SUMMARY
Vertebrates express three small ubiquitin-related modifiers (SUMO-1, SUMO-2 and SUMO-3) that are conjugated in part to unique subsets of proteins and thereby regulate distinct cellular processes. Mechanisms regulating paralog-selective sumoylation, however, remain poorly understood. Despite being equally well modified by SUMO-1 and SUMO-2 in vitro, RanGAP1 is selectively modified by SUMO-1 in vivo. We have found that this paralog-selective modification is determined at the level of deconjugation by isopeptidases. Our findings indicate that, relative to SUMO-2 modified RanGAP1, SUMO-1 modified RanGAP1 forms a more stable, higher affinity complex with the nucleoporin Nup358/RanBP2 that preferentially protects it from isopeptidases. By swapping residues in SUMO-1 and SUMO-2 responsible for Nup358/RanBP2 binding, or by manipulating isopeptidase expression levels, paralog-selective modification of RanGAP1 could be affected both in vitro and in vivo. Thus, protection from isopeptidases, through interactions with SUMO-binding proteins, represents an important mechanism defining paralog-selective sumoylation.
INTRODUCTION
SUMOs (small ubiquitin-related modifiers) are posttranslationally conjugated to lysine residues of other proteins through an enzyme cascade similar to ubiquitination (Johnson, 2004). Sumoylation is mediated by the coordinated action of an E1 activating enzyme (a heterodimer consisting of Aos1 and Uba2), an E2 conjugating enzyme (Ubc9) and in some cases one of a variety of different E3 ligases. A family of SUMO-specific isopeptidases (also known as sentrin-specific proteases, or SENPs) deconjugate modified proteins, making sumoylation a dynamic process (Mukhopadhyay and Dasso, 2007). To date, hundreds of target proteins have been identified, implicating sumoylation as a regulator of a wide range of processes that include nucleocytoplasmic transport, DNA replication and repair, transcription, control of chromatin structure and apoptosis. Among its molecular consequences, sumoylation affects the activities, localizations and/or binding partners of modified proteins (Geiss-Friedlander and Melchior, 2007).
Although invertebrates express a single SUMO, vertebrates express three functional SUMO paralogs: SUMO-1, SUMO-2 and SUMO-3. SUMO-2 and SUMO-3 share ~96% sequence identity (and are referred to as SUMO-2/3) but they are only ~45% identical to SUMO-1. Several lines of evidence indicate that SUMO-1 and SUMO-2/3 are conjugated in part to unique subsets of proteins and that they represent unique signals regulating different cellular functions. Proteomic studies, for example, have identified proteins that are modified selectively by either SUMO-1 or SUMO-2/3 (Rosas-Acosta et al., 2005; Vertegaal et al., 2006). Consistent with these findings, studies indicate that SUMO-1 and SUMO-2/3 modified proteins localize to unique sub-cellular domains. In interphase cells, SUMO-1 and SUMO-2/3 localize throughout the nucleoplasm, however, only SUMO-1 is also present in nucleoli and at nuclear pore complexes (NPCs) (Ayaydin and Dasso, 2004). In addition, SUMO-2/3 modified proteins associate with mitotic chromosomes, whereas SUMO-1 modified proteins localize to the mitotic spindle and midzone (Zhang and Sarge, 2008). Lastly, an association between SUMO1 haploinsufficiency and cleft lip and palate in humans has been reported, indicating that SUMO-1 and SUMO-2/3 regulate distinct aspects of vertebrate development (Alkuraya et al., 2006). Surprisingly, mice lacking SUMO-1 develop normally and without any overt phenotype (Evdokimov et al., 2008; Zhang et al., 2008). Thus, humans and mice may differ in their specific requirements for SUMO paralogs. It also remains to be determined whether more subtle phenotypes accompany loss of SUMO-1 expression in mice.
Despite evidence for its importance, mechanisms regulating paralog-selective sumoylation are only beginning to be defined. Recent studies have demonstrated a role for SUMO-interacting motifs (SIMs), with preferences for SUMO-1 or SUMO-2/3 binding, in determining paralog-selective modification (Meulmeester et al., 2008; Zhu et al., 2008). To examine other possible mechanisms, we have used RanGAP1 as a model substrate. RanGAP1 is the GTPase activating protein for Ran, and thus an important regulator of nucleocytoplasmic transport (Stewart, 2007). Notably, RanGAP1 is among the best-characterized SUMO substrates and it is preferentially modified by SUMO-1 in vivo (Mahajan et al., 1997; Matunis et al., 1996). SUMO-1 modification of RanGAP1 targets it to cytoplasmic filaments of the NPC where it forms a complex with Nup358 (also known as RanBP2) and Ubc9 (Reverter and Lima, 2005; Zhang et al., 2002; Zhu et al., 2006). Interactions between Nup358 and SUMO-1 modified RanGAP1 are mediated in part by SIMs in Nup358 (Reverter and Lima, 2005; Song et al., 2004). Here, we demonstrate that the SUMO-binding activity of Nup358 plays an important role in determining the paralog-selective modification of RanGAP1. Specifically, we found that SUMO-1 modified RanGAP1 interacts with Nup358 with a higher affinity relative to SUMO-2 modified RanGAP1. This more stable interaction with Nup358 enables SUMO-1 modified RanGAP1 to be preferentially protected from isopeptidase-mediated deconjugation.
RESULTS
In vivo and in vitro sumoylation of RanGAP1
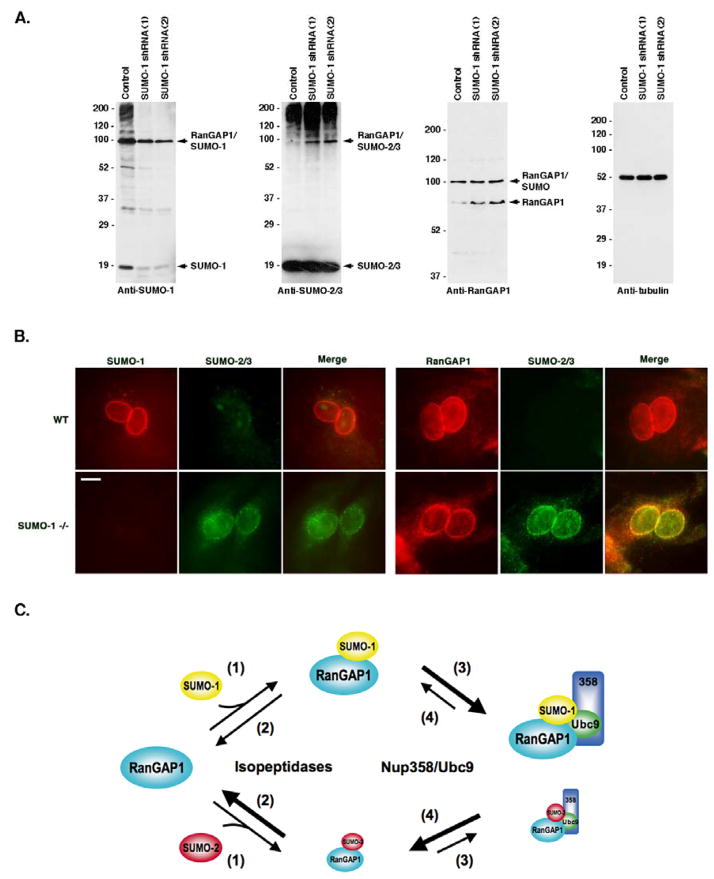
Subtle differences in high molecular mass SUMO-1 and SUMO-2/3 conjugates are detected by immunoblot analysis of whole cell lysates. However, the most prominent difference is the presence of a band migrating at ~90 kDa and detected primarily by antibodies to SUMO-1 (Figure 7A, Control lanes). As previously determined, this band corresponds to sumoylated RanGAP1, which is preferentially modified by SUMO-1 in vivo (Mahajan et al., 1997; Matunis et al., 1996).
Figure 7.
SUMO-1 and SUMO-2/3 modified RanGAP1 compete for Nup358 binding and protection from isopeptidases. A. Whole cell lysates were prepared from control 293T cells and 293T cells stably expressing shRNAs specific for SUMO-1. Lysates were analyzed by immunoblotting with antibodies specific for SUMO-1, SUMO-2/3, RanGAP1 and tubulin. B. Wild type and SUMO-1 deficient MEFs were analyzed by immunofluorescence microscopy. Cells were either co-labeled with SUMO-2/3 and SUMO-1 specific antibodies, or SUMO-2/3 and RanGAP1 specific antibodies. C. A model illustrating the roles of isopeptidases and Nup358 binding in determining SUMO-1 selective modification of RanGAP1. (1) SUMO-1 and SUMO-2/3 are equally well conjugated to RanGAP1. (2) SUMO-1 and SUMO-2/3 modified RanGAP1 are differentially recognized and deconjugated by isopeptidases. (3) SUMO-1 and SUMO-2/3 modified RanGAP1 compete for formation of a ternary complex with Nup358 and Ubc9 at NPCs. Due to a higher affinity for Nup358, SUMO-1 modified RanGAP1 competes more effectively for binding. Formation of the ternary complex protects both SUMO-1 and SUMO-2/3 modified RanGAP1 from isopeptidases. (4) SUMO-1 and SUMO-2/3 modified RanGAP1 dissociate from Nup358/Ubc9. Due to tighter association with Nup358, SUMO-1 modified RanGAP1 is more effectively protected from deconjugation.
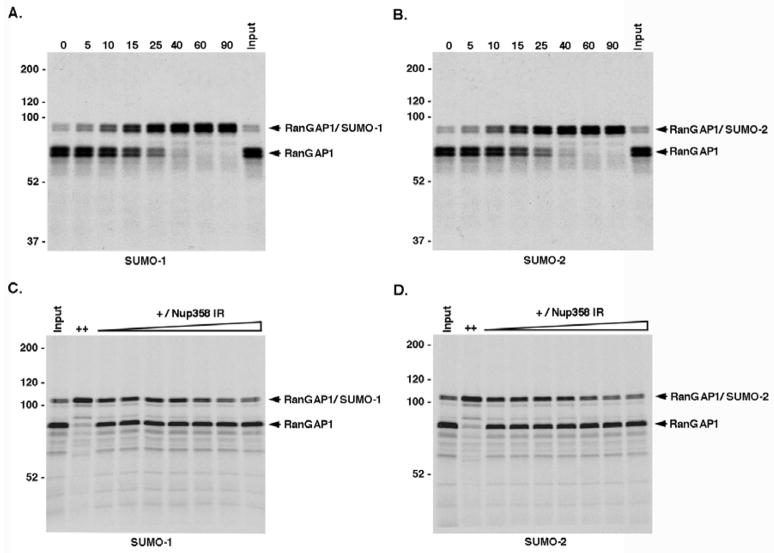
To investigate mechanisms regulating its paralog-selective modification, we characterized the ability of RanGAP1 to be modified by SUMO-1 and SUMO-2 in vitro. Due to direct high affinity interactions with Ubc9, RanGAP1 is efficiently sumoylated in reactions containing only E1 activating enzyme and Ubc9 (Bernier-Villamor et al., 2002). In these reactions, SUMO-1 and SUMO-2 are both equally well activated, and their abilities to form thiolester intermediates with Ubc9 are indistinguishable (data not shown) (Tatham et al., 2003). However, paralog-selective modification could result if differences existed in the efficiency of interactions between SUMO-1 or SUMO-2 charged Ubc9 and RanGAP1, and/or in the rates of isopeptide bond formation. We therefore analyzed the modification of in vitro expressed full-length RanGAP1. Under assays conditions containing exogenously added recombinant E1 activating enzyme and Ubc9, RanGAP1 was modified by SUMO-1 and SUMO-2 at indistinguishable rates (Figures 1A and 1B). We also characterized the modification of a purified carboxyl-terminal domain of RanGAP1 (RanGAP1-NΔ419) and found that it too was modified equally well by SUMO-1 and SUMO-2 in reactions containing only purified proteins (Figures S1A and S1B).
Figure 1.
RanGAP1 is equally well modified by SUMO-1 and SUMO-2 in vitro and modification is not affected by Nup358 E3 ligase activity. RanGAP1 was expressed in vitro and incubated with recombinant E1 activating and E2 conjugating enzymes together with either SUMO-1 or SUMO-2 and in the presence or absence of Nup358-IR. A and B. Reactions were incubated for the indicated times (min), resolved by SDS-PAGE and visualized by autoradiography. C and D. Reactions were incubated with high concentrations of E1 and E2 enzymes (++) or low concentrations of E1 and E2 enzymes (+) together with increasing concentrations of Nup358-IR (0, 3, 10, 30, 100, 300 and 600 ng). Reactions were terminated after 10 min and analyzed by SDS-PAGE and autoradiography.
In vivo, SUMO-1 modified RanGAP1 associates with Nup358 at NPCs. Because Nup358 is a SUMO E3 ligase (Pichler et al., 2002), we investigated whether its ligase activity could affect paralog-selective sumoylation of RanGAP1. Full-length RanGAP1 was expressed in vitro and its modification by SUMO-1 or SUMO-2 was assayed in reactions containing limiting concentrations of E2 enzyme and increasing concentrations of the Nup358 internal repeat/E3 ligase domain (Nup358-IR) (Figures 1C and 1D). Under these assay conditions, Nup358-IR had no effect on the rate of RanGAP1 sumoylation by either SUMO-1 or SUMO-2 and inhibited modification at high concentrations, as previously observed (Reverter and Lima, 2005). In contrast, Nup358-IR had similar stimulatory effects on SUMO-1 and SUMO-2 modification of Sp100 (Figure S2). Thus, paralog-selective sumoylation of RanGAP1 is not determined by an intrinsic ability to be preferentially modified by SUMO-1, nor does the Nup358-IR E3 ligase domain affect its modification.
Nup358/Ubc9 binding and protection from isopeptidases
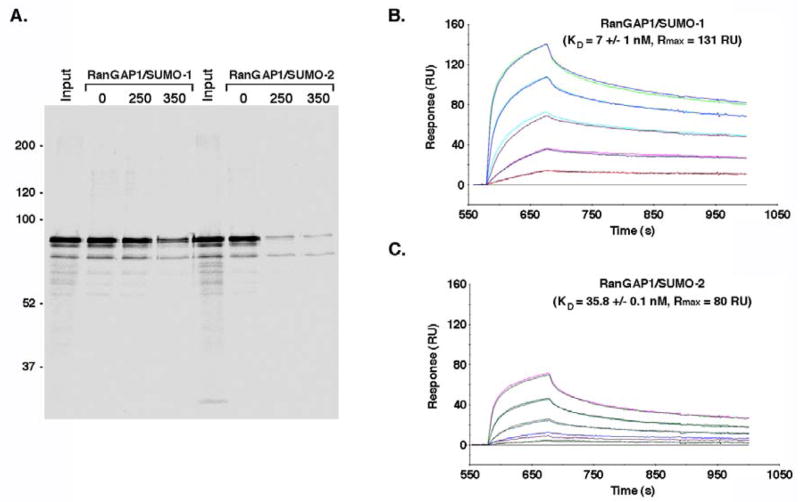
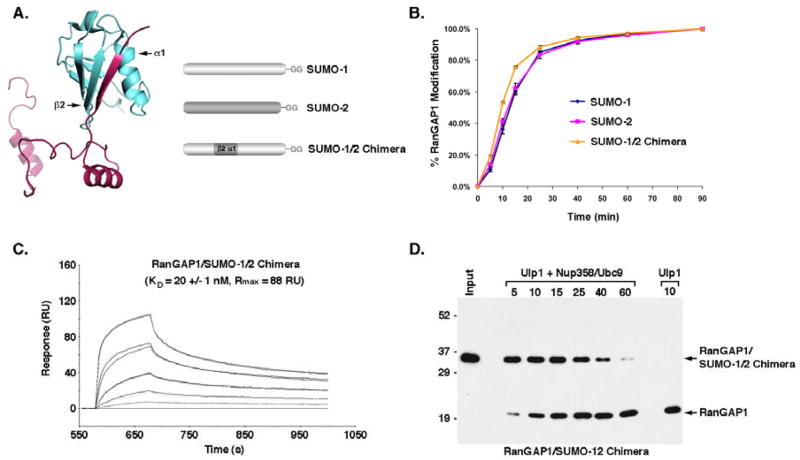
Nup358 and Ubc9 form a complex with SUMO-1 modified RanGAP1 that protects it from isopeptidases (Zhang et al., 2002). In addition, yeast two-hybrid assays and in vitro binding reactions indicate that Nup358 interacts preferentially with SUMO-1 relative to SUMO-2/3 (Hecker et al., 2006; Tatham et al., 2005). We therefore investigated the hypothesis that SUMO-1 modified RanGAP1 may form a more stable, high affinity complex with Nup358, and thus be preferentially protected from deconjugation. We first characterized the interactions of SUMO-1 and SUMO-2 modified RanGAP1 with Nup358-IR using in vitro binding assays (Figure 2A). The internal repeat domain contains binding sites for SUMO-1 modified RanGAP1 and Ubc9 (Zhang et al., 2002). Both SUMO-1 and SUMO-2 modified RanGAP1 associated with Nup358-IR under low salt binding conditions. However, only SUMO-1 modified RanGAP1 also bound Nup358-IR under high salt conditions, indicating paralog-dependent differences in binding. We next used surface plasmon resonance (Biacore) analysis to determine the relative binding affinities between SUMO-1 and SUMO-2 modified RanGAP1 and Nup358-IR (Figure 2B and 2C). GST-tagged Nup358-IR was immobilized on sensor chips and SUMO-1 or SUMO-2 modified RanGAP1 was passed over the chips in the presence of Ubc9. Binding in the absence of Ubc9 was insufficient to obtain reliable results (data not shown). This analysis indicated that SUMO-1 modified RanGAP1 binds Nup358-IR with an apparent KD of ~7 nM compared with an apparent KD of ~36 nM for SUMO-2 modified RanGAP1. The maximal responses observed for SUMO-1 and SUMO-2 modified RanGAP1 binding were also notably different (~131 and 80 resonance units respectively), implying potentially unique binding stoichiometries. Thus, SUMO-1 modified RanGAP1 may interact simultaneously with both internal repeats of Nup358-IR and it binds with a higher affinity relative to SUMO-2 modified RanGAP1.
Figure 2.
SUMO-1 modified RanGAP1 has a higher affinity for Nup358 relative to SUMO-2 modified RanGAP1. A. RanGAP1 was expressed in vitro and modified with either SUMO-1 or SUMO-2. Reaction products were incubated with glutathione beads containing GST-tagged Nup358-IR. Beads were washed with buffer containing the indicated MgCl2 concentrations (mM) and bound proteins were eluted and analyzed by SDS-PAGE and autoradiography. B. Biacore analysis was performed using sensor chips coated with GST-tagged Nup358-IR. SUMO-1 modified RanGAP1-NΔ419 was injected at multiple concentrations (8, 20, 50, 125 and 312 nm) together with Ubc9 and the resulting association and dissociation events were recorded. C. Biacore analysis of interactions between SUMO-2 modified RanGAP1-NΔ419 and Nup358-IR. SUMO-2 modified RanGAP1-NΔ419 was injected at multiple concentrations (10, 24, 61, 152 and 380 nm) together with Ubc9. RU=resonance units.
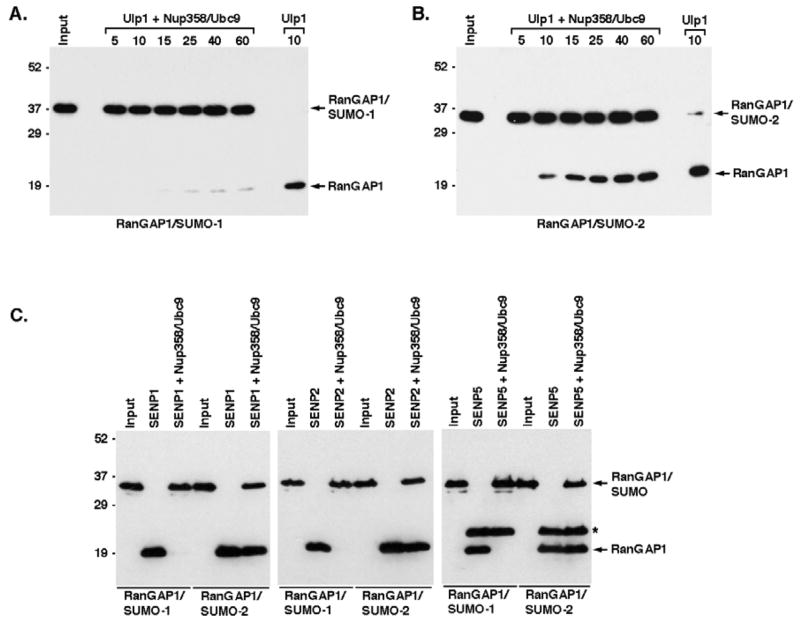
We next investigated whether differences in affinity for SUMO-1 and SUMO-2 modified RanGAP1 could affect the ability of Nup358 binding to prevent isopeptidase-mediated deconjugation (Figure 3A and 3B). In the absence of Nup358-IR and Ubc9, SUMO-1 and SUMO-2 modified RanGAP1 were both nearly completely deconjugated after a 10 min incubation in the presence of Ulp1. Ulp1 is a yeast isopeptidase with an inherent preference for SUMO-1 modified RanGAP1 (Figure S3). In the presence of Nup358-IR and Ubc9, deconjugation of SUMO-1 modified RanGAP1 was inhibited, with only a relatively small fraction of RanGAP1 being deconjugated after 60 minutes (Figure 3A). In contrast, although partial protection was observed, a significantly greater fraction of SUMO-2 modified RanGAP1 was deconjugated after a similar 60-minute incubation (Figure 3B). Given the SUMO-1 preference of Ulp1, these results clearly demonstrate that Nup358 and Ubc9 binding confer selective protection on SUMO-1 modified RanGAP1.
Figure 3.
Nup358 binding differentially protects SUMO-1 and SUMO-2/3 modified RanGAP1 from isopeptidases. A. SUMO-1 modified His-RanGAP1-NΔ419 (input) was incubated with Ulp1 alone for 10 min (Ulp1) or incubated with Ulp1 for the indicated times (min) after pre-incubation with Nup358 and Ubc9 (Ulp1 + Nup358/Ubc9). Reactions were analyzed by immunoblotting with an anti-His antibody. B. Similar Ulp1 protection assays were performed with SUMO-2 modified His-RanGAP1-NΔ419. C. SUMO-1 or SUMO-2 modified His-RanGAP1-NΔ419 (input) were incubated with the catalytic domains of human SENPs alone (SENP1, SENP2, SENP) or after pre-incubation with Nup358 and Ubc9 (SENP1 + Nup358/Ubc9, SENP2 + Nup358/Ubc9, SENP5 + Nup358/Ubc9). Reactions were analyzed by immunoblotting with an anti-His antibody. SUMO modified and unmodified RanGAP1 are indicated. An asterisk indicates His-tagged SENP5.
We also performed protection assays using the catalytic domains of three human isopeptidases, SENP1, SENP2 and SENP5 (Figure 3C). All three isopeptidases efficiently deconjugated both SUMO-1 and SUMO-2 modified RanGAP1 in the absence of Nup358 and Ubc9. However, under similar assay conditions but in the presence of Nup358 and Ubc9, SUMO-1 modified RanGAP1 was nearly completely protected from deconjugation by SENP1, SENP2 and SENP5. SUMO-2 modified RanGAP1, in contrast, was only partially protected by the addition of Nup358 and Ubc9. Thus, both SUMO-1 and SUMO-2 modified RanGAP1 are protected from multiple different isopeptidases through interactions with Nup358 and Ubc9. Importantly, however, SUMO-1 modified RanGAP1 is protected more effectively.
In vitro and in vivo analysis of a SUMO-1/2 chimera
To further test the hypothesis that paralog-selective modification of RanGAP1 is affected by Nup358 binding and protection from isopeptidases, we produced a SUMO-1/2 chimera predicted to have interactions with Nup358 that are comparable to SUMO-2. To make this chimera, we replaced the region of SUMO-1 encompassing the first α-helix and second β-strand with the same region from SUMO-2 (Figure 4A). Structural studies indicate that this region of SUMO-1 is critical for non-covalent interactions with Nup358 (Reverter and Lima, 2005; Song et al., 2004). The ability of the SUMO-1/2 chimera to be conjugated to RanGAP1 in vitro was indistinguishable from conjugation with wild type SUMO-1 and SUMO-2, indicating that the domain swap did not adversely affect protein folding or function (Figure 4B). To determine the effect of the domain swap on Nup358 binding, we performed Biacore analysis. As predicted, the domain swap lowered the affinity for Nup358, reducing the apparent KD from ~7 nM for SUMO-1 modified RanGAP1 to ~20 nM for SUMO-1/2 chimera modified RanGAP1. The maximal response also dropped from ~130 to 88 resonance units (Figure 2B and 4C). Finally, we tested the effect of the domain swap on the ability of Nup358 to bind to and protect RanGAP1 from deconjugation. In the absence of Nup358, Ulp1 deconjugated RanGAP1 modified with the SUMO-1/2 chimera at a rate similar to that observed with wild type SUMO-1 modified RanGAP1 (Figure S3). In the presence of Nup358 and Ubc9, deconjugation was partially inhibited, but to a much lower extent compared to wild type SUMO-1 modified RanGAP1 (Figures 4D and 3A). Rather, the level of inhibition was similar to that observed with SUMO-2 modified RanGAP1 (Figure 3B). Thus, replacing the first α-helix 1 and second β-strand of SUMO-1 with the similar region from SUMO-2 reduced the affinity of modified RanGAP1 for Nup358 and also its protection from deconjugation.
Figure 4.
Generation of a SUMO-1/2 chimera with Nup358-binding properties comparable to SUMO-2. A. A crystal structure illustrating interactions between the SIM of Nup358 and SUMO-1 (Reverter and Lima, 2005). A SUMO-1/2 chimera was produced by replacing the first α helix and second β strand of SUMO-1 with the same region from SUMO-2. B. In vitro sumoylation assays were performed using fluorescently-tagged GST-RanGAP1-NΔ419 and SUMO-1, SUMO-2 or SUMO-1/2 chimera. Reactions were terminated at different time points, resolved by SDS-PAGE and the fraction of RanGAP1-NΔ419 modified at each time point was quantified by fluorescence imaging and plotted. C. Biacore analysis was performed using sensor chips coated with GST-tagged Nup358-IR. RanGAP1-NΔ419 modified with the SUMO-1/2 chimera was injected at multiple concentrations (10, 25, 64, 159 and 397 nm) together with Ubc9 and the resulting association and dissociation events were recorded. RU=resonance units. D. SUMO-1/2 chimera modified His-RanGAP1-NΔ419 (input) was incubated with Ulp1 alone for 10 min (Ulp1) or incubated with Ulp1 for the indicted times (min) after pre-incubation with Nup358 and Ubc9 (Ulp1 + Nup358/Ubc9). Reactions were analyzed by immunoblotting with an anti-His antibody.
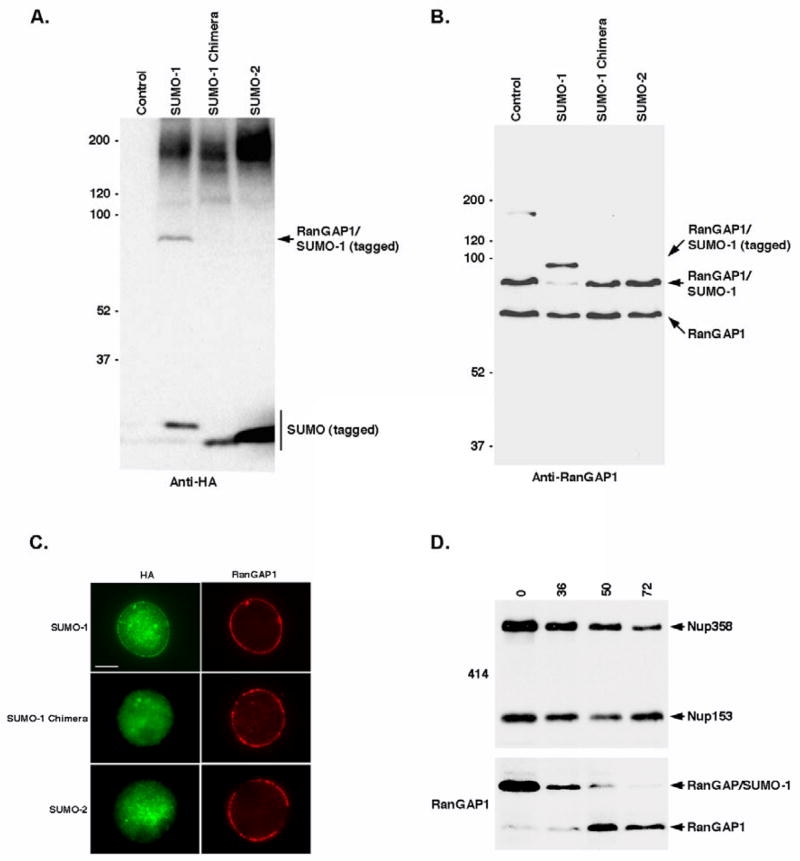
We next investigated the ability of the SUMO-1/2 chimera to be stably conjugated to RanGAP1 in vivo by generating stable cell lines that express tagged SUMO-1, SUMO-2 and the SUMO-1/2 chimera under control of inducible promoters. Following induction of protein expression, cell lysates were prepared and analyzed. Blots probed with an anti-HA antibody revealed that SUMO-1 and the SUMO-1/2 chimera were expressed at similar levels and that both proteins were functional, as they were detected in association with high molecular mass conjugates (Figure 5A). In contrast, however, a band migrating at ~95 kDa and corresponding to the predicted molecular mass of modified RanGAP1, was only detected in cells expressing tagged SUMO-1 but not the SUMO-1/2 chimera. This band was also absent from cells expressing tagged SUMO-2, despite considerably higher SUMO-2 expression. Blots probed with a RanGAP1-specific antibody confirmed that the ~95 kDa band detected in tagged SUMO-1 expressing cells corresponds to modified RanGAP1 (Figure 5B). Consistent with the anti-HA blot, forms of RanGAP1 modified by the SUMO-1/2 chimera and SUMO-2 were not detected.
Figure 5.
Stable SUMO modification of RanGAP1 in vivo correlates with high affinity Nup358 binding. A. Stable cell lines were induced to express HA-FKBP tagged SUMO-1, SUMO-1 chimera and SUMO-2. Whole cell lysates were resolved by SDS-PAGE and immunoblots were probed with anti-HA antibodies. Lysate from untransfected parental cells was included as a control. B. Cell lysates were probed with an antibody specific for RanGAP1. C. Stable cell lines expressing HA-FKBP tagged SUMO-1, SUMO-1 chimera and SUMO-2 were co-labeled with anti-HA and RanGAP1 specific antibodies and analyzed by immunofluorescence microscopy. D. Cells were transfected with Nup358-specific siRNAs for the indicated times (hrs) and whole cell lysates were prepared and resolved and SDS-PAGE. Immunoblots were probed with mAb 414 (recognizing Nup358 and Nup153 which serves as a loading control) and RanGAP1 specific antibodies.
We also investigated the sub-cellular localizations of tagged SUMO-1, SUMO-2 and the SUMO-1/2 chimera by immunofluorescence microscopy (Figure 5C). Consistent with stable conjugation to RanGAP1 and association with Nup358, tagged SUMO-1 was detected at the nuclear envelope as a discontinuous ring that co-localized with RanGAP1. Consistent with less stable conjugation to RanGAP1, SUMO-2 and the SUMO-1/2 chimera were detected throughout the nucleoplasm, but did not co-localize with RanGAP1 at NPCs. Thus, the SUMO-1/2 chimera is less stably conjugated to RanGAP1 in vivo relative to SUMO-1, supporting the hypothesis that paralog-selective modification is affected by Nup358 binding and protection from isopeptidases.
Effects of Nup358 and isopeptidase expression on RanGAP1 sumoylation
To test the prediction that Nup358 is critical for the accumulation of SUMO-1 modified RanGAP1 in vivo, we inhibited its expression using RNA interference (RNAi). Cells were transfected with siRNAs specific for Nup358 for increasing lengths of time and lysates were prepared and analyzed (Figure 5D). Blots probed with mAb 414, which recognizes multiple nucleoporins including Nup358 and Nup153, indicated a gradual decrease in Nup358 expression between 36 and 72 hrs post-transfection. Blots probed with antibodies specific for RanGAP1 revealed a parallel decrease in SUMO-1 modified RanGAP1, consistent with Nup358 being critical for its accumulation.
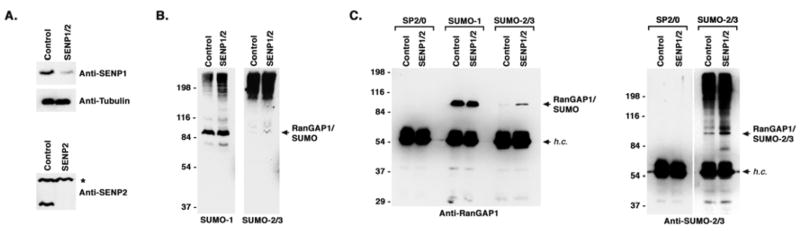
We also used RNAi to inhibit expression of individual SENPs and thereby test the prediction that isopeptidases influence the sumoylation status of RanGAP1. We specifically focused on SENP1 and SENP2, as these isopeptidases associate with NPCs (Bailey and O’Hare, 2004; Hang and Dasso, 2002; Zhang et al., 2002). 293T cells were co-transfected with siRNAs specific for SENP1 and SENP2 using conditions under which their expression levels were reduced by >90% (Figure 6A). The effect of SENP1 and SENP2 knockdown on overall sumoylation levels was investigated by immunoblot analysis (Figure 6B). The levels of SUMO-1 conjugates detected in SENP1 and SENP2 depleted cells were slightly higher than those detected in controls cells, although the pattern of conjugates did not differ significantly. In contrast, overall SUMO-2/3 conjugate levels did not differ significantly between control cells and SENP1 and SENP2 depleted cells. However, a band migrating with the expected molecular mass of SUMO-2/3 modified RanGAP1 (~90 kDa) was noticeably increased in knockdown cells.
Figure 6.
Isopeptidases affect paralog selective RanGAP1 sumoylation in vivo. A. 293T cells were either mock transfected (Control) or co-transfected with SENP1 and SENP2 specific siRNAs (SENP1/2). 48 hr after transfection, whole cell lysates were prepared and analyzed by immunoblotting with SENP1 or tubulin specific antibodies (top panel). SENP2 knockdown was confirmed by co-transfecting FLAG-tagged SENP2 expressing cells (Control) with SENP2 specific siRNAs (SENP2) and immunoblotting with a SENP2 specific antibody (bottom panel). A non-specific cross-reacting band (*) served as a loading control. B. Whole cell lysates were prepared from mock-transfected cells (Control) or cells co-transfected with SENP1 and SENP2 specific siRNAs (SENP1/2). Lysates were analyzed by immunoblotting with SUMO-1 or SUMO-2/3 specific antibodies. C. Immunopurifications with SP2/0 control antibodies or SUMO-1 or SUMO-2/3 specific antibodies were performed using lysates from mock-transfected cells (Control) or cells co-transfected with SENP1 and SENP2 specific siRNAs (SENP1/2). Immunopurified SUMO conjugates were analyzed by immunoblotting with RanGAP1 or SUMO-2/3 specific antibodies. SUMO modified RanGAP1 and antibody heavy chains (h.c.) are indicated.
To determine whether levels of SUMO-2/3 modified RanGAP1 are elevated in SENP1 and SENP2 depleted cells, we performed immunopurifications with SUMO-1 and SUMO-2/3 specific antibodies and analyzed purified conjugates by immunoblot analysis with SUMO-1, SUMO-2/3 or RanGAP1 antibodies (Figure 6C). Nearly equivalent levels of SUMO-1 modified RanGAP1 were immunopurified from both control cells and from cells depleted of SENP1 and SENP2. In contrast, levels of SUMO-2/3 modified RanGAP1 were significantly higher in SENP1 and SENP2 depleted cells compared to control cells. No SUMO modified conjugates were immunopurified with the control antibody, SP2/0. Only modest effects on SUMO-2/3 modification of RanGAP1 were observed upon depletion of SENP1 or SENP2 individually (Figure S4). These findings demonstrate that SUMO-2/3 is conjugated to RanGAP1 in vivo and that accumulation of SUMO-2/3 modified RanGAP1 is limited by the activities of SENP1 and SENP2.
Modification of RanGAP1 in SUMO-1 deficient cells
Lastly, we investigated the effects of inhibiting SUMO-1 expression on RanGAP1 sumoylation. Two cell lines were developed that stably express SUMO-1 specific short hairpin RNAs (shRNAs). Immunoblot analysis was performed using cell lysates derived from these cells and control cells (Figure 7A). Analysis with a SUMO-1 specific antibody revealed that levels of free SUMO-1 and high molecular mass SUMO-1 conjugates were significantly reduced in the shRNA expressing cells compared to control cells (Figure 7A). Notably, a band corresponding in molecular mass to SUMO-1 modified RanGAP1 was detected in shRNA expressing cells, but at a level equal to ~one-third of the level detected in control cells. Despite the reduction in levels of SUMO-1 modified RanGAP1, however, immunoblot analysis with a RanGAP1-specific antibody revealed that the levels of modified RanGAP1 were similar between control and shRNA expressing cells. An explanation for this apparent discrepancy was provided by analysis of with a SUMO-2/3 specific antibody. As expected, there was minimal evidence of SUMO-2/3 modification of RanGAP1 in control cells. However, bands migrating at the expected molecular mass of SUMO-2/3 modified RanGAP1 were detected in both shRNA expressing cell lines.
The above findings are consistent with reports indicating that levels of SUMO-2/3 modified RanGAP1 are elevated in mouse embryonic fibroblasts (MEFs) derived from SUMO-1 knockout mice (Evdokimov et al., 2008; Zhang et al., 2008). We obtained SUMO-1 deficient MEFs and confirmed the effects on RanGAP1 sumoylation by immunoblot analysis (data not shown). To investigate whether SUMO-2/3 modification of RanGAP1 in SUMO-1 deficient MEFs correlates with an association with Nup358 at NPCs, we performed immunofluorescence microscopy using cells fixed and permeabilized with digitonin (Figure 7B). In control cells, SUMO-1 and RanGAP1 signals appeared as punctate rings around the nuclear periphery, consistent with NPC localization. No SUMO-2/3 signal was detected in control cells. In SUMO-1 deficient MEFs, RanGAP1 staining was still detected as a ring around the nuclear periphery, even though SUMO-1 staining was no longer visible. Significantly, SUMO-2/3 staining was also detected as a punctate ring at the nuclear periphery, indicating that the accumulation of SUMO-2/3 modified RanGAP1 in these cells correlates with NPC localization.
DISCUSSION
Although increasing evidence indicates that SUMO-1 and SUMO-2/3 are conjugated to different subsets of proteins and may act as unique signals specifying unique fates, information about the mechanisms regulating paralog-selective sumoylation is limited. RanGAP1 is among the best-studied SUMO substrates and it has been known that it is preferentially modified by SUMO-1 for more than 10 years (Mahajan et al., 1997; Matunis et al., 1996). We therefore used RanGAP1 as a model substrate to gain insights into the molecular mechanisms regulating paralog-selective sumoylation. Surprisingly, we found no evidence that SUMO-1 modification of RanGAP1 is regulated at the level of conjugation; rather, we found that paralog-selective modification is determined at the level of protection from deconjugation.
Several lines of evidence indicated that RanGAP1 is effectively modified by both SUMO-1 and SUMO-2/3. First, we found that RanGAP1 is equally well modified by both paralogs in vitro, indicating that it does not have an innate propensity for selective modification like the recently characterized BLM and USP25 proteins (Meulmeester et al., 2008; Zhu et al., 2008). Second, two lines of evidence indicated that SUMO-1 and SUMO-2/3 are both effectively conjugated to RanGAP1 in vivo. First, we found that SUMO-2/3 modified RanGAP1 accumulates in cells depleted of SENP1 and SENP2, indicating that isopeptidases deconjugate SUMO-2/3 modified RanGAP1 and contribute to SUMO-1 selectivity. We also confirmed the recent findings that SUMO-2/3 modified RanGAP1 accumulates in cells depleted of SUMO-1 (Evdokimov et al., 2008; Zhang et al., 2008). Notably, the accumulation of SUMO-2/3 modified RanGAP1 in SUMO-1 deficient cells correlated with its localization to NPCs. Finally, we found that Nup358, the SUMO E3 ligase most likely to affect RanGAP1 sumoylation in vivo, has no effect on the rate or paralog selectivity of RanGAP1 sumoylation in vitro. This result is consistent with previous findings that RanGAP1 is a unique substrate that interacts tightly with Ubc9 through surfaces adjacent to the conjugation site and is thus modified efficiently in the absence of E3 ligases (Bernier-Villamor et al., 2002; Reverter and Lima, 2005). RNAi-mediated depletion of Nup358 in cultured cells, nonetheless, resulted in a parallel decrease in levels of SUMO-1 modified RanGAP1. Although we could not definitively conclude that this decrease in sumoylated RanGAP1 is due in part to lose of Nup358 E3 ligase activity, our findings provided evidence for an alternative mechanism of action.
Rather than affecting preferential conjugation through E3 ligase activity, our findings indicated that Nup358 binds SUMO-1 modified RanGAP1 preferentially and thereby protects it from isopeptidases. The carboxyl-terminus of Nup358 contains two internal repeats (IR1 and IR2) whose core elements consist of SIMs that independently facilitate interactions with SUMO-1 modified RanGAP1 (Reverter and Lima, 2005; Song et al., 2004; Tatham et al., 2005). Evidence that SUMO-1 modified RanGAP1 is protected from isopeptidases through interactions with Nup358 was first suggested by studies in which deconjugation of NPC-associated RanGAP1 occurred only after solubilization with detergent (Matunis et al., 1996). In the current study, we used in vitro assays containing purified recombinant proteins to demonstrate that SUMO-1 and SUMO-2 modified RanGAP1 form complexes with Nup358 and Ubc9 that protect them from isopeptidases. Importantly, however, we also found that SUMO-1 modified RanGAP1 forms a higher affinity complex with Nup358 and Ubc9 that is more effectively protected from deconjugation. Based on yeast two hybrid results and in vitro binding assays, Nup358 binds preferentially to SUMO-1 (Hecker et al., 2006; Tatham et al., 2005). This preferential SUMO-1 binding likely determines the higher affinity interactions that we observed between Nup358 and SUMO-1 modified RanGAP1, an interpretation that is supported by our findings that swapping the SIM-binding residues of SUMO-1 with residues from SUMO-2 affected Nup358 binding. Future structural studies are required to understand the exact molecular details that determine higher affinity interactions between SUMO-1 modified RanGAP1 and Nup358. Notably, IR2 appears to bind SUMO-1 modified RanGAP1 with a higher affinity than IR1 (Tatham et al., 2005). Thus structures containing IR2, or the entire internal repeat domain, could be particularly insightful.
Relying on structural insights alone, it is difficult to explain how Nup358 and Ubc9 binding might preferentially protect SUMO-1 modified RanGAP1 from isopeptidases. The crystal structure of SUMO-1 modified RanGAP1 complexed with Nup358 and Ubc9 reveals that the six C-terminal residues of SUMO-1, as well as the isopeptide bond between the C-terminal glycine of SUMO-1 and lysine 524 of RanGAP1, are situated within a groove near the Ubc9 active site (Reverter and Lima, 2005). Other structural studies indicate that cleavage of SUMO-1 from RanGAP1 is dependent on interactions between isopeptidases and the same C-terminal residues of SUMO-1 as well as the buried isopeptide bond (Reverter and Lima, 2006; Shen et al., 2006). Thus, for cleavage to occur, SUMO-1 modified RanGAP1 must dissociate from Nup358 and Ubc9. Although there is no structural information available for SUMO-2/3 modified RanGAP1 complexed with Nup358 and Ubc9, it is likely that the isopeptide bond and carboxyl-terminal tail of SUMO-2/3 are similarly inaccessible. This is supported by our findings that SUMO-2 modified RanGAP1 is partially protected from isopeptidases by Nup358 and Ubc9 binding in vitro and that SUMO-2/3 modified RanGAP1 accumulates at NPCs in cells depleted of SUMO-1. Based on these considerations, we favor a model whereby SUMO-1 and SUMO-2/3 modified RanGAP1 are both protected from isopeptidases when associated with Nup358 and Ubc9. However, SUMO-1 and SUMO-2/3 modified RanGAP1 would be differentially protected due to their different affinities for Nup358 and Ubc9 and propensities to exist free of the protective complex. Mammalian SENPs have inherent preferences for deconjugating SUMO-2/3 modified RanGAP1, due in part to higher affinities for SUMO-2/3 (Mikolajczyk et al., 2007; Reverter and Lima, 2006). Thus, preferential interactions with isopeptidases may also contribute to a more rapid turn over of SUMO-2/3 modified RanGAP1. This is supported by our SENP1 and SENP2 knockdown studies demonstrating that isopeptidases play an important role in determining both the paralog selectivity of RanGAP1 sumoylation.
Based on the above considerations, we propose a model in which several factors, all operating at the level of deconjugation, contribute to paralog-selective modification of RanGAP1 (Figure 7C). According to this model, RanGAP1 is modified by both SUMO-1 and SUMO-2/3. Following conjugation, it is proposed that isopeptidases differentially affect the levels of free SUMO-1 and SUMO-2/3 modified RanGAP1. Mammalian cells express six isopeptidases, most of which deconjugate SUMO-2/3 from RanGAP1 more effectively than SUMO-1 (Mikolajczyk et al., 2007; Mukhopadhyay and Dasso, 2007). Thus, independent of Nup358 binding, SUMO-1 modified RanGAP1 is predicted to be more stable based on the intrinsic activity of isopeptidases. Nup358 binding, however, confers additional stability and selectivity. Following association with Nup358 at NPCs, it is proposed that SUMO-1 and SUMO-2/3 modified forms of RanGAP1 are differentially protected from isopeptidases. Higher affinity interactions between SUMO-1 modified RanGAP1 and Nup358 result in its preferential protection; SUMO-1 modified RanGAP1 is able to more effectively compete for Nup358 binding, and SUMO-1 modified RanGAP1 is also more effectively protected once complexed with Nup358 and Ubc9 due to its tighter association. Thus, paralog-selective isopeptidases, more effective competition for Nup358 binding, and more effective protection once bound, collectively contribute to the SUMO-1 selective modification of RanGAP1 in vivo.
Notably, SIMs related to those in Nup358 exist in multiple other proteins, and a subset display paralog-selective binding (Hecker et al., 2006; Kerscher, 2007; Song et al., 2004). Therefore, it can be anticipated that protection from isopeptidases, through interactions between SUMO-modified proteins and proteins containing paralog-selective SIMs, represents a common mechanism for determining paralog-selective sumoylation.
EXPERIMENTAL PROCEDURES
Plasmid constructs
Plasmids for full-length mouse RanGAP1 have been previously described (Matunis et al., 1998), as have plasmids for expressing recombinant SUMO-1 (amino acids 1–97), SUMO-2 (amino acids 1–92), E1 activating enzyme (Aos1/Uba2), E2 conjugating enzyme (Ubc9), Nup358-IR (amino acids 2569–2836) and RanGAP1-NΔ419 (amino acids 420–589 of mouse RanGAP1) (Zhang et al., 2002). The Ulp1 (amino acids 403–621) expression construct was a gift from Dr. Chris Lima (Mossessova and Lima, 2000). Plasmids for production of SUMO-1 and SUMO-2 modified proteins in bacteria (pT-E1E2S1 and pT-E1E2S2) were gifts from Dr. Hisato Saitoh (Uchimura et al., 2004). The SUMO-1 chimera was produced by replacing the coding region for amino acids 31–51 of SUMO-1 in the pGEXT-4T1 expression plasmid (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) with the coding region for amino acids 26–46 of SUMO-2 using PCR-based mutagenesis followed by standard cloning procedures. For expression of SUMO-1 chimera modified proteins in bacteria, the SUMO-1 coding sequence in pT-E1E2S1 was replaced with the SUMO-1 chimera sequence to produce pT-E1E2S1/chimera.
Plasmids for transient in vivo expression of HA-FKBP tagged SUMO-1 and SUMO-2 in mammalian cells have been previously described (Zhu et al., 2006). An HA-FKBP tagged SUMO-1 chimera expression construct was similarly generated by sub-cloning the SUMO-1 chimera cDNA into the pC4EN-F1 plasmid (Ariad Pharmaceuticals Inc., Cambridge, MA). Plasmids for stable transfection were generated by sub-cloning HA-FKBP tagged SUMO-1, SUMO-2 and SUMO-1 chimera cDNAs into pCDNA5/FRT/TO (Invitrogen Inc., Carlsbad, CA).
Protein expression
Full-length RanGAP1 was expressed in rabbit reticulocyte lysate in the presence of [35S]methionine according to the manufacturer’s instructions (Promega Corp., Madison, WI). Recombinant SUMO-1, SUMO-2, E1 activating enzyme, E2 conjugating enzyme, GST-tagged RanGAP1-NΔ419, GST-tagged Nup358-IR (amino acids 2596–2836) and Ulp1 were expressed and purified as described (Mossessova and Lima, 2000; Sampson et al., 2001; Zhang et al., 2002). Thiol-reactive Alexa Fluor 488 was covalently coupled to GST-tagged RanGAP1-NΔ419 according to the manufacture’s instructions (Invitrogen Inc., Carlsbad, CA). Recombinant human isopeptidases were expressed and purified as previously described (Mikolajczyk et al., 2007).
To prepare SUMO-1, SUMO-2 and SUMO-1 chimera modified RanGAP1, pET21RanGAP1-NΔ419 (encoding for His-tagged RanGAP1-NΔ419) was transformed into E. Coli BL21(DE3) together with pT-E1E2S1, pT-E1E2S2, or pT-E1E2S1/chimera. Modified RanGAP1-NΔ419 was purified by nickel-affinity column chromatography followed by MonoQ anion exchange chromatography (GE Healthcare Bio-Sciences Corp., Piscataway, NJ).
In vitro sumoylation assays
In vitro sumoylation assays were performed in 20 μl reactions containing 20 mM HEPES-KOH (pH 7.3), 2 mM DTT, 110 mM KOAc, 2 mM Mg(OAc)2, 4 mM ATP, 0.5 μg SUMO, 150 ng E1 activating enzyme, 15 ng Ubc9 and either 4 μl of in vitro expressed RanGAP1 or 1.5 μg of fluorescently labeled GST-tagged RanGAP1-NΔ419. Where indicated, reactions also contained 10 ng Nup358-IR. Reactions were incubated at 37°C for the indicated times and stopped by the addition of SDS sample buffer. Reactions were separated by SDS-PAGE and visualized by autoradiography or fluorescence detection using a Fuji FLA-7000 bio-imaging analyzer (Fujifilm Corp., Tokyo, Japan).
Nup358 binding and Ulp1 protection assays
To analyze interactions with Nup358, full-length RanGAP1 was expressed in rabbit reticulocyte lysate and modified with either SUMO-1 or SUMO-2. Reaction products were incubated with glutathione beads containing GST-tagged Nup358-IR in buffer containing 20 mM HEPES-KOH (pH 7.3), 1 mM DTT, 110 mM KOAc, and 2 mM Mg(OAc)2. Beads were washed 5 times with binding buffer supplemented with 0, 250 or 350 mM MgCl2 and proteins remaining bound were eluted with SDS sample buffer. Samples were resolved by SDS-PAGE and visualized by autoradiography.
Ulp-1 protection assay were performed in 20 μl reactions containing 25 mM Tris (pH 8.0), 150 mM NaCl, 2 mM DTT, 0.1% Tween-20, 5% glycerol, 400 ng SUMO-1, SUMO-2 or SUMO-1 chimera modified RanGAP1, 250 ng Ubc9, and 500 ng GST-tagged Nup358-IR. Reactions were pre-incubated for 10 min at RT, followed by the addition of 42 ng Ulp1 and further incubation for the indicated times. Control reactions were performed under identical reaction conditions in the absence of Ubc9 and Nup358-IR. Reactions were stopped by the addition of SDS sample buffer and analyzed by immunoblotting with an anti-His antibody.
Biacore analysis
Binding affinities between Nup358 and SUMO modified RanGAP1 were analyzed using a Biacore 3000 (GE Healthcare Biosciences Corp., Piscataway, NJ). Anti-GST antibodies were immobilized onto CM5 sensor chips via amine coupling according to the manufacture’s instructions (GE Healthcare Biosciences Corp., Piscataway, NJ). GST-tagged Nup358-IR, or GST alone as a reference, was subsequently captured on the chips and non-specific binding sites blocked by injection of BSA. Binding experiments were performed in HSP-EP/BSA buffer (10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, 0.005% v/v Surfactant P20 and 0.1mg/ml BSA) at 25°C. SUMO-1, SUMO-2 or SUMO-1 chimera modified RanGAP1-NΔ419 was mixed with Ubc9 at a molar ratio of 2:1 in HBS-EP/BSA buffer and serial dilutions of the mixture were injected for 125 sec at a flow rate of 75 μl/sec followed by 4 min of HBS-EP/BSA buffer at the same flow rate. Sensor chips were washed twice with regeneration buffer (10 mM glycine (pH1.5), 0.05% Surfactant P20) following each injection.
Kinetic constants were calculated by nonlinear fitting to the association and dissociation curves using BIAevaluation v4.4 software (GE Healthcare Biosciences Corp., Piscataway, NJ). Apparent experimental equilibrium responses (KD) and Rmax values were obtained using a “two-state reaction model with conformation change”. All binding assays were performed a minimum of three times.
Stable cell lines, cell culture and RNA interference
Stable cell lines expressing HA-FKBP tagged SUMO-1, SUMO-2 and SUMO-1 chimera under control of a tetracycline inducible promoter were established using the Flp-In T-REx system and maintained essentially as described by the manufacturer (Invitorgen Inc. Carlsbad, CA). Flp-In T-Rex-293 parent and stably transfected cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% tetracycline-free fetal bovine serum, 10 mM HEPES, 1% penicillin and streptomycin, 200 μg/ml Zeocin and 15 μg/ml Blasticidin S at 37 °C, 5% CO2.
To produce cells stably expressing SUMO-1 specific short hairpin RNAs, HEK-293 cells were transfected with shRNA constructs TRCN0000012889 or TRCN0000012890 (Sigma-Aldrich, St. Louis, MO). Stably transfected cells were selected for and maintained by growth in medium containing 2μg/ml puromycin. SUMO-1 knockout and wild type MEFs were generously provided by Dr. Olli Janne (University of Helsinki) and cultured as described (Zhang et al., 2008).
To knockdown Nup358 expression, HeLa cells were transfected with Nup358-specific siRNAs (Saitoh et al., 2006) (140 pmol/3.5 cm well) using Lipofectamine 2000, according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Cell lysates were prepared at the indicated times following addition of the siRNAs and analyzed by immunoblotting. For isopeptidase knockdowns, siRNAs specific for SENP1 (5′-UCCUUUACACCUGUCUCGAUGUCUU-3′) and/or SENP2 (5′-GCCCAUGGUAACUUCUGCUUGUAAU-3′) (Invitrogen, Carlsbad, CA) were transfected into 293T cells and cell lysates for immunopurifications were prepared 72 hrs following transfection. Knockdown efficiencies were verified by either direct analysis of endogenous SENP1 using SENP1 specific antibodies, or by evaluating knockdown of the expression of a co-transfected Myc-tagged SENP2.
Immunoblotting and immunopurification
Immunoblot analysis was performed using standard enzyme-linked chemiluminescence procedures. Primary antibodies were obtained from the following sources: RanGAP1 monoclonal antibody 19C7 and SUMO-1 monoclonal antibody 21C7 (Matunis et al., 1996), SUMO-2/3 monoclonal antibody 8A2 (Zhang and Sarge, 2008), rabbit anti-HA (Santa Cruz Biothechnology, Inc., Santa Cruz, CA), mouse anti-tubulin (SIGMA, St. Louis, MO), and mouse anti-His (GE Healthcare Biosciences Inc., Piscataway, NJ).
RNAi-treated or mock-treated cells were lysed in 50 mM TrisHCl, pH 7.4, 150 mM NaCl, 1% Triton-X 100, 2 mM EDTA, 1% deoxycholate, 1% SDS, Benzonase (Novagen), protease inhibitors, and 20 mM NEM and sonicated on ice. Clarified cell lysates were diluted 1 to 10 in buffer containing 50 mM TrisHCl, pH7.4, 150 mM NaCl, 1% Triton-X 100, 2 mM EDTA, 1% deoxycholate and were incubated with SP2/0, SUMO-1 (21C7), or SUMO-2/3 (8A2) antibodies beads for 5 hrs at 4°C. Beads were washed 5 times in lysis buffer containing 0.1 % SDS. Immunopurified proteins were eluted using SDS sample buffer and resolved by SDS-PAGE.
Immunofluorescence microscopy
For stably transfected cells, HA-FKBP-tagged SUMO-1, SUMO-2 and SUMO-1 chimera protein expression was induced for 48 hr with doxycycline. Cells were spun onto glass slides and permeabilized for 7 min at RT with 20 ng/ml digitonin (Sigma, St. Louis, MI) in buffer containing 20 mM HEPES-KOH (pH 7.3), 110 mM KOAc and 2 mM Mg(OAc)2. Cells were washed with PBS followed by fixation for 45 min at RT in 2% formaldehyde in PBS. Cells were co-labeled with anti-HA and RanGAP1 antibodies followed by Alexa 488 and 594 labeled secondary antibodies (Invitrogen Inc., Carlsbad, CA).
SUMO-1 knockout and wild type MEFs were permeabilized using 40 μg/ml digitonin (Sigma, St. Louis, MI) in buffer containing 20 mM HEPES-KOH (pH 7.3) 110 mM KOAc and 2 mM Mg(OAc)2 for 6 min at RT and then fixed in 2% formaldehyde in PBS for 40 min. Cells were co-labeled with SUMO-2/3-specific antibodies (rabbit polyclonal) and anti-SUMO-1 (mAb 21C7) or anti-RanGAP1 (mAb 19C7) antibodies followed by Alexa 488 and 594 labeled secondary antibodies (Invitrogen Inc., Carlsbad, CA).
Acknowledgments
We thank members of the Matunis laboratory and Drs. Judith Bender and Val Culotta for insightful discussions during the course of this work. We are grateful to Joshua Sims and Yaun Lin for assistance with Biacore-related experiments and data analysis and Dr. Mary Dasso for providing SENP1 antibodies. This work was supported by a grant from the National Institutes of Health (GM060980 to M.J.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alkuraya FS, Saadi I, Lund JJ, Turbe-Doan A, Morton CC, Maas RL. SUMO1 haploinsufficiency leads to cleft lip and palate. Science. 2006;313:1751. doi: 10.1126/science.1128406. [DOI] [PubMed] [Google Scholar]
- Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell. 2004;15:5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D, O’Hare P. Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J Biol Chem. 2004;279:692–703. doi: 10.1074/jbc.M306195200. [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Hang J, Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem. 2002;277:19961–19966. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Kerscher O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Wu J, Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell. 2008;30:610–619. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk J, Drag M, Bekes M, Cao JT, Ronai Z, Salvesen GS. Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs. J Biol Chem. 2007;282:26217–26224. doi: 10.1074/jbc.M702444200. [DOI] [PubMed] [Google Scholar]
- Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat Struct Mol Biol. 2006;13:1060–1068. doi: 10.1038/nsmb1168. [DOI] [PubMed] [Google Scholar]
- Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol Cell Proteomics. 2005;4:56–72. doi: 10.1074/mcp.M400149-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N, Uchimura Y, Tachibana T, Sugahara S, Saitoh H, Nakao M. In situ SUMOylation analysis reveals a modulatory role of RanBP2 in the nuclear rim and PML bodies. Exp Cell Res. 2006;312:1418–1430. doi: 10.1016/j.yexcr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- Shen L, Tatham MH, Dong C, Zagorska A, Naismith JH, Hay RT. SUMO protease SENP1 induces isomerization of the scissile peptide bond. Nat Struct Mol Biol. 2006 doi: 10.1038/nsmb1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Tatham MH, Kim S, Jaffray E, Song J, Chen Y, Hay RT. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat Struct Mol Biol. 2005;12:67–74. doi: 10.1038/nsmb878. [DOI] [PubMed] [Google Scholar]
- Tatham MH, Kim S, Yu B, Jaffray E, Song J, Zheng J, Rodriguez MS, Hay RT, Chen Y. Role of an N-terminal site of Ubc9 in SUMO-1, -2, and -3 binding and conjugation. Biochemistry. 2003;42:9959–9969. doi: 10.1021/bi0345283. [DOI] [PubMed] [Google Scholar]
- Uchimura Y, Nakao M, Saitoh H. Generation of SUMO-1 modified proteins in E. coli: towards understanding the biochemistry/structural biology of the SUMO-1 pathway. FEBS Lett. 2004;564:85–90. doi: 10.1016/S0014-5793(04)00321-7. [DOI] [PubMed] [Google Scholar]
- Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- Zhang FP, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Janne OA. Sumo-1 Function is Dispensable in Normal Mouse Development. Mol Cell Biol. 2008 doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol Cell Biol. 2002;22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sarge KD. Mel-18 interacts with RanGAP1 and inhibits its sumoylation. Biochem Biophys Res Commun. 2008;375:252–255. doi: 10.1016/j.bbrc.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhu S, Guzzo CM, Ellis NA, Sung KS, Choi CY, Matunis MJ. Small Ubiquitin-related Modifier (SUMO) Binding Determines Substrate Recognition and Paralog-selective SUMO Modification. J Biol Chem. 2008;283:29405–29415. doi: 10.1074/jbc.M803632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Zhang H, Matunis MJ. SUMO modification through rapamycin-mediated heterodimerization reveals a dual role for Ubc9 in targeting RanGAP1 to nuclear pore complexes. Exp Cell Res. 2006;312:1042–1049. doi: 10.1016/j.yexcr.2005.12.031. [DOI] [PubMed] [Google Scholar]