Abstract
HMGB proteins are abundant non-histone components of eukaryotic chromatin. The biological function of DNA sequence nonspecific HMGB proteins is obscure. These proteins are composed of one or two conserved HMG box domains, each forming three alpha helices that fold into a sequence nonspecific DNA-binding module recognizing the DNA minor groove. Box A and box B homology domains have subtle sequence differences such that box B domains bend DNA strongly while DNA bending by isolated box A domains is weaker. Both box A and box B domains preferentially bind to distorted DNA structures. Here we show using DNA cyclization kinetics assays in vitro and E. coli DNA looping assays in vivo that an isolated HMG box A domain derived from human HMGB2 folds poorly and does not enhance apparent DNA flexibility. Surprisingly, substitution of a small number of cationic residues from the N-terminal leader of a functional yeast box B protein, Nhp6Ap, confers the ability to enhance DNA flexibility. These results demonstrate important roles for cationic leader amino acids in HMGB folding, DNA interaction, and DNA bending.
HMGB1 sequence non-specific DNA binding proteins are abundant non-histone proteins in eukaryotic cells (1-3). These small proteins are composed of one [e.g. S. cerevisiae Nhp6Ap (4-8), D. melanogaster HMG-D (9, 10)] or two [e.g. vertebrate HMGB1, HMGB2 (11-15), S. cerevisiae Hmo1p (16)] repeats of a conserved ~80-residue “HMG box” motif. Highly charged flanking sequences are also often present (3). Thus, HMGB proteins are a subset of the proteins that carry one or two domains homologous to the ancestral HMG box. HMGB proteins accumulate to greater than micromolar concentrations in eukaryotic cells such that there is approximately one HMGB protein for every 5-10 nucleosomes in chromatin.
Among the striking features of HMGB proteins is their ability to induce strong DNA bending and, consequentially, their preference for binding to distorted DNA structures such as cruciforms (11, 16-18) and chemical lesions (19-23). DNA bending in these complexes can exceed 90°. In this sense, HMGB proteins are functionally (though not structurally) reminiscent of the prokaryotic nucleoid protein HU (24). There is evidence that HU functions by inducing a flexible hinge in DNA (25, 26), and HMGB proteins appear to alter DNA structure through formation of hinges with enhanced flexibility (27-29).
The biological functions of HMGB proteins are not understood. Initial insight into HMGB function came from pioneering studies by Johnson and co-workers (6, 7, 12). These authors found that HMGB proteins could substitute for prokaryotic HU proteins in transactions involving tightly bent DNA, such as recombination (12). These observations were extended to living E. coli where defects associated with loss of the HU nucleoid protein were suppressed by HMGB expression (7). There is also evidence that HMGB proteins facilitate the DNA binding of some sequence-specific DNA binding transcription factors and recombinases [reviewed in (3)]. In some cases this HMGB participation appears to be catalytic (30). The Nhp6A protein of budding yeast has been shown to enhance transcription activation at least in part through facilitation of Gal4p binding to DNA (6). Similarly, lethal hypoglycemia in newborn mice lacking HMGB1 may be due to defective glucocorticoid receptor binding to DNA (31). DNA bending to facilitate protein-protein interactions may be an important role of HMGB proteins in the function of enhancesomes (32, 33). On the other hand, HMGB proteins participate as subunits of the FACT complex required for efficient transcription of chromatin templates in yeast (34). In addition, the chromatin of yeast cells lacking the two-box HMGB protein Hmo1p becomes sensitive to micrococcal nuclease digestion (35). These results suggest multiple roles for HMGB proteins in chromatin structure and function (36).
We became interested in HMGB proteins after observing that the activator-dependence of in vitro transcription from plasmid DNA templates in HeLa cell nuclear extract was not consistent with the expectations of DNA looping over short distances (37). We subsequently confirmed that mammalian nuclear extracts are rich in a heat-resistant activity that dramatically enhances the apparent flexibility of DNA (38). This work also showed that recombinant HMGB proteins could substitute for at least some of this activity. We hypothesized that HMGB proteins increase the apparent flexibility of DNA in regions not packaged with histone octamers. Recent studies using single molecule force spectroscopy confirmed that apparent DNA persistence length decreases substantially in the presence of low concentrations of HMGB proteins, while higher HMGB concentrations can produce a more rigid protein-DNA filament (28, 29).
To study apparent DNA physical properties in vivo, we have recapitulated classic experiments of Record (39-41) and Müller-Hill (42-46) involving DNA looping in the E. coli lac operon. Our work has confirmed that the physical properties of DNA measured in vivo suggest enhanced bending and twisting flexibilities relative to in vitro expectations, and that nucleoid proteins modify these properties (47, 48). We have recently shown that certain eukaryotic HMGB proteins can complement the loss of bacterial HU (49). Thus, effects on in vivo DNA looping in E. coli can provide a sensitive bioassay for HMGB function.
The HMG box sequence encodes three alpha-helices that fold into an “L”-shaped structure (Fig. 1A). This module interacts with the DNA minor groove to both widen the groove and allow partial intercalation of one or more amino acid side chains between stacked base pairs (3). The combination of these perturbations can cause strong DNA bending away from the engaged DNA surface. HMG box A of human HMGB2 (Fig. 1A, left) is shown bound to a DNA segment distorted by platinum cross-linking (21). Also illustrated in Fig. 1A (center) is the structure of the yeast single-box protein Nhp6Ap complexed with unmodified DNA (4). Putative intercalating residues are shown in blue. Importantly, Nhp6Ap also contains a strongly cationic leader sequence (Fig. 1A, center, magenta) disposed in the major groove to asymmetrically neutralize the compressed sugar-phosphate backbone. There is evidence that this interaction contributes significantly to DNA bending (50). Thus, sharp DNA bending by HMGB proteins can involve minor groove widening, amino acid wedging, and asymmetric charge neutralization. Favorable electrostatic forces have been shown to dominate other interactions in both DNA binding and bending by sequence non-specific HMGB proteins (50). It is noteworthy that strong and sequence-nonspecific bending by the prokaryotic nucleoid protein HU involves minor groove interactions, but within a completely different scaffold (Fig. 1A, right).
Figure 1.
Proteins analyzed in this work. A. Molecular models of sequence nonspecific architectural proteins: mammalian HMGB domain HMGB2(box A) [left, PDB code 1ckt, (21)], yeast HMGB protein Nhp6Ap [center, PDB code 1j5n (4)], and E. coli HU [right, PDB code 1p51,(25)]. Bent DNA is rendered as cyan spheres, protein is shown as a red ribbon. Potential intercalating residues in the HMGB proteins are shown in blue. The cationic N-terminal leader of Nhp6Ap is shown in magenta. B. Sequence alignments of WT and chimeric HMGB proteins. Helical regions I-III are indicated above. Potential intercalating residues are indicated by vertical shading. Residues derived from HMGB2(box A) and Nhp6Ap are shown in plain text and bold text, respectively. The c-Myc epitope tag decapeptide is indicated by black rectangles. Proteins 1-5 and 16 were tested in vitro for their ability to increase the J-factor of 200-bp DNA fragments. Proteins 6-15 were tested in E. coli for their ability to complement the lac repression defect of hupA/BΔ cells.
Though similar in sequence and structure, it has been noted that subtle features of the “A box” domain of the two-box HMGB1 protein distinguish it from the “B box” domain (3). The A box domain is distinct in the shape and orientation of helix I, and the identity of potential intercalating residues. Isolated A box domains are reportedly less effective at DNA bending (12). The sequence of a his6-tagged version of human HMGB2(box A) is illustrated as construct 1 in Fig. 1B. Whereas the intercalating phenylalanine in helix II is present in both box A and box B domains of 2-box HMGB proteins, the alanine in helix I does not intercalate strongly (3). Furthermore, box A and box B domains are distinguished by the relative lengths and geometries of helices I and II, with helix I being short and straight in box A, but bent in box B (3). The single box HMGB protein Nhp6Ap conforms to the box B family and is shown as construct 16 in Fig. 1B. Nhp6Ap is distinguished from HMGB2(box A) by the identity of the intercalating residue in helix I (methionine in Nhp6Ap), a longer helix I, and a highly-cationic leader sequence.
In the present study we apply in vitro and in vivo assays to understand what minimal features of an HMG B box confer on an A box the ability to enhance apparent DNA flexibility.
MATERIALS AND METHODS
Bacterial strains and gene disruptions
The hupA and hupB genes in parental E. coli strain FW102(51) were disrupted as described (47, 52). Diagnostic PCR amplification following conjugation and selection was used to confirm strain deletion status and the presence of the proper looping assay episomes.
Expression and purification of HMGB proteins
Plasmid pJ583 encodes human HMGB2(box A) cloned in-frame with a his6 tag in plasmid pET-15b (Novagen). Mutations were introduced using the QuickChange site-directed mutagenesis kit. (Stratagene). Plasmids encoding HMGB2(box A) and variants were transformed into E. coli BL21-Gold (Stratagene) and 6-mL overnight cultures were prepared from single colonies in LB medium containing carbenicillin. Five mL of overnight culture were washed with fresh LB to remove secreted β-lactamase and the cells were used to seed a 250-mL LB culture. Cells were grown at 37°C with shaking until the culture reached a cell density corresponding to an OD600 of 0.6. IPTG was added to a final concentration of 1 mM and cells were grown at 37°C for three h with shaking, pelleted by centrifugation at 6000 g, and stored at -80°C. The induced cell pellet was then resuspended in 10 mL binding buffer (Novagen) per 250 mL pelleted cells, sonicated on ice with 15-s pulses separated by 1-min cooling intervals. The lysate was then subjected to centrifugation at 15000 g for 20 min at 4°C and the supernatant was recovered. His6-tagged protein was purified using Ni-NTA agarose resin (Qiagen) per the manufacturer's recommendations. Briefly, washed Ni-NTA agarose resin was added in a 1:4 (v:v) ratio to the lysate, gently rotated at 4°C for 1 h, and the resin collected by centrifugation at 2000 rpm for 2 min at 4°C in a clinical centrifuge. Beads were washed three times with wash buffer (10 mL), and protein was eluted from the resin twice by gentle rotation in 10 mL elution buffer for 5 min at 4°C. Eluted sample volume was reduced to 2 mL using 5000 molecular weight cut-off centrifugal cartridges (Vivaspin) and proteins were dialyzed in 3500 molecular weight cut-off cassettes (Slide-a-Lyzer, Pierce) at 4°C against 1 L buffer containing 20 mM HEPES pH 7.5, 100 mM KCl, 1 mM EDTA and 1 mM DTT, followed by a second dialysis against the same buffer containing 5% glycerol. Proteins were then purified by reverse phase HPLC on a Jupiter C18 column (250 × 21.2 mm, 15 μm; Phenomenex, Torrance, CA) in 0.1% TFA/water with a 50 min gradient from 10-70% B, where B was 80% aqueous acetonitrile/0.1% TFA. Lyophilized proteins were resuspended and characterized by mass spectrometry.
HMGB protein characterization in vitro
CD spectra were recorded using a Jasco 810 spectropolarimeter in continuous mode, collecting measurements every 1 nm between 200 and 260 nm at 295 K with an averaging time of 5 s. Protein sample concentrations were 2-10 μM in 20 mM Tris-HCl pH 8.0, 30 mM KCl, 10 mM MgCl2 with or without addition of 400 μM (base pairs) duplex DNA. All buffer and nucleic acid contributions to ellipticity were subtracted after data collection, and raw ellipticity data were converted to mean residue ellipticity (MRE).
Fluorescence anisotropy measurements were used to characterize DNA/protein interactions and the fraction of active protein. A 22-bp fluorescein-labeled duplex DNA probe was created by the annealing of oligonucleotides LJM-1100 (5′-AGT2GAG4ACT3C3AG2C-F; F indicates fluorescein) and LJM-1101 (5′-GC2TG3A3GTC4TCA2CT-3′). Fluorescein labeling efficiency was confirmed by comparing absorbance measurements at 495 nm and 260 nm. 10 nmol of complementary DNA strands were mixed in 48.5 μL TE buffer pH 8.0, 1.5 μL 1M NaCl was added, and the sample annealed (92°C, 10 min; 65°C, 5 min; 37°C, 1.5 h; 22°C, 30 min). The resulting duplex was purified by electrophoresis through a 12% native polyacrylamide gel in 0.5X TBE buffer. The fluorescent probe duplex was cut from the gel after detection over a TLC indicator plate under ultraviolet illumination and eluted overnight in 300 mM NaOAc, pH 4.5, precipitated from ethanol, suspended in TE buffer, and quantitated. Fluorescence anisotropy measurements for equilibrium dissociation constant estimation were performed in 96-well microtiter plates with 200-μL binding reactions containing fluorescent probe (5 nM) and protein dilutions in 20 mM Tris-HCl pH 8.0, 30 mM KCl, 10 mM MgCl2, and 100 μg/mL BSA. Protein dilutions were prepared in binding buffer with 10% glycerol and 100 μg/mL BSA. The Safire2 instrument (TECAN) was controlled with X-Fluor4 software. Data were fit to equation 1 to estimate the equilibrium dissociation constant.
| 1 |
where A indicates sample anisotropy, subscripts obs, min and max indicate observed, minimum, and maximum (respectively), Pt is the total concentration of added protein, and Kd is the fit equilibrium dissociation constant. Pt ~ Pfree under these binding conditions of large protein excess over fluorescent DNA probe. Estimation of the protein active fraction employed similar methods, except titration was performed with the fluorescent duplex DNA probe concentration at 250 nM (⪢Kd), and the discontinuity in the titration plot indicated the point of protein/DNA concentration equivalence.
J-factor estimate by ligase-mediated DNA cyclization assay
Plasmids pJ823, pJ825, pJ827, pJ829, pJ831, and pJ833 contain intrinsically straight ~200-bp sequences and were the kind gifts of A. Vologodskii (53). Probe sequences were amplified by PCR using primers LJM-3222 (5′-G3TA2CGC2AG3T4-3′) and LJM-3223 (5′-TGTGAGT2AGCTCACTCAT2AG2-3′) such that an additional ~100 bp was included upstream and downstream of the HindIII sites defining the probe. Polymerase chain reactions included plasmid template (20 ng), primers (400 nM), BSA (100 μg/mL), Taq DNA polymerase buffer (Invitrogen), MgCl2 (4 mM), 200 μM each dNTP, 50 μCi [α-32P]-dATP, and 3.2 U/uL Taq DNA polymerase (Invitrogen). Cycle parameters were: 3 min at 98°C; 30 cycles of 30 s at 94°C, 30 s at 55°C, 45 s at 72°; 5 min at 72°C; 4°C hold. PCR products were treated with 50 ng/μL proteinase K in the presence of 5mM EDTA, 10mM Tris-HCl pH8.0, and 0.5% SDS at 37°C for 1 h. The DNA was extracted with phenol:chloroform (1:1) and precipitated from ethanol. The DNA was digested overnight at 37°C with 40 U HindIII (New England Biolabs) in the presence of 100 μg/mL BSA. Digested product was loaded onto a 5% native polyacrylamide (29:1 acrylamide:bisacrylamide) gel and visualized by exposure of the wet gel to BioMAX XR film. The desired ~200 bp DNA product was cut from the gel and eluted overnight at 22°C in 50 mM NaOAc buffer, pH 7.0. The final DNA concentration was obtained by determining the fraction of total original radiolabel present in the purified product, multiplying this fraction by the total concentration of dATP in the labeling reaction, and dividing this result by the number of dA residues in each duplex DNA product.
DNA cyclization reactions (10 μL) were performed with 10 nM DNA probe and the indicated concentration of HMGB protein in T4 DNA ligase buffer (20 mM Tris-HCl pH 8.0, 30 mM KCl, 100 μg/mL BSA, 1.8 mM ATP, 10 mM MgCl2), and T4 DNA ligase (20-50 U/μL; New England Biolabs) at 22°C. Reactions were stopped at the indicated times by adjusting EDTA to 20 mM. DNA was purified using QIAQuick PCR clean up columns (Qiagen). DNA was eluted in 30 μL of elution buffer and 15 μL samples loaded onto 5% (29:1 acrylamide:bisacrylamide) native polyacrylamide gels. After electrophoresis in 0.5X TBE buffer, gels were dried and exposed to storage phosphor screens. Circular DNA products were identified in preliminary experiments by their resistance to Bal31 exonuclease (New England Biolabs).
Image quantitation was performed with ImageQuant software. Intensities of the monomer circle (MC) product and the sum of linear dimers and all other ligation products (LD) were determined for each time point, corrected for background signal, and their ratio plotted against time. The J-factor was determined using equation 2:
| (2) |
where M0 is the initial DNA concentration and indicated limit is the y-axis intercept of the plot. Conditions of time, ligase concentration and initial DNA concentration were empirically determined for each DNA probe and HMGB protein so that labeled starting material was not significantly depleted and linear fits were obtained.
Cloning and expression of HMGB proteins in bacteria
HMG protein expression constructs were created by inserting purified PCR products into plasmid pJ1035, a modified version of pLX20 (51) containing a promoter for a moderate level of protein expression and an inactive lac operator. In addition, a 10-amino acid c-Myc epitope tag (EQKLISE2DL) was added either to protein N-termini (constructs 6 and 7), or C-termini (constructs 8-13) to facilitate analysis of accumulation. To create plasmid construct 8 (pJ1491), forward primer LJM-3314 (GCTCTAGA2TG3TA3G2AGAC4A2C) and reverse primer LJM-3318 (CGA2GCT2GCAGCTACAGATCT2CT2CAGA3TA2GT5GT2CAC2T3G3AG2A2C; c-Myc tag in bold italics) were used to amplify from M1 to G83 of HMGB2(box A). Plasmid construct 6 (pJ1493) was created using forward primer LJM-3315 (GCTCTAGA2TGGA2CA5CT2AT3CTGA2GA2GATCTGG2TA3G2AGAC4A2C), and reverse primer LJM-3327 (CGA2GCT2GCAGCTA2C2T3G3AG2A2CGTA2T5C). Plasmid construct 10 was created by PCR amplification using forward primer LJM-3621 (GCTCTAGA2TGAGA3GA2GA2G2AC4A2CA2GC2G) and reverse primer LJM-3318. Plasmid construct 12 was created with forward primer LJM-3620 (GCTCTAGA2TGA2GA2GAGA2C2ACTAGA3GA2GA2G2AC4A2CA2GC2G) and reverse primer LJM-3318. All PCR products were inserted between XbaI and HindIII restriction sites of pJ1035. Site-directed mutagenesis (Stratagene) employed primers LJM-3613 (CA4TGTC2TCGTACATGT2CT2CGTGCAGAC) and LJM-3614 (GTCTGCACGA2GA2CATGTACGAG2ACAT4G) to introduce the A17M mutation in constructs 7, 9, 11, and 13 from 6, 8, 10 and 12, respectively. Accumulation of epitope-tagged HMGB proteins was monitored by western blotting of E. coli extracts using primary antibody op10T (CalBiochem) as previously described (49).
In vivo DNA looping assay
DNA looping constructs were based on plasmid pJ992, created by modifications of pFW11-null (51) as previously described (47-49). Reporter gene assays were performed as previously described (49).
RESULTS
Experimental strategy
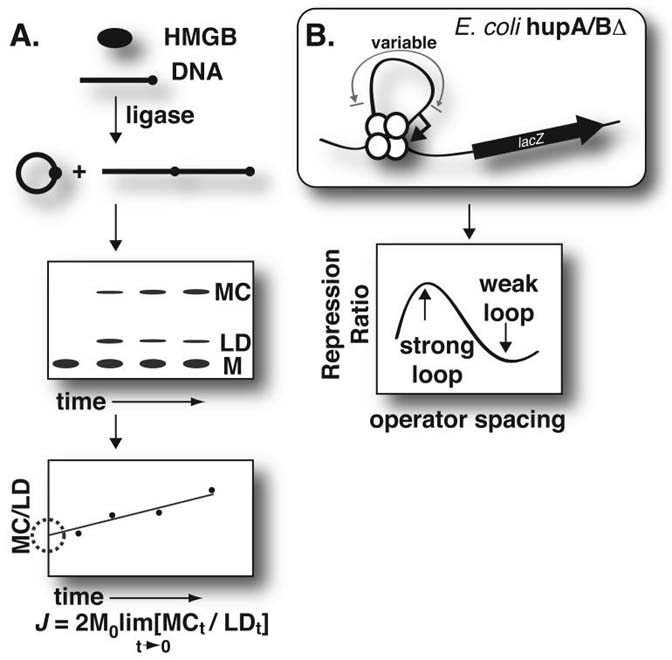
The experimental strategy for comparing activities of single HMGB box chimeric proteins is shown in Fig. 2. In vitro analyses of DNA flexibility enhancement were performed as shown in panel A. Purified his6-tagged HMGB protein was mixed with a radiolabeled sample of 200-bp intrinsically straight duplex DNA containing HindIII cohesive termini (53) and incubated with T4 DNA ligase for different times to generate a mixture of labeled products (Fig. 2A, top). These DNAs were separated by native polyacrylamide gel electrophoresis and quantitated by storage phosphor technology (Fig. 2A, middle). The ratio of monomer circles (MC) to all other species including (and derived from) linear dimers (LD) is extrapolated to zero time to estimate the J-factor [Fig. 2A, bottom; (54)]. The J-factor, in turn, expresses the effective local concentration of one terminus of the 200-bp DNA with respect to the other, a parameter that should increase with increasing longitudinal DNA flexibility. The J-factor also measures DNA twist because cohesive termini must be in proper rotary alignment for ligation. Our simulations of ring closure kinetics in the presence of HMGB proteins (L. Czapla, E. Bystry, W. Olson, L.J. Maher, in preparation) have shown that J-factors can be influenced by protein-induced changes in both DNA twist and DNA bending at the site of the bound protein. In addition, we have shown that bound HMGB proteins cause enhanced anisotropic DNA flexibility in the protein/DNA complex (27). This combination of effects (changes in DNA twist, DNA bending, and anisotropic DNA flexibility) increases the J-factor associated with the ensemble of DNA complexes present during the ligation reaction.
Figure 2.
Experimental strategy. A. In vitro measurement of HMGB protein enhancement of apparent DNA flexibility by ligase-catalyzed DNA cyclization kinetics. Top: HMGB protein was incubated for various times with ~200-bp radiolabeled DNA (M) in the presence of T4 DNA ligase to produce a mixture of intramolecular monomer circles (MC) and products derived from linear dimers (LD). Middle: electrophoretic analysis. Bottom: quantitation and extrapolation estimating the J-factor. B. Top: In vivo measurement of HMGB protein complementation of lac repression loop defect in hupA/Bδ E. coli cells. Lac operator spacing was varied between 75-85 bp in an E. coli hupA/B deletion strain that is disabled in lac repression looping. Bottom: the strength of DNA repression looping as a function of operator spacing and HMGB protein expression is monitored by the repression ratio.
As shown in Fig. 2B, in vivo studies of single HMGB box chimeric proteins were performed by expression of the epitope-tagged proteins in E. coli. The experimental strains harbored F′ episomes engineered to carry a lacZ reporter gene driven by a promoter flanked by two lac operators (47). The upstream operator strongly binds lac repressor, while the downstream operator is weaker. Operator spacing is varied so that the stability of the lac repression loop (Fig. 2B, top) varies with DNA looping energy. This relationship can be monitored as the ratio of induced to uninduced β-galactosidase activities as shown schematically in Fig. 2B, bottom.
In vitro characterization of single HMGB box chimeric proteins
HMGB protein variants 1-5 and 16 (Fig. 1B) were expressed in E. coli and purified. The proteins represent HMGB2(box A) [construct 1], Nhp6Ap [construct 16], and chimeras differing in terms of the identity of the primary intercalating residue in helix I [constructs 4 and 5], and the amount of basic leader sequence substituted from Nhp6Ap [constructs 2, 3 and 5]. Proteins were initially characterized by circular dichroism spectroscopy to monitor folding in the absence of DNA (Fig. 3 and Table I). Nhp6Ap (construct 16) showed a moderate ellipticity signal consistent with α-helix formation in the absence of DNA (Fig. 3A). The corresponding α-helical signal from construct 5 (7 cationic leader substitutions and methionine as the primary intercalating residue) was particularly strong (Fig. 3A). Interestingly, signals from constructs 1-4 were all weak (Fig. 3A), though clearly α-helical (Fig. 3B). Surprisingly, CD signals did not substantially change in the presence of excess DNA (Table I), suggesting that the various similar HMG box proteins strongly differed in their folding stabilities.
Figure 3.
HMGB protein characterization in vitro. A. Analysis of protein folding by circular dichroism spectroscopy showing mean residue ellipticity (MRE) as a function of wavelength for protein constructs 1-5 and 16. B. Rescaled plot of CD data for construct 4 showing weak α-helical folding signature. C. Example HMGB protein DNA binding data obtained by fluorescence anisotropy. Raw anisotropy values are fit to a simple binding isotherm to estimate the equilibrium dissociation constant as described in methods. Symbols for protein constructs: 1, filled circles; 2, open squares; 3, open circles; 5, filled squares.
TABLE 1.
In vitro characteristics of HMGB chimeric proteins
| Θ222c |
Anisotropyd |
||||||
|---|---|---|---|---|---|---|---|
| constructa | Lengthb | pI | -DNA | +DNA | Kd, nM | % active | J-factor (nM)e |
| 1 | 116 | 9.69 | -614 | -711 | 570 ± 40 | Ndg | 1.5 ± 0.7 |
| 2 | 116 | 9.9 | -619 | -398 | 268 ± 27 | nd | 9.6±3.2 |
| 3 | 116 | 10.1 | -730 | -311 | 31± 6 | 40 | 28.2 ± 4.7 |
| 4 | 116 | 9.69 | -600 | -660 | 103 ± 20 | nd | 2.2± 0.6 |
| 5 | 116 | 10.1 | -6354 | -2567 | 11± 2 | 100 | 14.0 ± 2.5 |
| 16 | 93 | 9.72 | -1585 | -1488 | 10f | 40 | 8.2±0.7 |
See Fig. 1
amino acid residues
MRE by CD (deg cm2 dmol-1)
fluorescence anisotropy measurements as described in materials and methods. Kd estimates (average ± standard error) were based on three determinations.
determined for a 200-bp DNA fragment (intrinsic J-factor 1.5 ± 0.3 nM) as described in materials and methods.
determined previously from pooled data.
nd: not determined because of insufficient DNA affinity.
DNA binding was further characterized by fluorescence anisotropy measurements. Example data are shown in Fig. 3C, with compiled results shown in Table I. Equilibrium dissociation constants for DNA binding that ranged from 10 nM (Nhp6Ap; construct 16) to greater than 500 nM [HMGB(Box A), construct 1) with affinities following the ranking 16 ~ 5 > 3 > 4 > 2 > 1 (Table I). In the cases where strong DNA binding activity was detected, the active protein fraction was measured by binding titration in the presence of DNA duplex concentrations much higher than Kd and found to be in the range of 40-100% (Table I). Overall, these data suggest that both intrinsic protein folding stability and DNA interaction are strongly correlated with the protein isoelectric point (Table 1).
In vitro enhancement of apparent DNA flexibility by single HMGB box chimeric proteins
Results of in vitro cyclization kinetics experiments are shown in Fig. 4. Examples of electrophoretic data fitting results for J-factor assays of DNA probe alone (—) or in the presence of 40 nM HMGB proteins 16 and 5 (Fig. 1B) are shown in panels A and B of Fig. 4. Estimated J-factors are given in Fig. 4C and Table I. Consistent with previous studies (53), the naked 200-bp DNA duplex is characterized by a J-factor of 1.5 nM. This DNA flexibility is significantly enhanced by incubation with 40 nM construct 16 (Nhp6Ap) but not by incubation with human HMGB2(box A), even when the primary intercalating residue of helix I is changed from alanine to methionine (Fig. 4C, constructs 1 and 4). Interestingly, certain chimeras between Nhp6Ap and HMGB2(box A) showed activities as least as strong as Nhp6Ap. For example, substitution of the leader residues of HMGB2(box A) [construct 1] with a small number of the corresponding cationic residues from Nhp6Ap (Fig. 4C, constructs 2 and 3) resulted in strong enhancement of apparent DNA flexibility, independent of whether the primary intercalating residue of helix I was alanine (constructs 2 and 3) or methionine (construct 5). These results strongly correlate with the predicted protein isoelectric point and observed DNA binding affinity, indicating that a cationic leader is sufficient to endow any of these HMGB2(box A) variants with the ability to enhance apparent DNA flexibility. Asymmetric phosphate neutralization of the compressed major groove (as in Nhp6Ap) may play a role in this effect.
Figure 4.
In vitro assay of DNA flexibility enhancement by HMGB proteins and chimeras. A. Example data from T4 DNA ligase cyclization assay for 200-bp DNA probe in the absence (—) and presence of 40 nM HMGB constructs 16 and 5 (see Fig. 1). B. Graphical analysis and extrapolation to estimate J-factor. Data for DNA probe alone (open circles), construct 16 (closed circles) and construct 5 (closed squares) are shown. C. Summary data for in vitro measurement of effects of HMGB proteins and chimeras on J-factor of 200-bp DNA. Residue at potential intercalating position 17 (alanine vs. methionine) and upstream cationic chimeras (RKKK “+4” vs. KKRTTRKKK “+7”) are highlighted schematically (see Fig. 1B). Black vs. white regions indicate sequences derived from Nhp6Ap vs. HMGB2(box A), respectively.
In vivo characterization of single HMGB box chimeric proteins
The results of in vitro experiments showed surprisingly strong effects of N-terminal cationic leader sequences on protein folding, DNA binding, and enhancement of DNA flexibility by HMG domains. We wished to rule out that these effects were due to artifacts of protein purification and/or refolding. The same chimeric single HMG box proteins were therefore evaluated in vivo for their abilities to complement the lac repression looping defect observed in living hupA/Bδ E. coli cells (47, 48). Accumulation of epitope-tagged HMGB constructs 6-15 (Fig. 1B) was first assessed by western blotting as shown in Fig. 5. Panel A of Fig. 5 indicates that epitope tagging of the N terminus supports increased HMGB protein accumulation (constructs 6 vs. 8 are otherwise identical). The exposure in panel B of Fig. 5 is shorter, allowing visualization of the comparable accumulation levels of constructs 6,7 and 10-13. Panel C of Fig. 5 compares accumulation of constructs 14 and 15. Importantly, although construct 15 (C-terminal epitope-tagged Nhp6Ap) accumulates to the lowest level of all tested HMGB constructs, even this level of accumulation has previously been shown to be sufficient for strong complementation of the hupA/Bδ repression looping defect (49). Thus, all of the tested HMGB proteins and chimeras accumulate within E. coli cells at levels with the potential to complement the hupA/Bδ defect.
Figure 5.
In vivo expression of HMGB proteins and chimeras. Accumulation of the indicated proteins (see Fig. 1B) was analyzed by western blotting. A. Exposure comparing expression of HMGB2(box A) variants carrying C-terminal Myc tags (constructs 8 and 9) vs. N-terminal tag (construct 6) or vector control (v). B. Shorter exposure showing expression levels of other HMGB2(box A)/Nhp6Ap chimeric HMGB proteins. C. Comparison of expression levels of Nhp6A protein carrying N-terminal (construct 14) or C-terminal (construct 15) epitope tag.
Lac repression looping in hupA/Bδ E. coli cells was measured for operator spacings in the range 75-85 bp. The results are shown in Fig. 6. Top panels show repression ratio data (ratio of induced to uninduced β-galactosidase activity). Higher peak values indicate favorable DNA loop geometries. Middle and bottom panels display δ-galactosidase output (E′) in the presence (middle panel) or absence (bottom panel) of IPTG inducer. E′ normalizes raw reporter gene signal in a given genetic strain background to the output from an unlooped promoter lacking a downstream lac operator in that same strain. Higher E′ values indicate more gene expression (less energetically favorable DNA repression loops). Fig. 6A compares repression in WT and hupA/Bδ genetic backgrounds. Shaded regions highlight the increased promoter “leakiness” in cells lacking HU (open symbols).
Figure 6.
Complementation analysis of lac repression looping defect in hupA/Bδ cells by expression of HMGB proteins and chimeras. Top panels: repression ratio. Shaded reference region illustrates the difference in repression ratio behavior for WT and mutant E. coli. The shaded region is replicated in all upper panels for reference. Lower panels: reporter gene expression (E′) in the presence (middle) or absence (bottom) of IPTG. The shaded regions are replicated in all bottom panels for reference to contrast reporter gene expression in WT and hupaA/Bδ E. coli cells.
Graphs B-F of Fig. 6 compare complementation of the hupA/Bδ looping defect by various single HMG box chimeric proteins. The bottom panels of Fig. 6 (reporter activity under repressed conditions) are especially informative. Box A derived from HMGB2 could not complement the hupA/Bδ looping defect, regardless of epitope tag location (Fig. 6B). In contrast, both N- and C-terminal epitope-tagged versions of yeast Nhp6Ap were active in complementation, with the more highly expressed N-terminal epitoope-tagged construct 14 being particularly effective (Fig. 6C). Regardless of epitope tag location, replacement of alanine by methionine as primary intercalating residue in helix I of HMGB2(box A) did not confer complementation activity (Fig. 6D). When the Nhp6Ap contribution to the HMGB2(box A) construct was limited to four basic amino acids in the leader (RKKK) complementation depended on the identity of the primary intercalating amino acid in helix I. A protein with alanine in this position (Fig. 6E, construct 10) had no detectable function. In contrast, the combination of RKKK leader and primary intercalating methionine (Fig. 6E, construct 11, note bottom panel) supported complementation even though the methionine substitution was ineffective when tested alone (Fig. 6D, construct 9). Finally, as had been observed for DNA flexibility enhancement in vitro (Fig. 3), substitution of the Nhp6Ap leader sequence KKRTTRKKK endowed the otherwise nonfunctional HMGB2(box A) construct with the ability to fully complement DNA looping in hupA/Bδ E. coli, independent of whether alanine or methionine was in the primary intercalating position of helix I (Fig. 6F).
DISCUSSION
We are interested in understanding features of HMG boxes A and B that distinguish their abilities to enhance apparent DNA flexibility in vitro and in vivo. Discussion has focused on differentiating characteristics of helix I and the loop between helices I and II (3, 10). For example, the Drosophila single HMG box B protein HMGD has been carefully analyzed (10). A non-sequence-specific HMGB protein, HMGD resembles yeast Nhp6Ap in its bulky methionine residue at the primary intercalating position of helix I, where HMGB(box A) has alanine in this position. Mutation of methionine to alanine in HMGD somewhat reduced the thermal stability of the protein (2°) and its DNA binding affinity (6-fold) (10). Importantly, these authors found that residues near the primary intercalating amino acid in helix I also played roles in DNA binding and bending. These results clearly point to the importance of the primary intercalating residue for tuning function. On the other hand, these mutations of the primary intercalating residue of HMGD caused relatively subtle effects on DNA binding and bending (10). In fact, detailed thermodynamic analysis indicates that it is electrostatic interactions that drive sequence nonspecific HMGB proteins onto DNA (50, 55). In addition, results of fluorescence resonance energy transfer experiments showed that the positively charged leader sequence of yeast Nhp6Ap (Fig. 1A, magenta spheres; Fig. 1B construct 16) contributes significantly to DNA bending (50). Strongly cationic sequences are found N-terminal to helix I in Nhp6Ap, and C-terminal to helix III in HMGD and HMGB1/2. The HMG box fold places both N-terminal and C-terminal sequences in approximately the same position near the major groove of DNA where both release of condensed counterions and asymmetric phosphodiester charge neutralization could enhance binding affinity and DNA bending. We therefore wished to compare the functional significance of i) a primary intercalating residue in helix I and ii) a cationic leader sequence for HMGB function. We have studied HMGB2(box A) as an example.
We previously showed that budding yeast Nhp6Ap (a member of the box B family of HMGB domains) could complement loss of HU protein in an E. coli assay of DNA looping in the lac operon, while human HMGB2(box A) could not (49). This result suggested an experimental approach to understand functional features distinguishing Nhp6Ap from HMGB2(boxA). In the present study we constructed a small number of chimeric proteins substituting the primary intercalating methionine residue of Nhp6Ap helix I, and /or cationic segments of the Nhp6Ap leader for corresponding sequences in HMGB2(box A). Our in vitro and in vivo results show that substitution of cationic leader residues of Nhp6Ap can determine protein folding stability and DNA binding affinity. In fact, a 9-amino acid segment (7 cationic residues) of the Nhp6Ap leader is sufficient to confer full activity on HMGB2(box A), whereas substitution of HMGB2(box A) with the primary intercalating methionine of Nhp6Ap makes a detectably favorable contribution only when a smaller number of Nhp6Ap cationic leader residues is included.
The similarity of our results obtained in vitro and vivo suggests a remarkably dominant effect of leader cationic residues in HMGB folding, DNA binding, and DNA bending. The cationic leader (and thus the higher protein isoelectric point) correlate with more stable protein folding and stronger DNA binding. It has previously been shown that truncation of the basic residues of Nhp6Ap reduces binding affinity and DNA bending (50). These authors estimated that each ionic contact in the major groove contributes ~6° of DNA bending (50). The present data are consistent with these results. Sequence-non-specific HMGB proteins asymmetrically neutralize DNA charges at their random DNA binding sites, with a corresponding contribution to DNA bending at each site. Analogy can therefore be drawn between this natural DNA bending strategy and the ability of sequence-specific DNA binding proteins with engineered basic domains to induce directed DNA bending at specific binding sites (56-59).
ACKNOWLEDGMENT
We thank A. Vologodskii and J. Kahn for materials and advice related to cyclization kinetics assays, J. Peterson and the Mayo Clinic Proteomics Research Center for assistance with protein purification, W. and B. Owen and R. Weinshilboum for assistance with fluorescence anisotropy, and L. Sikkink and M. Ramirez-Alvarado for assistance with CD spectroscopy.
Supported by the Mayo Foundation and NIH Grant GM 75965 to L.J.M.
Footnotes
Abbreviations: BSA, bovine serum albumin; box A, N-terminal high mobility group homology domain; box B, C-terminal high mobility group homology domain; EDTA, ethylenediaminetetraacetic acid; FACT, facilitator of chromatin transactions; HEPES, (4-(2-hydroxyethyl)-1-piperazineethylsulfonic acid); HMG, high mobility group; HMGB, high mobility group B protein family; HMGB1, mammalian member 1 of HMGB protein family; HMGB2, mammalian member 2 of HMGB protein family; HPLC, high performance liquid chromatography; IPTG, isopropyl β-D-1-thiogalactopyranoside; J-factor, concentration of one DNA end in the vicinity of the other; LB, Luria-Bertani broth; Ni-NTA, Ni2+-nitrilotriacetic acid; TE, Tris-HCl, EDTA buffer; TLC, thin layer chromatography.
REFERENCES
- 1.Crothers DM. Architectural elements in nucleoprotein complexes. Curr. Biol. 1993;3:675–676. doi: 10.1016/0960-9822(93)90065-v. [DOI] [PubMed] [Google Scholar]
- 2.Travers AA, Ner SS, Churchill MEA. DNA chaperones: A solution to a persistence problem. Cell. 1994;77:167–169. doi: 10.1016/0092-8674(94)90306-9. [DOI] [PubMed] [Google Scholar]
- 3.Thomas JO, Travers AA. HMG1 and 2, and related 'architectural' DNA-binding proteins. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 4.Allain FH, Yen YM, Masse JE, Schultze P, Dieckmann T, Johnson RC, Feigon J. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 1999;18:2563–2579. doi: 10.1093/emboj/18.9.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masse JE, Wong B, Yen YM, Allain FH, Johnson RC, Feigon J. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J. Mol. Biol. 2002;323:263–284. doi: 10.1016/s0022-2836(02)00938-5. [DOI] [PubMed] [Google Scholar]
- 6.Paull TT, Carey M, Johnson RC. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes & Dev. 1996;10:2769–2781. doi: 10.1101/gad.10.21.2769. [DOI] [PubMed] [Google Scholar]
- 7.Paull TT, Johnson RC. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. J. Biol. Chem. 1995;270:8744–8754. doi: 10.1074/jbc.270.15.8744. [DOI] [PubMed] [Google Scholar]
- 8.Yen YM, Wong B, Johnson RC. Determinants of DNA binding and bending by the Saccharomyces cerevisiae high mobility group protein NHP6A that are important for its biological activities. Role of the unique N terminus and putative intercalating methionine. J. Biol. Chem. 1998;273:4424–4435. doi: 10.1074/jbc.273.8.4424. [DOI] [PubMed] [Google Scholar]
- 9.Churchill ME, Jones DN, Glaser T, Hefner H, Searles MA, Travers AA. HMG-D is an architecture-specific protein that preferentially binds to DNA containing the dinucleotide TG. EMBO J. 1995;14:1264–1275. doi: 10.1002/j.1460-2075.1995.tb07110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klass J, Murphy F. V. t., Fouts S, Serenil M, Changela A, Siple J, Churchill ME. The role of intercalating residues in chromosomal high-mobility-group protein DNA binding, bending and specificity. Nucleic Acids Res. 2003;31:2852–2864. doi: 10.1093/nar/gkg389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bianchi ME, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 12.Paull TT, Haykinson MJ, Johnson RC. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes & Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 13.Pil PM, Chow CS, Lippard SJ. High-mobility-group 1 protein mediates DNA bending as determined by ring closure. Proc. Natl. Acad. Sci. USA. 1993;90:9465–9469. doi: 10.1073/pnas.90.20.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weir HM, Kraulis PJ, Hill CS, Raine ARC, Laue ED, Thomas JO. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993;12:1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolffe AP. Architectural regulations and Hmg1. Nat. Genet. 1999;22:215–217. doi: 10.1038/10267. [DOI] [PubMed] [Google Scholar]
- 16.Kamau E, Bauerle KT, Grove A. The Saccharomyces cerevisiae high mobility group box protein HMO1 contains two functional DNA binding domains. J. Biol. Chem. 2004;279:55234–55240. doi: 10.1074/jbc.M409459200. [DOI] [PubMed] [Google Scholar]
- 17.Grasser KD, Teo SH, Lee KB, Broadhurst RW, Rees C, Hardman CH, Thomas JO. DNA-binding properties of the tandem HMG boxes of high-mobility-group protein 1 (HMG1) Eur. J. Biochem. 1998;253:787–795. doi: 10.1046/j.1432-1327.1998.2530787.x. [DOI] [PubMed] [Google Scholar]
- 18.Zimmerman J, Maher LJ., 3rd Transient HMGB protein interactions with B-DNA duplexes and complexes. Biochem. Biophys. Res. Commun. 2008;371:79–84. doi: 10.1016/j.bbrc.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Q, Ohndorf UM, Lippard SJ. Intercalating residues determine the mode of HMG1 domains A and B binding to cisplatin-modified DNA. Biochemistry. 2000;39:14426–14435. doi: 10.1021/bi001700j. [DOI] [PubMed] [Google Scholar]
- 20.Jung Y, Lippard SJ. Nature of full-length HMGB1 binding to cisplatin-modified DNA. Biochemistry. 2003;42:2664–2671. doi: 10.1021/bi026972w. [DOI] [PubMed] [Google Scholar]
- 21.Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999;399:708–712. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- 22.Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ. Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature. 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 23.Churchill ME, Changela A, Dow LK, Krieg AJ. Interactions of high mobility group box proteins with DNA and chromatin. Meth. Enzymol. 1999;304:99–133. doi: 10.1016/s0076-6879(99)04009-4. [DOI] [PubMed] [Google Scholar]
- 24.Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: Formation of flexible hinges and rigid filaments. Proc. Natl Acad. Sci. USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, McCauley MJ, Maher LJ, 3rd, Williams MC, Israeloff NE. Mechanism of DNA flexibility enhancement by HMGB proteins. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkn1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCauley MJ, Zimmerman J, Maher LJ, 3rd, Williams MC. HMGB binding to DNA: Single and double box motifs. J. Mol. Biol. 2007;374:993–1004. doi: 10.1016/j.jmb.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCauley M, Hardwidge PR, Maher LJ, 3rd, Williams MC. Dual binding modes for an HMG domain from human HMGB2 on DNA. Biophys. J. 2005;89:353–364. doi: 10.1529/biophysj.104.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayaraman L, Moorthy NC, Murthy KG, Manley JL, Bustin M, Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes & Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calogero S, Grassi F, Aguzzi A, Voigtlander T, Ferrier P, Ferrari S, Bianchi ME. The lack of chromosomal protein HMG1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nature Genetics. 1999;22:276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 32.Ellwood KB, Yen YM, Johnson RC, Carey M. Mechanism for specificity by HMG-1 in enhanceosome assembly. Mol. Cell. Biol. 2000;20:4359–4370. doi: 10.1128/mcb.20.12.4359-4370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitsouras K, Wong B, Arayata C, Johnson RC, Carey M. The DNA architectural protein HMGB1 displays two distinct modes of action that promote enhanceosome assembly. Mol. Cell. Biol. 2002;22:4390–4401. doi: 10.1128/MCB.22.12.4390-4401.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeRoy G, Orphanides G, Lane WS, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 35.Lu J, Kobayashi R, Brill SJ. Characterization of a high mobility group 1/2 homolog in yeast. J. Biol. Chem. 1996;271:33678–33685. doi: 10.1074/jbc.271.52.33678. [DOI] [PubMed] [Google Scholar]
- 36.Travers AA. Priming the nucleosome: a role for HMGB proteins? EMBO Rep. 2003;4:131–136. doi: 10.1038/sj.embor.embor741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross ED, Keating AM, Maher LJ., 3rd DNA constraints on transcription activation in Vitro. J. Mol. Biol. 2000;297:321–334. doi: 10.1006/jmbi.2000.3562. [DOI] [PubMed] [Google Scholar]
- 38.Ross ED, Hardwidge PR, Maher LJ., 3rd HMG proteins and DNA flexibility in transcription activation. Mol. Cell. Biol. 2001;21:6598–6605. doi: 10.1128/MCB.21.19.6598-6605.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bellomy G, Mossing M, Record M. Physical properties of DNA in vivo as probed by the length dependence of the lac operator looping process. Biochemistry. 1988;27:3900–3906. doi: 10.1021/bi00411a002. [DOI] [PubMed] [Google Scholar]
- 40.Law SM, Bellomy GR, Schlax PJ, Record MT., Jr. In vivo thermodynamic analysis of repression with and without looping in lac constructs. Estimates of free and local lac repressor concentrations and of physical properties of a region of supercoiled plasmid DNA in vivo. J. Mol. Biol. 1993;230:161–173. doi: 10.1006/jmbi.1993.1133. [DOI] [PubMed] [Google Scholar]
- 41.Mossing MC, Record MT., Jr. Upstream operators enhance repression of the lac promoter. Science. 1986;233:889–892. doi: 10.1126/science.3090685. [DOI] [PubMed] [Google Scholar]
- 42.Kramer H, Amouyal M, Nordheim A, Müller-Hill B. DNA supercoiling changes the spacing requirements of two lac operators for DNA loop formation with lac repressor. EMBO J. 1988;7:547–556. doi: 10.1002/j.1460-2075.1988.tb02844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kramer H, Niemoller M, Amouyal M, Revet B, von Wilcken-Bergmann B, Müller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987;6:1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller J, Oehler S, Müller-Hill B. Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J. Mol. Biol. 1996;257:21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- 45.Oehler S, Amouyal M, Kolkhof P, von Wilcken-Bergmann B, Müller-Hill B. Quality and position of the three lac operators of E. coli define efficiency of repression. EMBO J. 1994;13:3348–3355. doi: 10.1002/j.1460-2075.1994.tb06637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oehler S, Eismann ER, Kramer H, Müller-Hill B. The three operators of the lac operon cooperate in repression. EMBO J. 1990;9:973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Becker NA, Kahn JD, Maher LJ., 3rd Bacterial repression loops require enhanced DNA flexibility. J. Mol. Biol. 2005;349:716–730. doi: 10.1016/j.jmb.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 48.Becker NA, Kahn JD, Maher LJ., 3rd Effects of nucleoid proteins on DNA repression loop formation in Escherichia coli. Nucleic Acids Res. 2007;35:3988–4000. doi: 10.1093/nar/gkm419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Becker NA, Kahn JD, Maher LJ., 3rd Eukaryotic HMGB proteins as replacements for HU in E. coli repression loop formation. Nucleic Acids Res. 2008;36:4009–4021. doi: 10.1093/nar/gkn353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dragan AI, Read CM, Makeyeva EN, Milgotina EI, Churchill ME, Crane-Robinson C, Privalov PL. DNA binding and bending by HMG boxes: energetic determinants of specificity. J. Mol. Biol. 2004;343:371–393. doi: 10.1016/j.jmb.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 51.Whipple FW. Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in Escherichia coli. Nucleic Acids Res. 1998;26:3700–3706. doi: 10.1093/nar/26.16.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vologodskaia M, Vologodskii A. Contribution of the intrinsic curvature to measured DNA persistence length. J. Mol. Biol. 2002;317:205–213. doi: 10.1006/jmbi.2001.5366. [DOI] [PubMed] [Google Scholar]
- 54.Hagerman PJ, Ramadevi VA. Application of the method of phage T4 DNA ligase-catalyzed ring-closure to the study of DNA structure. I. Computational analysis. J. Mol. Biol. 1990;212:351–362. doi: 10.1016/0022-2836(90)90130-E. [DOI] [PubMed] [Google Scholar]
- 55.Stros M. Two mutations of basic residues within the N-terminus of HMG-1 B domain with different effects on DNA supercoiling and binding to bent DNA. Biochemistry. 2001;40:4769–4779. doi: 10.1021/bi002741i. [DOI] [PubMed] [Google Scholar]
- 56.Strauss-Soukup JK, Maher LJ., 3rd Electrostatic effects in DNA bending by GCN4 mutants. Biochemistry. 1998;37:1060–1066. doi: 10.1021/bi972146p. [DOI] [PubMed] [Google Scholar]
- 57.Hardwidge PR, Kahn JD, Maher LJ., 3rd Dominant effect of protein charge rather than protein shape in apparent DNA bending by engineered bZIP domains. Biochemistry. 2002;41:8277–8288. doi: 10.1021/bi020185h. [DOI] [PubMed] [Google Scholar]
- 58.McDonald RJ, Kahn JD, Maher LJ., 3rd DNA bending by bHLH charge variants. Nucleic Acids Res. 2006;34:4846–4856. doi: 10.1093/nar/gkl552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McDonald RJ, Dragan AI, Kirk WR, Neff KL, Privalov PL, Maher LJ., 3rd DNA bending by charged peptides: electrophoretic and spectroscopic analyses. Biochemistry. 2007;46:2306–2316. doi: 10.1021/bi061921a. [DOI] [PubMed] [Google Scholar]