complexes formed are indicated with brackets. GST-tagged SRp38 RBD (250 ng) was incubated with β-SRp38 RNA (lane 6), and increasing amounts of U1 or U2 snRNPs (150 ng, 300 ng or 600 ng) were added (lanes 7–9). SRp38 (150 ng) was incubated with β-control pre-mRNA alone (lane 13) or β-control pre-mRNA plus U1 or U2 snRNPs (300 ng; lane 14). (c) His-tagged ASF/SF2 was incubated with β-SRp38 in the absence (lane 2) or in the presence of U1 snRNP (lane 3) or U2 snRNP (lane 4). (d) Splicing reactions were performed with the β-SRp38 RNA. The (5′ ss-Py) oligonucleotide was preincubated with nuclear extract (NE) or with S100 plus NF40–60 and SRp38 before splicing reactions were carried out. Final concentrations of the oligonucleotide are 0 µM, 0.1 µM , 0.5 µM, 1 µM and 2 µM. Note that 300 ng ASF/SF2 was used in the splicing reactions to achieve similar splicing levels as in the NE.
We next set out to investigate whether the SRp38 interactions with U1 and U2 snRNPs are functionally important. To this end, we used a splicing inhibition assay designed to measure, albeit indirectly, recruitment of U1 and U2 snRNPs to the 5′ splice site and branch site, respectively, during splicing. The assay measures the sensitivity of splicing to inhibition by antisense RNA oligonucelotides. For example, an RNA oligonucleotide containing a polypyrimidine stretch was shown to block the U2 snRNP–branch site interaction by competing with the 3′ splice site for binding to U2AF30. We predicted that, if the interactions between SRp38 and U1 and U2 snRNPs were functionally relevant, then SRp38-activated splicing might show greater resistance to RNA oligonucleotides that interfered with 5′-splice-site and/or branch-site recognition than would splicing activated by other pathways. We measured the sensitivity of β globin-SRp38 splicing to several different RNA oligonucleotides in the nuclear extract, and in S100 plus NF40–60 and either 50 ng SRp38 or 300 ng ASF/SF2. Results showed that an RNA oligonucleotide containing both the 5′ splice site consensus and a polypyrimidine stretch (5′ ss-Py) strongly inhibited splicing in the nuclear extract and in S100 in the presence of ASF/SF2 but was much less effective in S100 plus SRp38 (Fig. 4d, compare lanes 1–5 and lanes 6–15). Other RNA oligonucleotides tested, such as anti-U2 (Supplementary Fig. 4a online) and anti-U6 (Supplementary Fig. 4b) oligonucleotides, led to equivalent splicing repression in nuclear extract and SRp38-activated splicing. Taken together, these results provide evidence that SRp38 stimulates splicing by facilitating recruitment of U1 snRNP and U2 snRNP to the 5′ splice site and branch site.
SRp38 activates an RNA containing SRp38 binding sites
Given that SRp38 functions as a sequence-specific splicing activator, we next wished to identify possible natural targets. One potential substrate is that the alternatively spliced transcript is encoded by the mouse AMPA receptor subunit GluR-B gene31. Exons 14 and 15 (referred to as Flip and Flop) are mutually exclusive and impart different properties on currents evoked by l-glutamate or AMPA32. Transient transfection experiments showed previously that SRp38 isoforms can differentially influence Flip and Flop splicing31.
In light of the above, we examined the sequences of both Flip and Flop for the presence of potential SRp38 binding sites. We found the motifs GACAAA in the Flop exon and AGACAA in the Flip exon, constituting good matches to the core of the SRp38 consensus sequence, AAAGACAAA (Fig. 5a). To determine whether these sequences might be functional, we performed gel-shift assays with both Flop and Flip RNAs and purified GST-tagged SRp38, and compared binding with an RNA containing three copies of the SRp38 consensus (Fig. 5b). The results show that both Flop and Flip RNAs bind SRp38 but with different affinities. The Flip RNA showed an affinity for SRp38 (~20 nM Kd) comparable to the SRp38 consensus RNA (compare lanes 5–8 with lanes 9–12), whereas binding to the Flop RNA was noticeably weaker (Kd ~60 nM; lanes 1–4). These RNAs showed no affinity for SC35 (Fig. 5c), indicating that the binding was specific for SRp38.
Figure 5.
SRp38 binds to and activates splicing of RNA substrates containing the GluR-B Flip or Flop exon. (a) Comparison of a putative SRp38 binding motif in Flip and Flop exons from the murine GluR-B transcript with the SRp38 consensus recognition sequence. (b) Flop and Flip exons of the GluR-B pre-mRNA bind SRp38 specifically. Gel-shift assays with the indicated radiolabeled RNAs were performed with increasing amounts of GST-SRp38 (25 ng, 75 ng and 225 ng). Complexes were resolved by nondenaturing PAGE. (c) Flop and Flip exons of the GluR-B pre-mRNA do not bind His-SC35. Increasing amounts of purified His-SC35 (25 ng, 75 ng and 225 ng) were added to gel-shift assays containing the identical RNAs. (d) SRp38 activates splicing of RNA substrates containing Flip or Flop exons. The downstream β-globin exon was replaced with Flip or Flop exons, and splicing reactions were performed in S100 with increasing amounts of GST-SRp38 (20 ng, 40 ng and 100 ng) in the presence or absence of NF40–60 as indicated.
We next asked whether the sequences in Flop and Flip could bring about SRp38-dependent splicing and whether the differential binding affinity for SRp38 might be functionally important in splicing activated by SRp38. Following the same strategy used to measure SRp38-dependent splicing described above, we constructed modified β-globin substrates containing Flop or Flip sequences in the second exon; we named these subtrates β-Flop and β-Flip (Fig. 5d). We tested these RNAs in S100 activated by SRp38 alone or by SRp38 plus NF40–60. Notably, the Flip-containing RNA was spliced efficiently, again in a manner dependent on NF40–60 (lanes 13–16). On the other hand, SRp38-activated splicing of the Flop-containing substrate was detectable but much weaker, consistent with its lower affinity for SRp38 (lanes 5–8).
SRp38 promotes inclusion of the Flip exon in vivo
We next investigated whether SRp38 favors inclusion of Flip in GluR-B pre-mRNA splicing in vivo. To address this, we constructed a modified GluR-B minigene plasmid in which GluR-B sequences were preceded by the chicken β-actin promoter and followed by an SV40 poly(A) site (Fig. 6a). This plasmid was stably transfected into chicken DT40 cells with the genetic background SRp38(+/+) or SRp38(−/−)25. Several stably transfected colonies were isolated and total RNAs were extracted and analyzed first by reverse-transcription PCR (RT-PCR). Three transcript variants are expected to be produced by alternative splicing from GluR-B minigene transcripts33: Flop (612 bp), Flip (612 bp) and Truncated (lacking both the Flop or Flip exons; about 500 bp) (Fig. 6a). The pattern and intensity of RT-PCR products were identical with RNA samples from SRp38-containing and SRp38-lacking DT40 cells (Fig. 6b, lanes 1 and 2, and data not shown). We then took advantage of the fact that there is a unique StuI site in the Flop exon and treated PCR products with StuI followed by agarose gel analysis. Notably, we observed substantially more of the Flop variant in the SRp38(−/−) cells (Fig. 6b, lanes 3 and 4), suggesting that loss of SRp38 promotes inclusion of Flop in GluR-B pre-mRNA splicing. To confirm this, we performed an RNase protection assay using an antisense RNA targeted against full-length exon 13–14 sequences as shown in Figure 6a. Consistent with the above RT-PCR results, we observed a noticeable difference between SRp38(+/+) and SRp38(−/−) cells, such that there was an increase in the amount of Flop exon in the absence of SRp38 and a decrease in Flop in the presence of SRp38 (Fig. 6c, lanes 1 and 2).
Figure 6.
Loss of SRp38 promotes inclusion of the Flop exon in vivo. (a) Diagram of reporter plasmids containing mouse GluR-B truncated genomic sequences and three alternatively spliced products. The pair of primers used in the RT-PCR in b are shown as two reverse arrows. The RNA probe used in RNase protection assay in c is indicated above the three products, and the length of protected probe is on the side. (b) Comparison of alternatively spliced products from SRp38(+/+) and SRp38(−/−) DT40 cells. RT-PCR was performed with RNAs extracted from stably transfected DT40 cells and analyzed directly on a 1.5% agarose gel (lanes 1 and 2), or digested with StuI followed by agarose gel analysis (lanes 3 and 4). (c) RNase protection assay. RNA was extracted from stably transfected DT40 cells in the background of SRp38(+/+), SRp38(−/−) and SRp38(−/−) cells expressing exogenous hemagglutinin (HA)-SRp38. RNase protection assay was performed using the radiolabeled RNA probe indicated in a. Products were resolved by 6% denaturing PAGE.
Finally, we wished to confirm that the increased Flop mRNA levels were caused directly by loss of SRp38 in the SRp38(−/−) cells. To this end, we transfected the GluR-B reporter plasmid into SRp38(−/−) cells that expressed hemagglutinin-tagged SRp38 (ref. 25). RNA was isolated and analyzed by RNase protection assay. Notably, expression of SRp38 reduced Flop mRNA to levels observed in the SRp38(+/+) cells (lane 3). For confirmation, we analyzed several additional hemagglutinin-SRp38 expressing cell lines that contain the stably transfected GluR-B minigene and found that all have decreased Flop mRNA levels (data not shown). These results demonstrate that SRp38 influences alternative splicing of the GluR-B Flop and Flip exons in vivo.
DISCUSSION
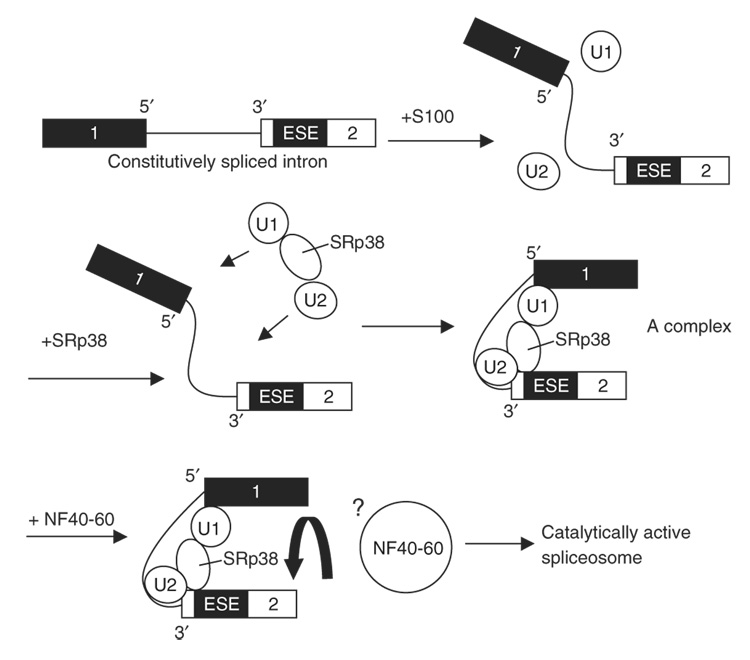
In this paper, we have shown that phosphorylated SRp38, unlike its dephosphorylated counterpart, activates splicing. Our data suggest a model in which U1 and U2 snRNP are unable to associate stably with certain pre-mRNA substrates in the absence of SRp38. Binding of these snRNPs to the pre-mRNA is stabilized by SRp38, but the A complex that is formed is stalled or inactive and unable to proceed in the splicing pathway in the absence of a specific coactivator (Fig. 7). This activator function of SRp38 is not only opposite to the function of dephosphorylated SRp38, which acts as a general splicing repressor28, but also distinct from the mechanism of standard SR or SR-related proteins in activating splicing12. Extending these findings, we observed that SRp38 affects the selection of mutually exclusive exons in the GluR-B pre-mRNA in a sequence-and affinity-dependent manner. Below, we discuss the features of SRp38-activated splicing compared to previously reported examples of activation and the role of SRp38 in the regulation of alternative splicing.
Figure 7.
Model for SRp38-dependent splicing activation. SRp38 binds the SRp38-dependent ESE in target transcripts and facilitates association of U1 and U2 snRNPs with the pre-mRNA to stabilize 5′-splice-site and branch-site recognition by interacting with U1 and U2 snRNPs, respectively. However, the spliceosomal A complex formed is stalled and requires an SRp38-specific cofactor to proceed through the splicing pathway.
We previously reported that SELEX-determined sequences optimal for ASF/SF2, SRp40 or TRA2 binding can function as ESEs when placed downstream of an enhancer-dependent intron9,34,35. Artificial tethering experiments have also shown that RS domains are sufficient to activate splicing in vitro when recruited to ESEs36,37. Therefore, it seems to be a general feature of SR proteins that they can interact with downstream ESEs and activate splicing of weak upstream introns. However, SRp38-mediated activation seems to have different requirements. In early experiments, we failed to detect any stimulation by SRp38 of substrates in which three copies of the SRp38 binding site were placed downstream of weak introns (Y.F. and J.L.M., unpublished data), suggesting that perhaps this sequence cannot function as an SRp38-dependent ESE. Additionally, this would be analogous to the behavior of SELEX-selected sequences for the related protein SC35, which cannot function as an ESE in vitro34. Alternatively, given that SRp38 is an atypical SR protein, we reasoned that the selected sequence might be able to function as an ESE but in a different way from sequences for standard SR proteins. This led us to test whether the SRp38 sequences might function with pre-mRNAs containing consensus splice sites, such as the β-globin and AdML substrates, which allowed us to demonstrate that the SRp38 consensus sequence can indeed function as an ESE. However, unlike standard SR proteins, it may be that SRp38 functions preferentially on substrates containing introns with strong splice sites. An intriguing possibility is that this property is related to the inability of SRp38 to function as a general splicing activator26.
What underlies the unusual properties of SRp38 described here and previously? All the available data indicate that an atypical RS domain28 and its response to phosphorylation25,26 are crucial. Whereas standard SR proteins become inactive in splicing when in an unphosphorylated state29,38, SRp38 is converted into a potent, general repressor26. This correlates with differences in protein-protein interactions. For example, the interaction of ASF/SF2 with a target protein, the U1 70K protein, is abolished by dephosphorylation38, whereas that of SRp38 is greatly enhanced but in a way that results in repression rather than activation25. But although SRp38 is a strong repressor, it seems to be a relatively weak activator when phosphorylated. SRp38 is unable to function as a general activator, as do other SR proteins25, and is able to function only as an ESE-dependent activator on substrates with strong splice sites. This correlates with the differences in snRNP recruitment: ASF/SF2 can form a ternary complex with U1 snRNP on an RNA lacking high-affinity binding sites16, whereas SRp38 cannot. It is again reasonable to speculate that this reflects difference in RS domain–mediated protein-protein interactions and the influence of phosphorylation16,25,38,39.
Our data demonstrate that SRp38 requires a specific cofactor (or cofactors) for activity. It was previously shown that a 100-kD non-SR protein, in addition to SR proteins, is required for activation of the weak 3′ splice site of a-tropomyosin exon 2 (ref. 40). Together with our results, this suggests that the activity of certain ESEs can be modulated by the assembly of additional proteins on the enhancer element, in addition to specific SR proteins. This is similar to the role of the TRA and TRA2 proteins in D. melanogster, which form an ESE-dependent complex with SR proteins and stimulate splicing of the female-specific exon of the doublesex transcript41. Likewise, human TRA2 requires the activity of SR proteins for sequence-specific activation9,42. Notably, this is distinct from the mechanism of SRp38, which does not require additional SR proteins.
We do not yet know the identity of the coactivator. However, SRp38 was not able to cooperate with general coactivators such as SRm160 and SRm300 (ref. 22) because no splicing inhibition was observed when SRm160/300 antibody was added to an SRp38-activated splicing reaction (M.C. and J.L.M, unpublished data). Another SR protein, 9G8, which has been shown to synergize with ASF/SF2 at lower concentrations43, was also ruled out as the coactivator because it was not present in partially purified fractions that retain cofactor activity. Consistent with the antibody data, SRm160 and SRm300 were also absent from this fraction (M.C. and J.L.M., unpublished data).
How might the SRp38 cofactor function? One possibility is that it recruits the U4/U6-U5 tri-snRNP to the preformed A complex. Standard SR proteins do in fact have a role in recruitment of the tri-snRNP into the spliceosome44,45. It is thus possible that SRp38 by itself is unable to recruit the tri-snRNP to the A complex. It is also possible that tri-snRNPs can bind transiently to the early spliceosome complex and form an unstable ‘B-like’ complex, but a conformational rearrangement brought about by the cofactor is necessary for stability. Another possibility is that the A complex formed by SRp38 is not functionally active. In this scenario, the cofactor may affect the conformation of the A complex formed with SRp38, so that the tri-snRNPs will be able to bind to it productively and allow splicing to proceed. Identification of the cofactor is likely to provide considerable insight into why SRp38-activated spliceosomes are stalled at the A complex and is of course an important goal of future experiment.
Consistent with its role as a sequence-specific splicing activator, SRp38 can act as a regulator of alternative splicing, influencing selection of mutually exclusive exons of the GluR-B pre-mRNA. The mutually exclusive Flip and Flop exons are both flanked by what seem to be strong splice sites. Although SRp38 binds to both exons, the affinities are different. Therefore, it is reasonable to propose that the differential binding by SRp38 influences the decision to include Flip or Flop, and may reflect in part intracellular concentrations of SRp38. SC35 did not bind to either exon but, consistent with the general activation function of SR proteins, could activate splicing of substrates containing either the Flop or Flip exon but without preference (Y.F. and J.L.M., unpublished data). We note that this contradicts previous data in which transiently overexpressed SC35 (as well as ASF/SF2) was found to increase the Flop to Flip ratio, and SC35-responsive elements in the Flop exon were identified46. Another group reported that the two splice variants of SRp38 (NSSR1 and NSSR2, or SRp38 and SRp38-2) had opposite effects on Flip inclusion31. However, our in vitro data indicate that both SRp38 variants have similar effects on Flip versus Flop splicing, although SRp38-2 showed reduced activity (Y.F. and J.L.M., unpublished data). The basis for these differences is unclear, but they may reflect the use of transient overexpression assays in the previous experiments.
In summary, SRp38 represents a distinct type of splicing regulatory protein. Phosphorylation and dephosphorylation switch the protein from a general splicing repressor to a sequence-specific activator. SRp38 activates splicing by facilitating formation of early splicing complexes that cannot proceed to active spliceosomes without the aid of a specific cofactor. Furthermore, SRp38 can regulate alternative splicing and potentially has important roles in various physiologically important processes. Future studies on both its mechanism and target transcripts should be informative.
METHODS
Plasmid constructions, cell culture and transfection
We constructed all plasmids (β-SRp38, β-control, β-Flop, β-Flip and AdML-SRp38) used to produce substrates for in vitro splicing by replacement of sequences between the AccI site and BamHI site in the second exon with the indicated sequences. We used site-directed mutagenesis to produce the β-SRp38 5′-splice-site mutant from the β-SRp38 plasmid, as described47. The forward primer was 5′-TGGTG AGGCCCTGGGCATACTGGTATCAAGGTTACAAGA-3′ and the reverse primer was 5′-TCTTGTAACCTTGATACCAGTATGCCCAGGGCCTCACCA-3′. For in vitro gel-shift assays and RNase protection assays, we constructed plasmids by insertion of indicated sequences into pBluescript SK(+) plasmid. The GluR-B reporter minigene was constructed as described previously31, except that pEXpress plasmid48 was used as the backbone. DT40 cells of different background including wild-type, SRp38(−/−) and SRp38(−/−) containing exogenously expressed hemagglutinin-tagged SRp38 were maintained essentially as described previously25. We also carried out transfection of the GluR-B reporter minigene into DT40 cells as described previously25.
Recombinant proteins
We prepared His-SRp38, His-SC35 and His-ASF/SF2 and GST-SRp38 from recombinant baculovirus-infected High Five cells (Invitrogen). His-tagged recombinant proteins were purified under denaturing conditions by Ni2+agarose chromatography and renatured by dialysis26,34. Dephosphorylated SRp38 was prepared by incubating recombinant SRp38 with calf intestinal phosphatase (CIP) and repurified by agarose chromatography26. GST-tagged SRp38 was purified by using glutathione–Sepharose 4B25. (His- and GST-tagged SRp38 behaved indistinguishably in all assays tested.) GST-SRp38 RBD and hnRNP G RBD proteins were prepared from E. coli JM101 using glutathione-Sepharose 4B, and his-SC35 RBD was prepared from E. coli BL21 (ref. 34). Purity and concentration of proteins were determined by Coomassie blue staining of SDS gels.
In vitro splicing and spliceosome-assembly assays
We carried out in vitro splicing assays essentially as described34. Native SR proteins purified from HeLa cells were obtained from T. Kashima (Columbia University). Spliceosome-assembly assays were also performed as described49. In the splicing-inhibition and spliceosome-assembly assays, the anti-snRNAs sequences used were: U11–14, 5′-UGCCAGGUAAGUAU-3′; U22–15, 5′-GGCCGAGAAGCGAU-3′; U568–88, 5′-UUGGGUUAAGACUCAGAGUUG-3′; U678–95, 5′-CGCUUCACGAAUUUGCGU-3′; 5′ss-Py, 5′-UCACAGGUAAGUACUUAUUUUCCCAGGCC-3′. Antisense RNAs were preincubated with S100 at 30 °C for 15 min before spliceosome assays.
Gel-shift assays
We carried out gel-shift assays essentially as described16. Briefly, radiolabeled pre-mRNA, U1 snRNP (300 ng and 600 ng) and SRp38 (15 ng µl−1) were incubated under splicing conditions: 30 °C for 5 min. We then added heparin to a concentration of 0.8 mg ml−1, and the reaction mixtures were incubated at 30 °C for an additional 5 min. Products were analyzed by 5% nondenaturing PAGE and autoradiography.
RT-PCR analysis and RNase protection assay
Total RNA was extracted by using Trizol (Invitrogen). We carried out RT-PCR analysis using Transcriptor Reverse Transcriptase (Roche) following the instructions provided by the supplier. RNase protection assays were performed as described50. Briefly, labeled RNA probes were synthesized by in vitro transcription with SP6 RNAP. After hybridization and RNase digestion, protected RNAs were resolved by 6% denaturing PAGE.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
ACKNOWLEDGMENTS
We thank R. Lührmann (Max Planck Institute for Biophysical Chemistry, Germany) for providing purified U1 and U2 snRNP, T. Kashima (Columbia University, New York) for SR proteins and C. Shin (Columbia University, New York) for His-dSRp38. We thank A. Yang for help with the manuscript. We also thank members of the Manley laboratory for helpful discussions and comments. This work was supported by the US National Institutes of Health grant NIH GM48259.
Footnotes
Published online at http://www.nature.com/nsmb/
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 2.Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 3.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 4.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 5.Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J. Biol. Chem. 2008;283:1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- 6.David CJ, Manley JL. The search for alternative splicing regulators: new approaches offer a path to a splicing code. Genes Dev. 2008;22:279–285. doi: 10.1101/gad.1643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Smith PJ, Krainer AR, Zhang MQ. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res. 2005;33:5053–5062. doi: 10.1093/nar/gki810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol. Cell. Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tacke R, Tohyama M, Ogawa S, Manley JL. Human TRA2 proteins are sequence-specific activators of pre-mRNA splicing. Cell. 1998;93:139–148. doi: 10.1016/s0092-8674(00)81153-8. [DOI] [PubMed] [Google Scholar]
- 10.Goren A, et al. Comparative analysis identifies exonic splicing regulatory sequences—the complex definition of enhancers and silencers. Mol. Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Zheng ZM. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 2004;11:278–294. doi: 10.1159/000077096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manley JL, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 13.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hastings ML, Krainer AR. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 2001;13:302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- 15.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 16.Kohtz JD, et al. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 17.Shen H, Green MR. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev. 2006;20:1755–1765. doi: 10.1101/gad.1422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertel KJ, Graveley BR. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem. Sci. 2005;30:115–118. doi: 10.1016/j.tibs.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Hoffmann HM, Grabowski PJ. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- 21.Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]
- 22.Eldridge AG, Li Y, Sharp PA, Blencowe BJ. The SRm160/300 splicing coactivator is required for exon-enhancer function. Proc. Natl. Acad. Sci. USA. 1999;96:6125–6130. doi: 10.1073/pnas.96.11.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Blencowe BJ. Distinct factor requirements for exonic splicing enhancer function and binding of U2AF to the polypyrimidine tract. J. Biol. Chem. 1999;274:35074–35079. doi: 10.1074/jbc.274.49.35074. [DOI] [PubMed] [Google Scholar]
- 24.Ryner LC, et al. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–1089. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- 25.Shin C, Feng Y, Manley JL. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427:553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- 26.Shin C, Manley JL. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111:407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y, Manley JL. A complex signaling pathway regulates SRp38 phosphorylation and pre-mRNA splicing in response to heat shock. Mol. Cell. 2007;28:79–90. doi: 10.1016/j.molcel.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 28.Shin C, Kleiman FE, Manley JL. Multiple properties of the splicing repressor SRp38 distinguish it from typical SR proteins. Mol. Cell. Biol. 2005;25:8334–8343. doi: 10.1128/MCB.25.18.8334-8343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prasad J, Colwill K, Pawson T, Manley JL. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol. Cell. Biol. 1999;19:6991–7000. doi: 10.1128/mcb.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruskin B, Zamore PD, Green MR. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- 31.Komatsu M, Kominami E, Arahata K, Tsukahara T. Cloning and characterization of two neural-salient serine/arginine-rich (NSSR) proteins involved in the regulation of alternative splicing in neurones. Genes Cells. 1999;4:593–606. doi: 10.1046/j.1365-2443.1999.00286.x. [DOI] [PubMed] [Google Scholar]
- 32.Sommer B, et al. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249:1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- 33.Chen X, et al. Tra2bl regulates P19 neuronal differentiation and the splicing of FGF-2R and GluR-B minigenes. Cell Biol. Int. 2004;28:791–799. doi: 10.1016/j.cellbi.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc. Natl. Acad. Sci. USA. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graveley BR, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- 37.Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 38.Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 39.Xiao SH, Manley JL. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 1998;17:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dye BT, Buvoli M, Mayer SA, Lin CH, Patton JG. Enhancer elements activate the weak 3′ splice site of α-tropomyosin exon 2. RNA. 1998;4:1523–1536. doi: 10.1017/s1355838298980360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Bell LR. The Sex-lethal amino terminus mediates cooperative interactions in RNA binding and is essential for splicing regulation. Genes Dev. 1994;8:2072–2085. doi: 10.1101/gad.8.17.2072. [DOI] [PubMed] [Google Scholar]
- 42.Tacke R, Manley JL. Functions of SR and Tra2 proteins in pre-mRNA splicing regulation. Proc. Soc. Exp. Biol. Med. 1999;220:59–63. doi: 10.1046/j.1525-1373.1999.d01-10.x. [DOI] [PubMed] [Google Scholar]
- 43.Li X, Shambaugh ME, Rottman FM, Bokar JA. SR proteins Asf/SF2 and 9G8 interact to activate enhancer-dependent intron D splicing of bovine growth hormone pre-mRNA in vitro. RNA. 2000;6:1847–1858. doi: 10.1017/s1355838200000674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roscigno RF, Garcia-Blanco MA. SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- 45.Tarn WY, Steitz JA. Modulation of 5′ splice site choice in pre-messenger RNA by two distinct steps. Proc. Natl. Acad. Sci. USA. 1995;92:2504–2508. doi: 10.1073/pnas.92.7.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crovato TE, Egebjerg J. ASF/SF2 and SC35 regulate the glutamate receptor subunit 2 alternative flip/flop splicing. FEBS Lett. 2005;579:4138–4144. doi: 10.1016/j.febslet.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 47.Fisher CL, Pei GK. Modification of a PCR-based site-directed mutagenesis method. Biotechniques. 1997;23:570–571. 574. doi: 10.2144/97234bm01. [DOI] [PubMed] [Google Scholar]
- 48.Arakawa H, Lodygin D, Buerstedde JM. Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol. 2001;1:7. doi: 10.1186/1472-6750-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das R, Reed R. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA. 1999;5:1504–1508. doi: 10.1017/s1355838299991501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Manley JL. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.