Abstract
Hippocampal volume change over time, measured with MRI, has huge potential as a marker for Alzheimer's disease. The objectives of this study were: (i) to test if constant and accelerated hippocampal loss can be detected in Alzheimer's disease, mild cognitive impairment and normal ageing over short periods, e.g. 6–12 months, with MRI in the large multicentre setting of the Alzheimer's Disease Neuroimaging Initiative (ADNI); (ii) to determine the extent to which the polymorphism of the apolipoprotein E (ApoE) gene modulates hippocampal change; and (iii) to determine if rates of hippocampal loss correlate with cerebrospinal fluid (CSF) biomarkers of Alzheimer's disease, such as the β-amyloid (Aβ1–42) and tau proteins (tau). The MRI multicentre study included 112 cognitive normal elderly individuals, 226 mild cognitive impairment and 96 Alzheimer's disease patients who all had at least three successive MRI scans, involving 47 different imaging centres. The mild cognitive impairment and Alzheimer's disease groups showed hippocampal volume loss over 6 months and accelerated loss over 1 year. Moreover, increased rates of hippocampal loss were associated with presence of the ApoE allele ɛ4 gene in Alzheimer's disease and lower CSF Aβ1–42 in mild cognitive impairment, irrespective of ApoE genotype, whereas relations with tau were only trends. The power to measure hippocampal change was improved by exploiting correlations statistically between successive MRI observations. The demonstration of considerable hippocampal loss in mild cognitive impairment and Alzheimer's disease patients over only 6 months and accelerated loss over 12 months illustrates the power of MRI to track morphological brain changes over time in a large multisite setting. Furthermore, the relations between faster hippocampal loss in the presence of ApoE allele ɛ4 and decreased CSF Aβ1–42 supports the concept that increased hippocampal loss is an indicator of Alzheimer's disease pathology and a potential marker for the efficacy of therapeutic interventions in Alzheimer's disease.
Keywords: MRI, mild cognitive impairment, ageing, human brain mapping, hippocampus
Introduction
Alzheimer's disease is the most common cause of dementia and a growing health problem globally, affecting 20% of the population over 80 years of age (Ferri et al., 2005). Currently, the definite diagnosis of Alzheimer's disease can only be made through autopsy to find the pathological hallmarks of the disease, microscopic amyloid plaques and neurofibrillary tangles. The development of biomarkers that can reliably indicate presence of the disease at the earliest possible stage is therefore an important public health goal. Macroscopically, Alzheimer's disease is associated with progressive brain tissue loss (Braak and Braak, 1998), which MRI can non-invasively visualize to some extent in-vivo (Thompson et al., 2007). Not surprisingly, MRI has attracted considerable interest as a tool to identify Alzheimer's disease biomarkers.
Histological studies have shown that the hippocampus is particularly vulnerable to Alzheimer's disease pathology and already considerably damaged at the time clinical symptoms first appear (Braak and Braak, 1998). The hippocampus has therefore become a primary target of MRI studies in Alzheimer's disease. In agreement with histological findings, longitudinal MRI studies have shown increased rates of hippocampal volume loss in Alzheimer's disease (Jack Jr et al., 2000, 2008b; Du et al., 2004; Jack et al., 2004; Fox et al., 2005; Hashimoto et al., 2005; van de Pol et al., 2005) and mild cognitive impairment (MCI, a transitional stage to Alzheimer's disease that may define a window for effective therapeutic intervention) (Jack et al., 2005; van de Pol et al., 2007), in comparison to normal ageing. Several MRI studies also found that the apolipoprotein E gene allele ɛ4 (ApoE4), a major risk factor for Alzheimer's disease, is associated with higher rates of hippocampal loss (Moffat et al., 2000; Jack Jr et al., 2008b; van de Pol et al., 2007), though the mechanism behind the ApoE4 induced variations remains obscure. Longitudinal MRI studies at multiple time-points further indicate that brain loss in general (Chan et al., 2003; Carlson et al., 2008) and hippocampus loss in particular (Ridha et al., 2006; Jack et al., 2008c) accelerate in patients with MCI and Alzheimer's disease though the specific trajectory of change remains unknown. To link MRI observations of hippocampal loss more firmly to the presence of Alzheimer's disease pathology, there has also been growing interest in using MRI together with biochemical markers of Alzheimer's disease in CSF, such as the proteins of tau (indicative of tangle formation) and amyloid Aβ1–42 (a major component of amyloid plaques) (Clark et al., 2008). Lastly, clinical trials of Alzheimer's disease and mild cognitive impairment have used the progression of hippocampal loss as a potential surrogate for the efficacy of therapeutic interventions (Jack Jr et al., 2003, 2008b).
Despite many promising results, several technical issues in measuring hippocampal disease progression remain unresolved, such as data variability due to non-uniform acquisition and image restoration processing. In this study, MRI was performed using uniform imaging sequences and a centralized setup for quality control and image restorations (Jack et al., 2008a). Furthermore, the vast majority of longitudinal MRI studies were carried out over a period of at least 1 year or much longer to ensure that the accumulated hippocampal loss exceeded the incurred measurement errors. Most studies were also performed at a single site to avoid site-to-site variations in MRI. However, studies over a long period at a single research centre represent a ‘best-case’ scenario, which can rarely be achieved in large studies, such as clinical trials, involving several hundred subjects. The power of MRI to measure hippocampal loss, especially acceleration, over short periods and in a multisite setting remains to be determined. Another issue is that many MRI investigations of biological effects on hippocampal change involved only a small number of subjects, especially those which involved CSF biomarkers, raising concerns about the generalization of the findings. Lastly, few studies so far reported findings of hippocampal changes, ApoE and CSF biomarkers together (Hampel et al., 2005; de Leon et al., 2006; Fjell et al., 2008). Although it was found that CSF biomarker levels and the ApoE profile are associated with morphometric changes in the hippocampus, the extent to which each factor independently contributes to the progression of hippocampal changes has not been investigated.
The ongoing Alzheimer's disease Neuroimaging Initiative (ADNI) study has been designed to address these issues (Mueller et al., 2005) (see also http://www.loni.ucla.edu/ADNI and http://www.ADNI-info.org). The ADNI is a large multisite longitudinal MRI and FDG-PET (fluorodeoxyglucose positron emission tomography) study of 200 cognitively normal (normal) elderly controls, 400 subjects with MCI and 200 patients diagnosed with Alzheimer's disease. At the time of writing this report, data collection for the ADNI project is still in progress. Here, we report an initial analysis of rates of hippocampal loss in 498 subjects, who completed three successive MRI scans and clinical evaluations at baseline, 6 and 12 months, involving 47 MRI sites.
Our main objectives in this study were: first, to determine if the rigorous methods of the ADNI to control site-to-site variations in MRI allow the detection of hippocampal change, including acceleration, over a short period, e.g. 6–12 months; second, to determine the extent to which the ApoE genotype modulates rates of hippocampal loss; and third, to test if rates of hippocampal loss correlate with CSF biomarkers.
Methods
Data used in the preparation of this article were obtained from the ADNI database (www.loni.ucla.edu\ADNI). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public–private partnership. The primary goal of ADNI has been to test whether serial MRI, positron emission tomography (PET), other biological markers and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early Alzheimer's disease. Determination of sensitive and specific markers of very early Alzheimer's disease progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principle Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California, San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the US and Canada. For up-to-date information see www.adni-info.org.
Subjects
The study reported here involved 498 subjects who had MRI scans at baseline, 6 and 12 months (0–6–12 m) and hippocampal volume change measured. Of those subjects, 11 were excluded because they had lumbar puncture procedures within <10 days prior to their MRI (a protocol violation), which may have induced artificial volume changes of the brain. The MRI data of an additional 38 subjects were excluded for technical reasons, such as major hardware upgrades during the study (at two sites), miscalibration of image resolution or failure to register images to a brain template for tracing the hippocampus. At the end, 127 normal, 226 MCI and 96 Alzheimer's disease subjects were considered. The main demographical and clinical data of this group are summarized in Table 1. Consent was obtained according to the Declaration of Helsinki (Br Med J 1991; 302: 1194) and the Ethical Committees of each Institution in which the work was performed approved the study.
Table 1.
Group demographics and clinical data at baseline
| Measures | Normal | Mild cognitive impairment | Alzheimer's disease | P-value |
|---|---|---|---|---|
| n | 127 | 226 | 96 | NA |
| Women (%) | 48 | 38 | 47 | NS |
| Age (years) | 76.3 ± 5.1 | 75.0 ± 7.1 | 75.8 ± 6.6 | NS |
| MMSEa | 29.1 ± 0.9 | 26.9 ± 1.8 | 23.3 ± 1.9 | <0.001 |
| ADAS-Cogb | 6.1 ± 3.3 | 11.6 ± 4.5 | 17.9 ± 5.6 | <0.001 |
| CDR Sum-of-boxesc | 0 ± 0.1 | 1.6 ± 0.9 | 4.4 ± 1.5 | <0.001 |
| GDSd | 0.8 ± 1.3 | 1.5 ± 1.3 | 1.7 ± 1.4 | <0.001* |
| ApoE4 carriers (%) | 26 | 52 | 67 | <0.001 |
a Mini-Mental State Examination; range 0–30 points.
b Alzheimer's disease Assessment Scale—Cognitive Subscale; range 0–70 points.
c Clinical Dementia Rating Scale; range 0–18 points.
d Geriatric Depression Scale; range 0–15 points.
*Significant difference in GDS scores between normal and mild cognitive impairment or Alzheimer's disease.
All subjects underwent thorough clinical and cognitive assessments at the time of each of their MRI scans. Each subject's cognitive evaluation included: (i) the Mini-Mental State Examination (MMSE) (Folstein et al., 1975) to provide a global measure of mental status; (ii) the Alzheimer's disease Assessment Scale-Cognitive Subscale (ADAS-Cog) (Mohs et al., 1997), which is the most used cognitive assessment battery in clinical dementia trials; and (iii) the Clinical Dementia Rating (CDR) sum-of-boxes scale (sum of individual CDR scales) (Morris, 1993) to stage severity of dementia. All subjects were also examined for depression using the Geriatric Depression Scale (GDS) questionnaire (Yesavage et al., 1982), in which subjects are asked to respond to 30-items with ‘yes’ or ‘no’ in reference to how they felt over the past week. More details about all the tests can be found on the ADNI website www.loni.ucla.edu/ADNI.
The normal subjects had on average MMSE scores of 29.1 ± 0.9, ADAS-Cog scores of 6.1 ± 3.3, CDR sum of boxes scores of 0 ± 0.1 and GDS scores of 0.8 ± 1.3. The MCI subjects had mild memory complaints, but had no symptoms of dementia. On average, they had MMSE scores of 26.9 ± 1.8, ADAS-Cog scores of 11.6 ± 4.5, CDR sum of boxes scores of 1.6 ± 0.9 and GDS scores of 1.5 ± 1.3. All Alzheimer's disease patients met NINCDS/ADRDA criteria for probable Alzheimer's disease (McKhann et al., 1984). On average, they had MMSE scores of 23.3 ± 1.9 and ADAS-Cog scores of 17.9 ± 5.6, CDR sum of boxes scores of 4.4 ± 1.5 and GDS scores of 1.7 ± 1.3. As such, these patients would be considered as having mild to moderate, but not severe Alzheimer's disease and no depression.
All subjects had their blood ApoE genotype determined. In addition, two-third of the subjects had lumbar puncture procedures performed for the collection of specific Alzheimer's disease biomarkers. The determinations of ApoE and biomarkers were performed by Drs Leslie Shaw and John Trojanowski of the ADNI Biomarker Core at the University of Pennsylvania School of Medicine, which collects and banks biological samples (DNA, blood, urine and CSF) from all participating sites, and conducts studies of selected Alzheimer's disease biomarkers, including total tau (t-tau), hyperphosphorylated tau (p-tau), β-amyloid-1–42 (Aβ1–42), isoprostanes and homocysteine levels (Shaw et al., 2007; Vanderstichele et al., 2008; Shaw et al., 2009) (see http://www.adni-info.org/for more details).
Detailed exclusion criteria, e.g. regarding concomitant cerebral vascular disease or concurrent use of psychoactive medications, can be found in the ADNI protocol (page 29, http://www.adni-info.org/images/stories/Documentation/adni_protocol_03.02.2005_ss.pdf). Briefly, subjects were excluded if they had any significant neurological disease other than incipient Alzheimer's disease, any history of significant brain lesions or head trauma, or psychoactive medication use (including antidepressants, neuroleptics, chronic anxiolytics or sedative hypnotics, etc.).
MRI acquisition and pre-processing
All subjects had MRI at 1.5T. The data were collected at multiple ADNI sites using a standardized MRI protocol (http://www.loni.ucla.edu/ADNI/Research/Cores/index.shtml), which was developed after a major effort evaluating and comparing 3D T1-weighted sequences for morphometric analyses (Jack et al., 2008a). In this study, the MRI data came from 47 centres. Details of MRI acquisition and processing are described in several publications (Leow et al., 2006; Jack et al., 2008a). Briefly, for each subject, two T1-weighted MRI scans were collected using a sagittal volumetric magnetization prepared rapid gradient echo (3D MP-RAGE) sequence with the following acquisition parameters: echo time (TE) of 4 ms, repetition time (TR) of 9 ms, flip angle of 8°, acquisition matrix size of 256 × 256 × 166 in the x-, y- and z-dimensions with a nominal voxel size of 0.94 × 0.94 × 1.2 mm. The ADNI MRI quality control centre at the Mayo Clinic (Rochester, MN, USA) selected the MP-RAGE image with higher quality based on standardized criteria (Jack Jr et al., 2008b). To enhance the standardization across MRI sites and scanner platforms, each site used the same customized imaging sequence for volumetric brain morphometry, which was developed and tested on phantoms and on 137 subjects during the preparation phase of the ADNI study. Second, each subject scan was accompanied by a scan of a custom built phantom that was centrally evaluated for signal-to-noise and geometrical fidelity to ensure performance of the MRI scanners remained within a specific tolerance limit. More details about MRI standardization in the ADNI study can be found in reference (Jack et al., 2008a). Furthermore, system specific corrections of certain image artefacts were performed centrally at the Mayo Clinic. The corrections included: (i) a ‘B1-Field correction’ to adjust for inhomogeneity of image intensity induced by non-uniform radiofrequency (RF) excitation using B1 calibration scans, (ii) ‘N3 bias field correction’ to further reduce intensity inhomogeneity caused by non-uniform sensitivity of the receiver coils, (iii) a geometrical distortion correction to offset non-linearity in frequency encoding of magnetic field gradients and (iv) global geometrical scaling, based on phantom measurements that accompanied every subject's MRI scan to adjust for gradient calibration errors and drifts.
Hippocampal volume estimation
Tracing of the anatomical boundaries of the left and right hippocampus was performed using a semi-automated brain mapping method based on a high-dimensional fluid transformation algorithm (Christensen et al., 1997), which combines a coarse and then a fine transformation of a carefully marked hippocampal MRI template from a reference brain to match the target images of each subject. A commercially available version of the algorithm was used (Medtronic Surgical Navigation Technologies, Louisville, CO). To guide the initial transformations, 22 control points are manually placed as rough local landmarks for hippocampal segmentation: one point at the hippocampal head, one at the tail, and four per image (i.e. at the superior, inferior, medial and lateral boundaries) on five equally spaced image slices perpendicular to the long axis of the ipsilateral hippocampus. The last step is repeated for the contralateral hippocampus. Using the landmarks for initial guidance, automated hippocampal segmentation is then performed by the iterative fluid transformation image matching algorithm. The procedure has previously been validated for hippocampal volume measurements in elderly subjects, including mild cognitive impairment and Alzheimer's disease patients (Hsu et al., 2002).
To minimize measurement variability and bias from manually placing the landmarks, a comprehensive procedure for assessing reader reliability was implemented. First, the readers were extensively trained and their reliability was evaluated by having each reader perform twice volume measurements of a ‘gold standard’ MRI dataset that consisted of 10 randomly selected subjects. To qualify, a reader had to achieve an intraclass correlation coefficient of at least 0.9 for both within and between reader consistencies. Furthermore, reliability of readings was monitored over time by blindly having each reader re-evaluate randomly selected sets from the ‘gold-standard’ sets as data processing progressed. The re-evaluations had to be within the 95% confidence limits to qualify. If a reader's performance dropped outside the limits, the last hippocampal markings by this reader were re-processed by a qualified reader. In addition, the disqualified reader had to complete training again to qualify.
Statistics
We employed a general linear mixed effects model for analysis of the longitudinal data in which the response variable (e.g. hippocampal volume or ADAS-Cog score) is regressed against the explanatory variable (time) separately from within subject correlations. To investigate the benefits of collecting data at more than two time-points, we extended the model by a transitional (i.e. Markov chain) model, in which past observations explicitly influence present measurements. More details are provided in the appendix. Other explanatory variables, e.g. diagnosis, age or ApoE4 profile, were added into the model as appropriate. To determine if the addition of explanatory variables, especially the inclusion of past observations in the Markov chain model, improved explanatory power, we designed paired models (with and without the additional explanatory variable), fitted the models by maximum likelihood (ML) and compared the resulting fits via F-tests. The level of significance was 0.05 for all tests.
To assess site-to-site variations in the MRI data, we bootstrapped the random effects residuals of the fits (i.e. the unexplained within-person variation and noise) and evaluated differences between the MRI centres by analysis of variance. To reduce the influence of group heterogeneity between centres, we first limited the analysis to mild cognitive impairment patients, which was usually the group with the most completed scans at each centre and second, we augmented bootstrapping by permutation tests.
Sample size calculations for the number of subjects needed in a hypothetical clinical trial of measuring a meaningful drug effect (i.e. slowing of rates of hippocampal loss or change in ADAS-Cog scores) between a treated and placebo group are outlined in the appendix. All statistical calculations were conducted using R (the R Project for Statistical Computing; http://www.r-project.org/).
Results
Data at baseline
Demographics and clinical data of the subjects at baseline are summarized in Table 1. The groups were comparable in age (P = 0.2) and sex distribution (P = 0.2, by χ2), but had markedly different MMSE, ADAS-Cog and CDR scores, as expected (all P < 0.001). While mild cognitive impairment and Alzheimer's disease subjects had similar GDS scores (P = 0.5), their scores were both markedly higher than those of normal subjects (P < 0.001). Mild cognitive impairment and Alzheimer's disease patients also had more than 2-fold higher rates of ApoE4 carriers than the normal subjects (P < 0.001, by χ2). Estimations of hippocampal volumes (in cubic millimetre, left and right averaged) at baseline are listed in Table 2 for each group. As expected, Alzheimer's disease patients had smaller hippocampal volumes than mild cognitive impairment subjects (P < 0.0001) and both had smaller volumes than normal subjects (P < 0.0001). Smaller baseline volumes were significantly associated with age (P < 0.01) in each group.
Table 2.
Hippocampal baseline volumes and rates of volume loss by group for different MRI scan intervals
| Normal | Mild cognitive impairment | Alzheimer's disease | |
|---|---|---|---|
| Baseline [mm3] | 2133 ± 25 (279) | 1846 ± 23 (348) | 1631 ± 34 (330) |
| Rates [mm3/year] | |||
| 0–6 month | −19.7 ± 17.0 [–89 to 51] (74) | –37.7 ± 10.6* [–144 to 74] (63) | –53.5 ± 15.6* [–152 to 62] (59) |
| 6–12 month | −8.3 ± 21.7 [−105 to 78] (85) | −54.2 ± 11.8* [−242 to 82] (60) | −91.3 ± 18.2* [−207 to 63] (62) |
| 6–12 montha | −6.7 ± 18.1 (74) | −56.1 ± 9.8* (53) | −93.0 ± 15.0* (50) |
| 0–12 month | −17.3 ± 10.5# [−97 to 53] (80) | −47.5 ± 6.5* [−153 to 79] (65) | −72.0 ± 9.0* [−162 to 50] (56) |
| Percent change/yearb | |||
| 0–6 month | −0.9 ± 0.8 | −2.0 ± 0.6* | −3.3 ± 0.9* |
| 6–12 month | −0.4 ± 1.0 | −2.9 ± 0.6* | −5.6 ± 1.1* |
| 6–12 montha | −0.3 ± 0.8 | −3.0 ± 0.5* | −5.7 ± 0.9* |
| 0–12 month | −0.8 ± 0.5# | −2.6 ± 0.3* | −4.4 ± 0.6* |
a Using 0–6 months as prior and a Markov chain model for analysis.
b Percent volume change relative to baseline volume.
*P < 0.0001; #P = 0.005.
Listed are mean ± SE; max/min rates in square brackets; random effect standard deviation in round brackets.
Rates of hippocampal volume loss
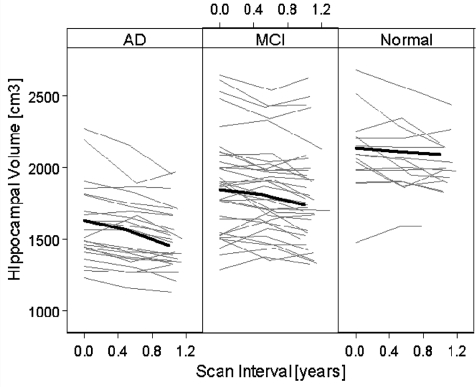
Differences in rates between the left and right hippocampus were not significant (P > 0.2) and therefore the values were averaged. Individual trajectories of hippocampal volume changes as a function of time are depicted in Fig. 1 for one-third of the subjects, randomly selected. The thick solid line in each panel represents the mean change in each respective group. This indicates that Alzheimer's disease patients had on average a smaller hippocampus and greater volume loss over time than normal subjects, whereas mild cognitive impairment patients had intermediate values between Alzheimer's disease and normal. Table 2 lists the estimated rates of hippocampal volume loss (in cublic millimetre/year) for each group, separately for 0–6 m, 6–12 m and 0–12 m scan intervals. The rates of volume loss are also given in annualized percentage change relative to baseline for comparison with other publications. The rates are adjusted for subject age at baseline. At 0–6 m, hippocampal loss in mild cognitive impairment and Alzheimer's disease was already highly significant (P < 0.0001), whereas the loss in normal became significant in the 0–12 m interval only (P = 0.005). In general, the estimations of hippocampal loss were more robust from measurements over the 0–12 m interval than over 0–6 m and 6–12 m, as indicated by smaller standard errors and correspondingly higher significance levels. For all scan intervals, Alzheimer's disease patients had a markedly higher average rate of hippocampal loss than normal subjects, whereas mild cognitive impairment patients had an intermediate rate between Alzheimer's disease and normal. There was no significant age by atrophy rates interaction for any group. Given the number of subjects in this study, e.g. mild cognitive impairment, the minimum detectable difference in atrophy rates was 25 mm3/year at 80% power (α = 0.05). Neither baseline volume nor rates were significantly associated with severity of depression (P = 0.8).
Figure 1.
Individual trajectories of hippocampal volume change from one-third of the subjects, randomly selected. The thick black lines indicate the mean trajectory of each group.
For comparison, rates derived from an analysis using the Markov chain approach, in which the observations from scan interval 0–6 m were explicitly used to estimate the rates in the 6–12 m interval are also listed in Table 2. Compared to the regular analysis of 6–12 m data, the Markov chain approach yielded markedly smaller random variations in addition to smaller standard errors in rates. The inclusion of the 0–6 m data made a significant contribution to the estimation of rates from the 6–12 m data for all groups [normal: likelihood ratio (LR) = 32.7; mild cognitive impairment: LR = 46.7; Alzheimer's disease: LR = 36.3; all P < 0.0001].
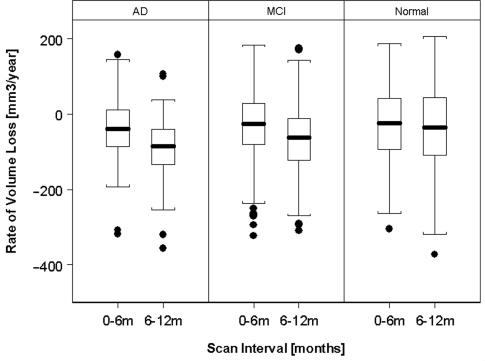
We further tested whether the rates of hippocampal loss accelerated. The box-and-whisker plots in Fig. 2 depict the change of hippocampal loss between 0–6 and 6–12 m scan intervals by group. In both mild cognitive impairment and Alzheimer's disease patients, the rates of hippocampal loss accelerated (P = 0.0001) but not in normal subjects (P = 0.2). Based on a quadratic expansion of time in the constant rate model, the hippocampal loss accelerated by 26.5 ± 4.5 mm3/year2 in Alzheimer's disease and 12.1 ± 3.2 mm3/year2 in mild cognitive impairment, equivalent to −1.6 ± 0.2%/year2 and 0.6 ± 0.2%/year2, respectively, relative to baseline volume.
Figure 2.
Accelerations of rates of hippocampal volume loss.
Correlations between rates of volume loss and cognitive decline
The rates of ADAS-Cog (log transformed) correlated with the rates of hippocampal loss in Alzheimer's disease and mild cognitive impairment patients (r = −0.27; P = 0.0005 by Spearman rank). Similarly, the rates of change in MMSE (log transformed) correlated with the rates of hippocampal loss in Alzheimer's disease and mild cognitive impairment patients (r = 0.18, P = 0.04 by Spearman rank).
ApoE profile and modulations of rates of hippocampal loss
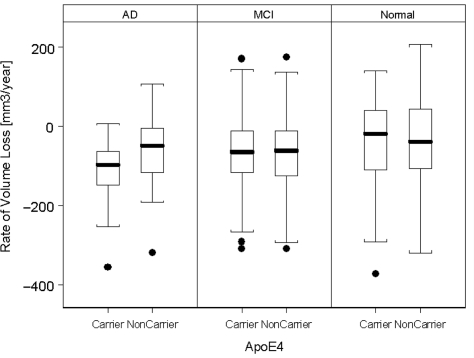
The effect of ApoE on rates of hippocampal loss is depicted in Fig. 3. Presence of the ApoE4 gene was generally associated with higher rates of hippocampal loss (LR = 4.4; P = 0.03) and smaller baseline volumes (LR = 27.2; P < 0.0001). The ApoE4 effect on rates was dominated by Alzheimer's disease patients with ApoE4, who lost on average 23.4 ± 8.4 mm3/year (1.4 ± 0.5%/year loss relative to baseline volume) more hippocampal volume than patients not carrying ApoE4 (LR = 7.8; P = 0.005), irrespective of the severity of cognitive impairments (based on ADAS-Cog). A direct comparison between ɛ3/3 with ɛ3/4 carriers (i.e. excluding those with alleles ɛ2/2, ɛ2/3, ɛ2/4 and ɛ4/4) yielded virtually, the same result. We also explored the dose effect of ApoE4 on hippocampal loss rates by comparing ɛ3/4 and ɛ4/4 carriers (ɛ2/4 carriers were not considered because there were only three) but found no significant effect. Interrelationships between explanatory variables (ApoE, age, ADAS-Cog) for rates of volume loss were weak (r < 0.2 for all). In normal and mild cognitive impairment subjects, ApoE4 was not significantly associated with higher hippocampal loss rates (P = 0.9).
Figure 3.
Rates of hippocampal volume loss in carriers and non-carriers of ApoE4.
Rates of hippocampal loss and CSF biomarkers concentrations
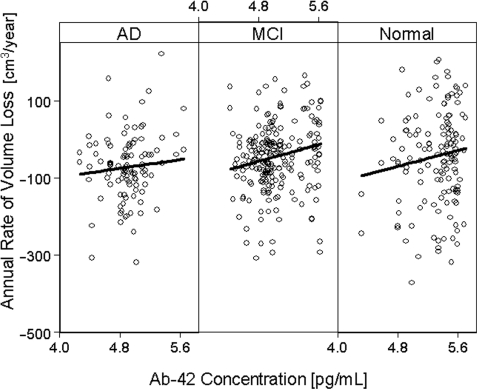
We also tested if the rates of hippocampal loss are correlated with the concentration of CSF biomarkers Aβ1–42, t-tau and p-tau at baseline on a smaller sample (68 normal, 109 mild cognitive impairment, 53 Alzheimer's disease), whose biomarker data were available at the time of the analysis. The baseline concentrations (in pg/ml) of Aβ1–42, t-tau and p-tau, respectively, were: 137 ± 39/119 ± 59/40 ± 17 in Alzheimer's disease; 159 ± 54/104 ± 52/36 ± 17 in mild cognitive impairment; and 211 ± 54/70 ± 30/25 ± 14 in control. Overall, higher rates of hippocampal loss were associated with decreased Aβ1–42 concentration (P < 0.01). The effect remained significant after accounting for ApoE genotype and level of cognitive impairment. Among the explanatory variables for rates of volume loss, Aβ1–42 and ApoE were the only ones that had a significant interrelationship (r = −0.58) with Aβ1–42 contributing 1.5 times as much to the variability of hippocampus atrophy rates than ApoE. The link between Aβ1–42 and hippocampal rates was driven by mild cognitive impairment subjects, who lost on average 0.3 ± 0.1 mm3/year hippocampal volume per pg/ml decrease of Aβ1–42 in CSF (P = 0.04), equivalent to 0.02 ± 0.01%/year loss relative to baseline volume.
No significant correlation between Aβ1–42 and MRI was seen in the Alzheimer's disease (P = 0.1) or normal group (P = 0.5). The regression plots in Fig. 4 depict the relationship between rates of hippocampal loss and Aβ1–42 concentration, separately by group. Note, Aβ1–42 concentration is plotted on a logarithmic scale to stabilize the regressions by imposing a more uniform distribution of the Aβ1–42 values. In contrast to Aβ1–42, the correlation between CSF t-tau concentrations and hippocampal loss rates was only a trend (P = 0.058) and this was limited to Alzheimer's disease patients. There were no significant correlations between CSF p-tau concentrations and hippocampal loss rates in any group.
Figure 4.
Associations between annual rates of hippocampal volume loss and Aβ1–42 concentration in CSF.
Sample size
To determine the power of MRI in detecting effects on hippocampal volume loss over time we estimated the sample size needed in a hypothetical treatment trial to measure a 25% slowing in the rate of volume loss with 90% confidence and α = 0.05 significance. We considered two separate strategies to maximize power: in the first strategy, we considered increasing either the inter-scan interval from 0–6 to 0–12 m or alternatively increasing the number of MRI scans from two to three, i.e. scanning at 0–6–12 m. In the second strategy, we considered gaining power by using prior information, such as exploiting correlations between successive observations by Markov chain analysis and/or by considering the ApoE4 profile and Aβ1–42 concentration in CSF at baseline. The powers, expressed as sample sizes estimations of the different strategies are summarized in Table 3, separately for Alzheimer's disease and mild cognitive impairment. This shows in general that prolonging the inter-scan interval from 6 to 12 months reduced the sample size, as expected. The use of three time-points instead of two lowered the sample size for mild cognitive impairment and accounting for ApoE4 lowered the sample size for Alzheimer's disease. By far the most effective reduction of sample size in this context was accomplished by exploiting correlations between observations, i.e. using Markov chain analysis. Accounting for ApoE4 status in the Markov chain analysis further reduced the sample size for Alzheimer's disease to finally 86 patients and for mild cognitive impairment to 341 patients per arm. Including the Aβ1–42 concentration in CSF at baseline did not lead to additional improvements. We also considered the rate of dropouts in power estimations of three versus two scans. For a Markov chain analysis, which requires at least three scans, the dropout rate has to exceed roughly 20% before the power benefit compared to a conventional mixed effects analysis with only two scans is lost. In contrast to Markov chain, a conventional mixed effects analysis with three scans holds a slight advantage over one with two scans as long as the dropout is less than about 5%. In this study, the dropout rate between two and three scans was about 5%.
Table 3.
Estimations of sample size per arm for a hypothetical trial to detect 25% rate slowing with 90% power and α = 0.05
| Study design | Alzheimer's disease | Mild cognitive impairment |
|---|---|---|
| MRI Hippocampal volume | ||
| Two scans, 0–6 ma | 462 | 949 |
| Two scans, 0–12 ma | 252 | 698 |
| Three scans, 0–6–12 ma | 255 | 673 |
| Three scans + ApoE4b | 196 | 672 |
| Three scans + MCc | 140 | 383 |
| Three scans + MC + ApoE4d | 86 | 341 |
| ADAS-Coge | ||
| Two tests, 0–6 m | 745 | 4663 |
| Two tests, 0–12 m | 814 | 9350 |
| Three tests, 0–6–12 m | 569 | 8354 |
| MMSEe | ||
| Two tests, 0–6 m | 1280 | 6300 |
| Two tests, 0–12 m | 1083 | 3900 |
| Three tests, 0–6–12 m | 780 | 3353 |
a Based on MRI scans at baseline (0 m), 6 (6 m) and/or 12 months (12 m).
b Accounting for ApoE4 profile.
c Using the Markov chain approach (see text for details).
d Using ApoE4 and Markov chain together.
e Using the Markov chain approach did not significantly improve sample size.
For comparison, the sample sizes based on rates of ADAS-Cog or MMSE are also listed in Table 3 for various study scenarios. In contrast to MRI, there was no benefit for sample size of ADAS-Cog and MMSE rates from using the Markov chain approach.
Variability between MRI sites
Lastly, we determined the extent to which MRI performed at multiple centres increases the variability compared with MRI at individual centres. Figure 5 depicts the variability of random effects errors in measurements of hippocampal rates between the centres. Data are shown for 13 MRI centres, which had MRI data collected at three time-points in at least seven mild cognitive impairment patients at the time this report was written. For comparison, the variability of random effects errors when all 47 ADNI centres were included is indicated by the black bar on the far right. Overall, variability of MRI performance between the centres was not significant (P = 0.5).
Figure 5.
Variability in measuring hippocampal rates between MRI centres. Centres are coded by numbers.
Discussion
Our main findings are: (i) in mild cognitive impairment and Alzheimer's disease, progression of hippocampal loss was detected over 6 months and accelerated over 1 year, whereas in the normal group hippocampal loss was detected over 1 year with no indication of acceleration; (ii) ApoE4 was associated with higher rates of hippocampal loss in Alzheimer's disease patients, irrespective of their level of cognitive impairments; (iii) higher rates of hippocampal loss correlated with a lower concentration of CSF Aβ1–42, irrespective of ApoE genotype, predominantly in mild cognitive impairment patients. Furthermore, we showed that the power of measuring hippocampal change can be improved by exploiting intrinsic correlations between successive MRI observations. In addition, we showed that site-to-site variations in MRI can effectively be brought to levels similar to single site settings using the rigorous methods of the ADNI.
We found significant hippocampal volume loss in mild cognitive impairment and Alzheimer's disease over only 6 months. All but one other MRI study reported hippocampal loss over such a short period and only for Alzheimer's disease (Barnes et al., 2008b). Furthermore, the prior study conducted MRI at a single centre only, yielding no conclusions for multicentre trials. Nonetheless, our results in Alzheimer's disease and those from this single centre study are intriguingly similar. Our results (average of 0–6 and 6–12 months rate values) expressed as percentage annual change yield 4.5% hippocampal loss in Alzheimer's disease (2–6% within 95% CI) compared with the range 4.35–5.04% that the single centre study reported. For mild cognitive impairment, our results over 6 months yield 2.5% annualized hippocampal loss (2.0–3.3% within 95% CI), in good agreement with reports of most other longitudinal MRI studies of mild cognitive impairment (Jack et al., 2000; Du et al., 2004; Fox et al., 2005; Hashimoto et al., 2005) that used much longer scan intervals, including two multicentre trials (Jack Jr et al., 2008b; van de Pol et al., 2007). The rates of hippocampal loss in our study are also within the range found in a large meta-analysis (Barnes et al., 2008a). It is furthermore re-affirming that our results obtained with a semi-automated method for tracing the hippocampus are comparable with those that employed entirely manual methods (Jack et al., 2000). Taken together, our results imply that an assessment of hippocampal loss over 6 months is possible and this extends to multicentre trials. However, measurement power is clearly sacrificed at shorter intervals as indicated by the larger standard errors for rates from measurements over 6 months compared to those over 12 months. Limited spatial resolution of MRI is likely the main reason for incurring errors at shorter scan intervals. Therefore, longitudinal studies of hippocampal loss should benefit from higher MRI resolution, if it can be afforded. Since a quarter of the participants in the ADNI will also be scanned at 3T parallel to 1.5T but at 20% higher resolution, the impact of image resolution on the power to measure brain volume loss can ultimately been tested. It is surprising that the rate of volume loss in mild cognitive impairment between 6 and 12 months is similar to that in Alzheimer's disease between 0 and 6 months, though some mild cognitive impairment subjects are destined to develop Alzheimer's disease. A possible explanation for this observation is that accelerated volume loss over time is a more prominent feature that separates Alzheimer's disease from mild cognitive impairment than constant volume loss. In addition, the annual percentage change from baseline is lower in mild cognitive impairment than in Alzheimer's disease.
The rates of hippocampal loss also correlated with rates of cognitive decline. In general, Alzheimer's disease and mild cognitive impairment patients with high hippocampal rates also had rapidly increasing ADAS-Cog scores, while the correlation with MMSE scores was weaker. It is possible that cognitive tests that are more specific for hippocampal function show stronger correlations with MRI. Additional analyses that are beyond the scope of this report are warranted to further explore the cognitive correlates of hippocampal volume changes.
We also found significant hippocampal loss in the normal group over 1 year, in agreement with several prior MRI studies (Fox and Schott, 2004; Du et al., 2005; Jack et al., 2005) as well as with autopsy findings of neuronal loss in the ageing hippocampus (West, 1993; Simic et al., 1997). However, to determine if hippocampal loss in normal subjects is already an indication of incipient Alzheimer's disease or other pathologies affecting the hippocampus requires clinical follow-up of the subjects to determine their cognitive decline and ultimate development of Alzheimer's disease.
We found accelerated hippocampal loss in the mild cognitive impairment and Alzheimer's disease groups over a period of 1 year. Accelerated rates of hippocampal loss have previously been reported for mild cognitive impairment (Jack et al., 2008c) and for familial Alzheimer's disease (Ridha et al., 2006), but over periods that ranged from 2 to 5 years. Our finding is also consistent with reports of accelerated loss of whole brain (Chan et al., 2003; Carlson et al., 2008) and clinical studies of accelerated cognitive decline in mild cognitive impairment and early Alzheimer's disease (Mungas et al., 2005; Boyle et al., 2006). To estimate accelerated hippocampal loss, we used a simple quadratic expansion for change that may not accurately reflect the true progression of hippocampal loss. Observations at much more than three time- points, as planned in the ADNI, should help to better characterize the trajectory of accelerated loss. Nonetheless, the finding of accelerating hippocampal loss is important for understanding the natural history of Alzheimer's disease and emphasizes the need for early diagnosis and therapeutic intervention. The fact that we were able to detect accelerated rates over 1-year period has consequences for longitudinal MRI studies with only two time-points. First, such studies may overestimate or underestimate hippocampal loss rates since two serial measurements are indifferent to accelerations. Second, the ability to detect differences may be limited if accelerations vary among subjects. The ADNI is funded to scan each subject multiple times over 3 years to provide more information on longitudinal change.
Our finding that ApoE4 is a modulator of hippocampal loss rates is consistent with other MRI studies (Moffat et al., 2000; Cohen et al., 2001; Jack Jr et al., 2008b; van de Pol et al., 2007), but some studies found no ApoE effect (Laakso et al., 2000). Furthermore, whereas others reported an ApoE4 effect on hippocampal rates for mild cognitive impairment (Jack Jr et al., 2008b), we found it limited to Alzheimer's disease. In a recent cross-sectional MRI study of 676 ADNI subjects using tensor-based morphometry (Hua et al., 2008), over half of the Alzheimer's disease and mild cognitive impairment subjects carried the ApoE4 gene, and they showed greater hippocampal and temporal lobe deficits than non-carriers. Around one-sixth of the controls carried the protective ApoE2 gene and showed reduced ventricular expansion, perhaps reflecting a lesser degree of overall brain atrophy. We did not find a significant dose effect of ApoE4 on hippocampal loss rates in contrast to another study (van de Pol et al., 2007). Given our sample size of 226 mild cognitive impairment patients and ∼10% within subject variation, we should have been able to detect about 5% difference between carriers and non-carriers with 80% power and at α = 0.05 significance. One possible explanation for the discrepant ApoE findings in mild cognitive impairment is the notorious heterogeneity of this group, which—in absence of histological evidence for Alzheimer's disease pathology—may include other causes of cognitive complaints, such as mood disorders or cerebrovascular disease that both can impact the hippocampus (Lloyd et al., 2004; Mungas et al., 2005). Nonetheless, our finding suggests that presence of ApoE4 exacerbates the impact of Alzheimer's disease on the hippocampus. Alzheimer's disease cohorts of future therapeutic trials could be enriched by including specifically patients who carry ApoE4 and are likely to have small hippocampi and high rates of hippocampal loss.
We also found a marked association between higher rates of hippocampal loss and decreased concentration of Aβ1–42 in CSF, predominantly in the mild cognitive impairment group, whereas for t-tau we found only a trend in the Alzheimer's disease group and for p-tau no significant association in any of the groups. Aβ1–42 and tau concentrations in CSF represent the earliest and most intensely studied biochemical markers of Alzheimer's disease (Frank et al., 2003; Grossman et al., 2005). How well these biomarkers reflect an autopsy-confirmed dementia diagnosis has intensely been studied (Clark et al., 2003). Both proteins are directly linked to the two hallmark lesions of Alzheimer's disease, Aβ1–42 with amyloid plaques and tau (t-tau and p-tau) with neurofibrillary tangles (Clark et al., 2006). Numerous studies have documented reduced Aβ1–42 and increased p-tau in CSF in Alzheimer's disease patients (see Clark et al., 2008 for review). But only two prior MRI studies compared CSF biomarkers with hippocampal loss rates in a small pool of subjects. Consistent with our findings, de Leon et al. (2006) reported greater hippocampal loss with greater Aβ1–42 decrease. In contrast to our results in mild cognitive impairment, they also found significantly greater hippocampal loss with increased p-tau but their study included only nine mild cognitive impairment subjects. Hampel et al. (2005), studying Alzheimer's disease, found increased hippocampal rates correlated with increased p-tau in 22 patients, while we found only a trend. Our finding that increased rates of hippocampal loss correlate with decreased Aβ1–42 in a large number of mild cognitive impairment patients is particularly interesting, because Aβ1–42 is directly related to Alzheimer's disease pathology. It is also important to note that the association between high hippocampal rates and reduced CSF Aβ1–42 was independent of the ApoE profile, implying that CSF Aβ1-42 levels and rates of hippocampal loss are directly linked. The results support the view that high rates of hippocampal loss in mild cognitive impairment indicate Alzheimer's disease pathology. Since the hippocampus is spared from early amyloid burden (Silbert et al., 2003), the correlation between high rates of hippocampal loss and decreased CSF Aβ1–42, further implies that the two measures provide complimentary information about the presence of Alzheimer's disease pathology. However, the diagnostic value of the measures used together remains unclear, because each measure can also change in other pathological conditions, such as Lewy body dementia (Clark et al., 2003).
Using a Markov chain model to analyse hippocampal change, we showed that intrinsic correlations between successive MRI observations exist and can be exploited to reduce within subject variability and consequently improve measurement power. The impact on power was substantial despite the fact that we performed hippocampal tracing independently for each time-point. The results imply that image processing algorithms that utilize correlations between observations, i.e. by using the hippocampal boundaries from past MRI scans as priors for tracing the boundaries in a new MRI scan, should be superior to those algorithms that do not employ priors. However, the approach also has limitations, as our results indicate. For instance, the Markov chain approach did not benefit the analysis of data from the normal group as much as the analysis of the mild cognitive impairment or Alzheimer's disease group, presumably because errors in assessing the small volume changes in normal subjects were random and dominated systematic biological changes. At the other end, the benefit of a Markov chain analysis was also less effective for Alzheimer's disease than for mild cognitive impairment data, presumably because the volume changes in Alzheimer's disease patients were sufficiently large to incur fewer errors to begin with. Since each subject in the ADNI study will ultimately have multiple successive MRI scans, it will be possible to evaluate the benefit of analysis using Markov chain models in more detail.
Our results demonstrate that for studies of hippocampal rates site-to-site variations in MRI can effectively be controlled using the rigorous methods of the ADNI. The result is important, because multiple MRI centre settings are indispensable for large studies, such as clinical trials with hundreds of subjects. Since additional variability is inevitably introduced in multiple MRI site settings compared to a single site setting, which is more common for investigational study use, we were also interested in comparing the powers of multisite and single site studies to detect a certain level of atrophy rates. Based on the site-to-site variations shown in Fig. 5, we estimated that 80% power of this multisite study of 47 MRI sites translates roughly into 87% power if the same study with the same population and number of subjects was conducted at the ‘best’ single MRI site in this study (#116 in Fig. 5) with the least variability of all sites. Although the result implies a benefit in power from a single site, as expected, the gain overall seems small, especially if one considers that the other participating sites in this study have smaller benefit margins since they show greater variability than the ‘best’ single site. In summary, the result attests to the effectiveness of the rigorous control methods developed by the ADNI. Our power estimations also show that MRI consistently provides greater power to measure progression than cognitive tests, such as ADAS-Cog or MMSE. Furthermore, some additional power can be gained by measuring hippocampal change at more time-points and by considering whether patients carry ApoE4.
Several limitations ought to be mentioned: First, mild cognitive impairment and normal subjects have not been followed long enough to determine the incidence of incipient Alzheimer's disease in each respective group. The rates and accelerations for these groups may therefore be biased toward higher values if many subjects with preclinical Alzheimer's disease were included. Furthermore, clinical criteria are always imperfect and some subjects in a mild stage of disease can be difficult to classify. However, the ADNI utilizes the rigorous diagnostic criteria of therapeutic trials which are one of the best available. The chance of a major bias of the results due to clinical misdiagnosis is small. Second, white matter lesions, an indication for cerebrovascular disease, were not accounted for. As previous studies showed white matter lesions can be associated with hippocampal atrophy (Fein et al., 2000), our results could at least in part be related to vascular disease. Third, since the algorithm to measure the hippocampus utilizes information from the rest of the brain, changes in other brain regions as well as image artefacts could have mimicked hippocampal variations. Completely manual measurements of the hippocampus might therefore lead to a different outcome.
In conclusion, the demonstration of hippocampal loss in mild cognitive impairment and Alzheimer's disease patients over 6 months and accelerated loss over 12 months illustrates the power of MRI to track morphological brain changes over time in a large multisite setting. Furthermore, our finding of higher hippocampal loss in presence of ApoE4 and reduced CSFAβ1–42 supports the concept that increased hippocampal loss is an indicator of Alzheimer's disease pathology and a potential marker to assess therapeutic interventions in Alzheimer's disease.
Supplementary material
Supplementary material is available at Brain online.
Funding
The ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and through generous contributions from the following: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, Alzheimer's Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, Innogenetics, NV, Ghent, Belgium, and the Institute for the Study of Aging, with participation from the U.S. Food and Drug Administration. Industry partnerships are coordinated through the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. The ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of California, Los Angeles. Funding to pay the Open Access publication charges for this article was provided by the Alzheimer's Disease Neuroimaging Initiative National Institutes of Health (U01 AG024904 to M.W.).
Supplementary Material
Acknowledgements
We are indebted to Drs Laurel Beckett, UC San Diego, Danielle Harvey, UC Davis and John Kornak, UC San Francisco for statistical support. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI; Principal Investigator: M.W.).
Glossary
Abbreviations
- ADAS-Cog
Alzheimer's disease Assessment Scale-Cognitive Subscale
- ADNI
Alzheimer's Disease Neuroimaging Initiative
- ApoE
apolipoprotein E
- CDR
Clinical Dementia Rating
- CSF
cerebrospinal fluid
References
- Barnes J, Bartlett JW, van de Pol LA, Loy CT, Scahill RI, Frost C, et al. A meta-analysis of hippocampal atrophy rates in Alzheimer's disease. Neurobiol Aging. 2008a doi: 10.1016/j.neurobiolaging.2008.01.010. [Epub ahead of print; doi:10.1016/j.neurobiolaging.2008.01.010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J, Scahill RI, Frost C, Schott JM, Rossor MN, Fox NC. Increased hippocampal atrophy rates in Alzheimer's disease over 6 months using serial MR imaging. Neurobiol Aging. 2008b;29:1199–203. doi: 10.1016/j.neurobiolaging.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Boyle PA, Wilson RS, Aggarwal NT, Tang Y, Bennett DA. Mild cognitive impairment: risk of Alzheimer disease and rate of cognitive decline. Neurology. 2006;67:441–5. doi: 10.1212/01.wnl.0000228244.10416.20. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Evolution of neuronal changes in the course of Alzheimer's disease. J Neural Transm Suppl. 1998;53:127–40. doi: 10.1007/978-3-7091-6467-9_11. [DOI] [PubMed] [Google Scholar]
- Carlson NE, Moore MM, Dame A, Howieson D, Silbert LC, Quinn JF, et al. Trajectories of brain loss in aging and the development of cognitive impairment. Neurology. 2008;70:828–33. doi: 10.1212/01.wnl.0000280577.43413.d9. [DOI] [PubMed] [Google Scholar]
- Chan D, Janssen JC, Whitwell JL, Watt HC, Jenkins R, Frost C, et al. Change in rates of cerebral atrophy over time in early-onset Alzheimer's disease: longitudinal MRI study. Lancet. 2003;362:1121–2. doi: 10.1016/S0140-6736(03)14469-8. [DOI] [PubMed] [Google Scholar]
- Christensen GE, Joshi SC, Miller MI. Volumetric transformation of brain anatomy. IEEE Trans Med Imaging. 1997;16:864–77. doi: 10.1109/42.650882. [DOI] [PubMed] [Google Scholar]
- Clark CM, Davatzikos C, Borthakur A, Newberg A, Leight S, Lee VM, et al. Biomarkers for early detection of Alzheimer pathology. Neurosignals. 2008;16:11–8. doi: 10.1159/000109754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CM, Pratico D, Shaw L, Leight S, Xie S, Gu A, et al. Biochemical biomarkers of late-life dementia. Alzheimer's & Dementia. 2006;2:287–93. doi: 10.1016/j.jalz.2006.05.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CM, Xie S, Chittams J, Ewbank D, Peskind E, Galasko D, et al. Cerebrospinal fluid tau and beta-amyloid: how well do these biomarkers reflect autopsy-confirmed dementia diagnoses? Arch Neurol. 2003;60:1696–702. doi: 10.1001/archneur.60.12.1696. [DOI] [PubMed] [Google Scholar]
- Cohen RM, Small C, Lalonde F, Friz J, Sunderland T. Effect of apolipoprotein E genotype on hippocampal volume loss in aging healthy women. Neurology. 2001;57:2223–8. doi: 10.1212/wnl.57.12.2223. [DOI] [PubMed] [Google Scholar]
- de Leon MJ, DeSanti S, Zinkowski R, Mehta PD, Pratico D, Segal S, et al. Longitudinal CSF and MRI biomarkers improve the diagnosis of mild cognitive impairment. Neurobiol Aging. 2006;27:394–401. doi: 10.1016/j.neurobiolaging.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Du AT, Schuff N, Chao LL, Kornak J, Jagust WJ, Kramer JH, et al. White matter lesions are associated with cortical atrophy more than entorhinal and hippocampal atrophy. Neurobiol Aging. 2005;26:553–9. doi: 10.1016/j.neurobiolaging.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Du AT, Schuff N, Kramer JH, Ganzer S, Zhu XP, Jagust WJ, et al. Higher atrophy rate of entorhinal cortex than hippocampus in Alzheimer's disease. Neurology. 2004;62:422–7. doi: 10.1212/01.wnl.0000106462.72282.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fein G, Di SV, Tanabe J, Cardenas V, Weiner MW, Jagust WJ, et al. Hippocampal and cortical atrophy predict dementia in subcortical ischemic vascular disease [In Process Citation] Neurology. 2000;55:1626–35. doi: 10.1212/wnl.55.11.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri CP, Prince M, Brayne C, Brodaty H, Fratiglioni L, Ganguli M, et al. Global prevalence of dementia: a Delphi consensus study. Lancet. 2005;366:2112–7. doi: 10.1016/S0140-6736(05)67889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell AM, Walhovd KB, Amlien I, Bjornerud A, Reinvang I, Gjerstad L, et al. Morphometric changes in the episodic memory network and tau pathologic features correlate with memory performance in patients with mild cognitive impairment. AJNR Am J Neuroradiol. 2008;29:1183–9. doi: 10.3174/ajnr.A1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–98. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Fox NC, Black RS, Gilman S, Rossor MN, Griffith SG, Jenkins L, et al. Effects of Abeta immunization (AN1792) on MRI measures of cerebral volume in Alzheimer disease. Neurology. 2005;64:1563–72. doi: 10.1212/01.WNL.0000159743.08996.99. [DOI] [PubMed] [Google Scholar]
- Fox NC, Schott JM. Imaging cerebral atrophy: normal ageing to Alzheimer's disease. Lancet. 2004;363:392–4. doi: 10.1016/S0140-6736(04)15441-X. [DOI] [PubMed] [Google Scholar]
- Frank RA, Galasko D, Hampel H, Hardy J, de Leon MJ, Mehta PD, et al. Biological markers for therapeutic trials in Alzheimer's disease. Proceedings of the biological markers working group; NIA initiative on neuroimaging in Alzheimer's disease. Neurobiol Aging. 2003;24:521–36. doi: 10.1016/s0197-4580(03)00002-2. [DOI] [PubMed] [Google Scholar]
- Grossman M, Farmer J, Leight S, Work M, Moore P, Van Deerlin V, et al. Cerebrospinal fluid profile in frontotemporal dementia and Alzheimer's disease. Ann Neurol. 2005;57:721–9. doi: 10.1002/ana.20477. [DOI] [PubMed] [Google Scholar]
- Hampel H, Burger K, Pruessner JC, Zinkowski R, DeBernardis J, Kerkman D, et al. Correlation of cerebrospinal fluid levels of tau protein phosphorylated at threonine 231 with rates of hippocampal atrophy in Alzheimer disease. Arch Neurol. 2005;62:770–3. doi: 10.1001/archneur.62.5.770. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Kazui H, Matsumoto K, Nakano Y, Yasuda M, Mori E. Does donepezil treatment slow the progression of hippocampal atrophy in patients with Alzheimer's disease? Am J Psychiatry. 2005;162:676–82. doi: 10.1176/appi.ajp.162.4.676. [DOI] [PubMed] [Google Scholar]
- Hsu YY, Schuff N, Du AT, Mark K, Zhu X, Hardin D, et al. Comparison of automated and manual MRI volumetry of hippocampus in normal aging and dementia. J Magn Reson Imaging. 2002;16:305–10. doi: 10.1002/jmri.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW, et al. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer's disease: An MRI study of 676 Alzheimer's disease, mild cognitive impairment, and normal subjects. Neuroimage. 2008;43:458–69. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, et al. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008a;27:685–91. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Grundman M, Jin S, Gamst A, Ward CP, et al. Longitudinal MRI findings from the vitamin E and Donepezil treatment study for mild cognitive impairment. Neurobiol Aging. 2008b;29:1285–95. doi: 10.1016/j.neurobiolaging.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu Y, O’Brien PC, Smith GE, Ivnik RJ, et al. Rates of hippocampal atrophy correlate with change in clinical status in aging and Alzheimer's disease. Neurology. 2000;55:484–9. doi: 10.1212/wnl.55.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Shiung MM, Weigand SD, O’Brien PC, Gunter JL, Boeve BF, et al. Brain atrophy rates predict subsequent clinical conversion in normal elderly and amnestic mild cognitive impairment. Neurology. 2005;65:1227–31. doi: 10.1212/01.wnl.0000180958.22678.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Weigand SD, Shiung MM, Przybelski SA, O'Brien PC, Gunter JL, et al. Atrophy rates accelerate in amnestic mild cognitive impairment. Neurology. 2008c;70:1740–52. doi: 10.1212/01.wnl.0000281688.77598.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C Jr, Petersen RC, Xu Y, O’Brien PC, Smith GE, Ivnik RJ, et al. Rates of Hippocampal Atrophy in Normal Aging, Mild Cognitive Impairment, and Alzheimer's Disease. Neurology. 2000;55:484–9. doi: 10.1212/wnl.55.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Shiung MM, Gunter JL. Comparison of different MRI brain atrophy rate measures with clinical disease progression in Alzheimer's disease. Neurology. 2004;62:591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C Jr, Slomkowski M, Gracon S, Hoover TM, Felmlee JP, Stewart K, et al. MRI as a Biomarker of Disease Progression in a Therapeutic Trial of Milameline fo Alzheimer's. Neurology. 2003;60:253–60. doi: 10.1212/01.wnl.0000042480.86872.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laakso MP, Lehtovirta M, Partanen K, Riekkinen PJ, Soininen H. Hippocampus in Alzheimer's disease: a 3-year follow-up MRI study. Biol Psychiatry. 2000;47:557–61. doi: 10.1016/s0006-3223(99)00167-5. [DOI] [PubMed] [Google Scholar]
- Leow Alzheimer's disease, Klunder Alzheimer's disease, Jack CR, Jr, Toga AW, Dale AM, Bernstein MA, et al. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31:627–40. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AJ, Ferrier IN, Barber R, Gholkar A, Young AH, O’Brien JT. Hippocampal volume change in depression: late- and early-onset illness compared. Br J Psychiatry. 2004;184:488–95. doi: 10.1192/bjp.184.6.488. [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS- Alzheimer's diseaseRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's disease. Neurology. 1984;34:939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Moffat SD, Szekely CA, Zonderman AB, Kabani NJ, Resnick SM. Longitudinal change in hippocampal volume as a function of apolipoprotein E genotype. Neurology. 2000;55:134–6. doi: 10.1212/wnl.55.1.134. [DOI] [PubMed] [Google Scholar]
- Mohs RC, Knopman D, Petersen RC, Ferris SH, Ernesto C, Grundman M, et al. Development of cognitive instruments for use in clinical trials of antidementia drugs: additions to the Alzheimer's disease Assessment Scale that broaden its scope. The Alzheimer's Disease Cooperative Study. Alzheimer Dis Assoc Disord. 1997;11(Suppl 2):S13–21. [PubMed] [Google Scholar]
- Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules [see comments] Neurology. 1993;43:2412–4. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- The Alzheimer's disease neuroimaging initiative. Neuroimaging Clin N Am. 2005;15:869–77. doi: 10.1016/j.nic.2005.09.008. xi–xii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungas D, Harvey D, Reed BR, Jagust WJ, DeCarli C, Beckett L, et al. Longitudinal volumetric MRI change and rate of cognitive decline. Neurology. 2005;65:565–71. doi: 10.1212/01.wnl.0000172913.88973.0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridha BH, Barnes J, Bartlett JW, Godbolt A, Pepple T, Rossor MN, et al. Tracking atrophy progression in familial Alzheimer's disease: a serial MRI study. Lancet Neurol. 2006;5:828–34. doi: 10.1016/S1474-4422(06)70550-6. [DOI] [PubMed] [Google Scholar]
- Shaw LM, Korecka M, Clark CM, Lee VM, Trojanowski JQ. Biomarkers of neurodegeneration for diagnosis and monitoring therapeutics. Nat Rev Drug Discov. 2007;6:295–303. doi: 10.1038/nrd2176. [DOI] [PubMed] [Google Scholar]
- Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen P, Petersen RC, et al. and the ADNI Investigators. Cerebrospinal fluid biomarker signature in Alzheimer's Disease Neuroimaging Initiative (ADNI) subjects. Ann Neurol. 2009 doi: 10.1002/ana.21610. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbert LC, Quinn JF, Moore MM, Corbridge E, Ball MJ, Murdoch G, et al. Changes in premorbid brain volume predict Alzheimer's disease pathology. Neurology. 2003;61:487–92. doi: 10.1212/01.wnl.0000079053.77227.14. [DOI] [PubMed] [Google Scholar]
- Simic G, Kostovic I, Winblad B, Bogdanovic N. Volume and number of neurons of the human hippocampal formation in normal aging and Alzheimer's disease. J Comp Neurol. 1997;379:482–94. doi: 10.1002/(sici)1096-9861(19970324)379:4<482::aid-cne2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, Dutton RA, Chiang MC, Leow Alzheimer's disease, Sowell ER, et al. Tracking Alzheimer's disease. Ann N Y Acad Sci. 2007;1097:183–214. doi: 10.1196/annals.1379.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pol LA, Hensel A, van der Flier WM, Visser P, Pijnenburg YA, Barkhof F, et al. Hippocampal atrophy on MRI in frontotemporal lobar degeneration and Alzheimer's disease. J Neurol Neurosurg Psychiatry. 2005;10:1136. doi: 10.1136/jnnp.2005.075341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pol LA, van der Flier WM, Korf ES, Fox NC, Barkhof F, Scheltens P. Baseline predictors of rates of hippocampal atrophy in mild cognitive impairment. Neurology. 2007;69:1491–7. doi: 10.1212/01.wnl.0000277458.26846.96. [DOI] [PubMed] [Google Scholar]
- Vanderstichele H, De Meyer G, Shapiro F, Engelborghs B, DeDeyn PP, Shaw LM, et al. Alzheimer's disease biomarkers: From concept to clinical utility. In: Galimberti D, Scarpini E, editors. Biomarkers for Early Diagnosis of Alzheimer s Disease. Hauppauge, NY: Nova Scinece Publishers, Inc.; 2008. pp. 81–122. [Google Scholar]
- West MJ. Regionally specific loss of neurons in the aging human hippocampus. Neurobiol Aging. 1993;14:287–93. doi: 10.1016/0197-4580(93)90113-p. [DOI] [PubMed] [Google Scholar]
- Yesavage JA, Brink TL, Rose TL, Lum O, Huang V, Adey M, et al. Development and validation of a geriatric depression screening scale: a preliminary report. J Psychiatr Res. 1982;17:37–49. doi: 10.1016/0022-3956(82)90033-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.