Abstract
Rapidly accumulating evidence shows that common T-cell transcription factor (TCF)7L2 polymorphisms confer risk of type 2 diabetes through unknown mechanisms. We examined the association between four TCF7L2 single nucleotide polymorphisms (SNPs), including rs7903146, and measures of insulin sensitivity and insulin secretion in 1,697 Europid men and women of the population-based MRC (Medical Research Council)-Ely study. The T-(minor) allele of rs7903146 was strongly and positively associated with fasting proinsulin (P = 4.55 × 10−9) and 32,33 split proinsulin (P = 1.72 × 10−4) relative to total insulin levels; i.e., differences between T/T and C/C homozygotes amounted to 21.9 and 18.4% respectively. Notably, the insulin-to-glucose ratio (IGR) at 30-min oral glucose tolerance test (OGTT), a frequently used surrogate of first-phase insulin secretion, was not associated with the TCF7L2 SNP (P > 0.7). However, the insulin response (IGR) at 60-min OGTT was significantly lower in T-allele carriers (P = 3.5 × 10−3). The T-allele was also associated with higher A1C concentrations (P = 1.2 × 10−2) and reduced β-cell function, assessed by homeostasis model assessment of β-cell function (P = 2.8 × 10−2). Similar results were obtained for the other TCF7L2 SNPs. Of note, both major genes involved in proinsulin processing (PC1, PC2) contain TCF-binding sites in their promoters. Our findings suggest that the TCF7L2 risk allele may predispose to type 2 diabetes by impairing β-cell proinsulin processing. The risk allele increases proinsulin levels and diminishes the 60-min but not 30-min insulin response during OGTT. The strong association between the TCF7L2 risk allele and fasting proinsulin but not insulin levels is notable, as, in this unselected and largely normoglycemic population, external influences on β-cell stress are unlikely to be major factors influencing the efficiency of proinsulin processing.
A common variant in the TCF7L2 gene was found to increase the risk of type 2 diabetes by 1.41 in heterozygotes and by 2.27 in homozygotes in an Icelandic population, a finding that was replicated in a Danish and a European-American population (1). The association is robust and has been consistently reproduced in geographically and ethnically diverse populations (2-12).
The mechanisms through which TCF7L2 affects the susceptibility to type 2 diabetes remain to be elucidated. It is known that TCF7L2 serves as a nuclear receptor for β-catenin, which mediates the Wnt signaling pathway (13). The bipartite transcription factor, cat/TCF7L2, activates many genes downstream of the Wnt signaling cascade (14). One of the genes transcriptionally regulated by cat/TCF7L2 is proglucagon, which encodes the insulinotropic hormone glucagon-like peptide 1 (GLP-1) (13). Because GLP1, in concert with insulin, plays a critical role in blood glucose homeostasis, it has been postulated that TCF7L2 gene variants may affect the susceptibility to type 2 diabetes by indirectly altering GLP-1 levels (13). Since the Wnt signaling pathway is involved in a variety of biological events and TCF7L2 transcriptionally regulates many genes, other mechanisms cannot be excluded.
Case-control studies have been instrumental in confirming the association between TCF7L2 variants and type 2 diabetes. However, by design, these studies provide limited insight into the biological pathways through which TCF7L2 exerts its effect because the biochemical profile in case subjects is likely to have been altered by the disease status or by treatment. Cohort studies with detailed phenotypic characterization can provide new insights to further elucidate the mechanisms that underlie the robust association.
The present study examines the association between four TCF7L2 polymorphisms and measures of fasting insulin sensitivity, β-cell function, and insulin secretion during a 120-min oral glucose tolerance test (OGTT) in 1,697 Europid men and women of the population-based Ely study.
RESEARCH DESIGN AND METHODS
The MRC Ely Study is a population-based cohort study of the etiology of type 2 diabetes. Study participants were randomly selected from people living in Ely and surrounding villages (East Anglia, U.K.), an ethnically homogenous Europid population. The current analyses included 1,697 men and women, aged 35–79 years, for whom genotypic and phenotypic data were available from phase 3 as previously described (15). None of the participants had diagnosed type 2 diabetes, although 139 individuals met the World Health Organization criteria for type 2 diabetes. The study design, methods, and measurements have been described in Supplementary Information (available in an online appendix at http://dx.doi.org/10.2337/db07-0055).
In brief, all participants attended a clinical examination for anthropometric measurements, medical questionnaires, and a 75-g OGTT. Glucose and insulin concentrations were measured at fasting and at 30-, 60-, and 120-min postglucose load. Proinsulin and 32,33 split proinsulin was measured at fasting. We calculated the insulin-to-glucose ratio (IGR) at 30 and 60 min as a measure of insulin secretion. Homeostasis model assessment of insulin sensitivity (HOMA-S) and β-cell function were estimated by the homeostasis model using Levy's computer model (16). Descriptive characteristics of the population are given in supplementary Table 1.
Single nucleotide polymorphism selection and genotyping
We genotyped the two single nucleotide polymorphisms (SNPs) (rs12255372 and rs7903146) that showed strongest association in the Icelandic population (1) and added two neighboring SNPs (rs4506565 and rs12243326) that were in high but not perfect linkage disequilibrium with the first two SNPs according to the CEU HapMap data (supplementary Fig. 1).
Genotyping was performed using Custom TaqMan SNP genotyping assays (Applied Biosystems, Warrington, U.K.) (see Supplementary Information). The genotyping success rate exceeded 98% for all SNPs, and genotype frequencies were in Hardy-Weinberg equilibrium (P > 0.05) (supplementary Table 2).
In silico analyses
In silico analyses were performed to identify TCF-binding sites in the proprotein convertase 1 (PC1) and 2 (PC2) and carboxypeptidase E (CPE) gene sequences. Genomic sequence and annotation of each gene, including 5 kb upstream, were obtained for a number of species. A global alignment for each gene was performed. The consensus sequence for the TCF-binding site (WWCAAWG) was then used to search for any conserved sites within the alignment. The positions for all candidate sites were added to the human annotation file and the whole annotated alignment viewed (see Supplementary Information).
Statistical analyses
A likelihood ratio test was performed to assess Hardy-Weinberg equilibrium. Linkage disequilibrium between SNPs was estimated using Haploview, version 3.2 (http://www.broad.mit.edu/mpg/haploview).
We tested for association between phenotypes and SNPs using a generalized linear model assuming an additive effect for the presence of each additional minor allele. All phenotypes were adjusted for age, sex, and BMI (except BMI, fat percentage, and waist circumference). An estimate of the composite β-cell function was obtained by adjusting baseline insulin secretion (IGR) to insulin sensitivity (HOMA-S) through linear regression of log-transformed variables, as previously described (17). All phenotypes were log transformed to obtain normal distributions, and tables represent geometric means. Statistical analyses were conducted using SAS/Genetics (version 9.1; SAS Institute, Cary, NC).
RESULTS
The four TCF7L2 SNPs were in high linkage disequilibrium (supplementary Fig. 1); thus, the results of the associations were similar. Here, we describe the results for rs7903146. More details on the other TCF7L2 SNPs can be found in supplementary Table 3.
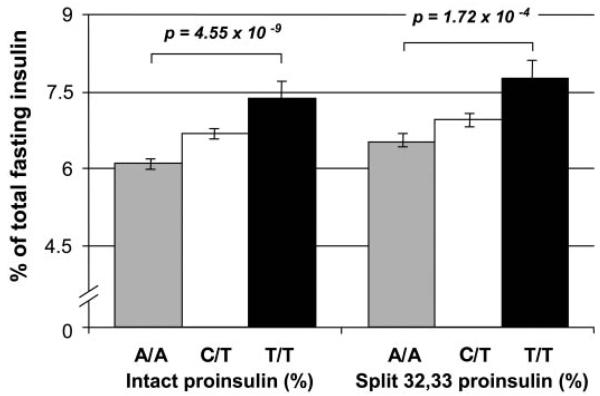
The strongest association was observed with fasting proinsulin concentrations (Table 1). The T-(minor) allele of rs7903146 showed a highly significant (P = 4.7 × 10−7) association with intact proinsulin concentrations; i.e., T/T homozygotes had, on average, 15% higher proinsulin concentrations compared with C/T heterozygotes, whose concentrations were 9% higher than those of C/C homozygotes. Similar associations were observed for fasting 32,33 split proinsulin concentrations (Table 1). Although the significance of the association was less (P = 2.8 × 10−3), the magnitude of the differences was similar; i.e., T/T homozygotes had a 15% higher 32,33 split proinsulin concentration than C/T heterozygotes, whose concentrations were 6% higher than those of C/C homozygotes. When intact proinsulin and 32,33 split proinsulin concentrations are expressed as a percentage of fasting total insulin (sum of insulin, intact proinsulin, and 32,33 split proinsulin), the associations were even stronger (intact proinsulin, P = 4.55 × 10−9; 32,33 split proinsulin, P = 1.72 × 10−4) (Fig. 1).
TABLE 1.
Means ± SEM for measures of body composition, fasting and OGTT insulin and glucose concentrations, and measures of insulin secretion, insulin sensitivity, and (β-cell function by genotype in 1,697 Europids for the rs7903146 TCF7L2 gene variant
| C/C | C/T | T/T | P | |
|---|---|---|---|---|
| n | 830 | 699 | 147 | |
| BMI (kg/m2) | 27.0 ± 0.2 | 26.8 ± 0.2 | 26.6 ± 0.4 | 0.24 |
| Fat percentage | 32.0 ± 0.2 | 31.8 ± 0.2 | 31.9 ± 0.5 | 0.61 |
| Waist circumference (cm) | 92.9 ± 0.4 | 92.5 ± 0.4 | 91.3 ± 0.9 | 0.21 |
| Fasting insulin (pmol/l) | 49.5 ± 0.9 | 48.9 ± 1.0 | 48.6 ± 2.1 | 0.60 |
| Insulin AUC (pmol/l)* | 616 ± 11 | 594 ± 12 | 621 ± 26 | 0.54 |
| Fasting glucose (pmol/l) | 5.49 ± 0.02 | 5.54 ± 0.03 | 5.61 ± 0.06 | 0.042 |
| Glucose AUC (pmol/l)* | 15.0 ± 0.1 | 15.3 ± 0.1 | 15.8 ± 0.3 | 0.013 |
| IGR at 30 min | 88.6 ± 2.5 | 86.1 ± 2.7 | 88.4 ± 5.9 | 0.70 |
| IGR at 60 min | 134.0 ± 4.5 | 120.2 ± 4.5 | 108.5 ± 8.6 | 0.0035 |
| Fasting intact proinsulin (pmol/l) | 3.56 ± 0.07 | 3.89 ± 0.08 | 4.46 ± 0.20 | 0.00000047 |
| Fasting 32,33 split proinsulin (pmol/l) | 3.85 ± 0.10 | 4.06 ± 0.11 | 4.69 ± 0.28 | 0.0028 |
| Fasting A1C (%) | 5.35 ± 0.02 | 5.38 ± 0.02 | 5.46 ± 0.04 | 0.012 |
| HOMA-B | 83.3 ± 1.1 | 81.0 ± 1.2 | 77.7 ± 2.4 | 0.028 |
| HOMA-S | 90.1 ± 1.6 | 91.3 ± 1.8 | 92.9 ± 4.0 | 0.45 |
Means are geometric means. All phenotypes were adjusted for age, sex, and BMI, except BMI and fat percentage, which were adjusted for age and sex. AUC, area under the curve; HOMA-B, homeostasis model assessment of β-cell function.
FIG. 1.
Percentage (means ± SEM) of fasting proinsulin and 32,33 split proinsulin of total fasting insulin concentration (intact proinsulin + 32,33 split proinsulin + insulin) by genotype in 1,697 Europids for the rs7903146 TCF7L2 gene variant.
IGR, as an estimate of insulin secretion, was significantly (P = 3.5 × 10−3) associated with the TCF7L2 variant but only when measured after 60-min OGTT; i.e., IGR at 60 min decreased by 10% for each additional T-allele in the genotype. No association was observed with IGR at a 30-min OGTT. Adjusting the IGR measures for insulin sensitivity (HOMA-S), as a measure of composite β-cell function (17), resulted in similar findings.
A1C concentration (P = 0.012) was increased, and β-cell function (homeostasis model assessment of β-cell function) was reduced (P = 0.028) in T/T homozygotes compared with C-allele carriers (supplementary Fig. 2). Also, fasting glucose concentrations (P = 0.04) and glucose area under the curve during an OGTT (P = 0.013) were significantly higher in T/T homozygotes compared with C-allele carriers. No associations were observed with insulin concentrations, insulin sensitivity (HOMA-S), BMI, fat percentage, or waist circumference.
Results were similar after excluding 139 individuals who met the World Health Organization criteria for type 2 diabetes (data not shown). No association or interactions with BMI were observed.
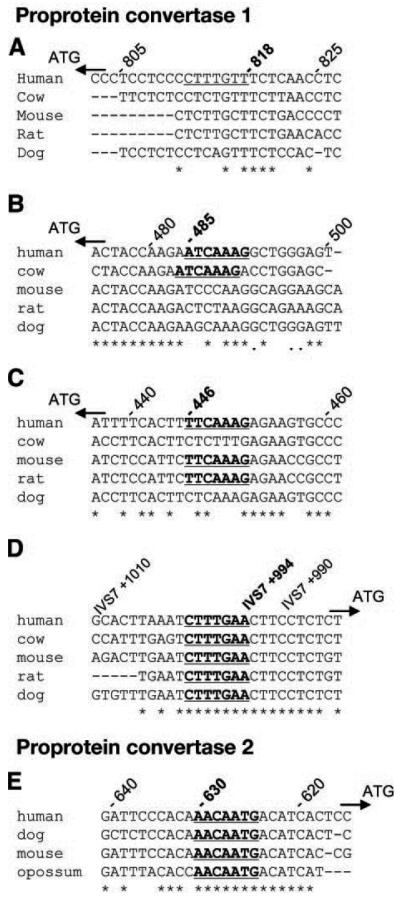
Because of the strong association between the TCF7L2 variants and insulin precursors, we theorized that TCF7L2 might act through transcription of PC1 and PC2 and/or CPE, which are required for processing proinsulin to insulin (18,19). We identified a TCF-binding site in PC1 between exon 7 and exon 8 (Fig. 2) and three more binding sites in the PC1 promoter. We also found a TCF-binding site in the PC2-promoter region that was conserved across human, dog, mouse, and opossum. Although potential consensus TCF-binding sequences were located in the 10-kb region upstream of the human CPE gene, none were fully conserved across species.
FIG. 2.
Consensus sequence (WWCAAWG) for TCF-binding sites in the PC1 (A–D) and PC2 (E) gene sequence. Four consensus sites were identified in PC1. A: Twenty-seven basepairs of the sequence upstream of start codon. The raw genomic position of the consensus site is 95795320–95795326 at −818 bp from the start codon. B: Twenty-seven basepairs of the sequence upstream of the start codon. The raw genomic position of the consensus site is 95794987–95794993 at −485 bp from the start codon. C: Twenty-seven basepairs of the sequence upstream of the start codon. The raw genomic position of the consensus site is 95794948–95794954 at −446 bp from start codon. D: Twenty-seven basepairs of the sequence between exons 7 and 8. The raw genomic position is 95772777–95772783 at 994 bp from the start of intron 7. One consensus site was identified in PC2. E: Twenty-seven basepairs of the sequence upstream of the start codon. The raw genomic position of the consensus site is 17155321–17155327 at −630 bp from the start codon. All alignments are composed of sequence taken 5′ → 3′ from the forward strand, and positions are as per the current build (NCBI 36 assembly) of the human genome (August 2006).
DISCUSSION
This population-based cohort study in British Europids provides new insights into the potential mechanisms through which common TCF7L2 gene variants increase the susceptibility to type 2 diabetes. Consistent with previous observations, we found that TCF7L2 variants might predispose to type 2 diabetes by impairing β-cell insulin secretion, rather than by changes in insulin sensitivity. In addition, our results suggest that TCF7L2 is associated, directly or indirectly, with impaired insulin synthesis and/or processing of insulin precursors. The minor allele carriers have substantially increased intact proinsulin and 32,33 split proinsulin relative to insulin and demonstrate impaired 60-min but not 30-min OGTT insulin secretion. The strong association between the TCF7L2 risk allele and proinsulin but not insulin is notable, as, in this unselected and largely normoglycemic population, external influences on β-cell stress are unlikely to be major factors influencing the efficiency of proinsulin processing.
Thus far, only four cohort studies have reported quantitative trait analyses (5,10,20,21), whereas others performed these analyses within a case-control (2,6,11,12) or intervention (3) design. The TCF7L2 polymorphisms were found to be associated with insulin secretion (2,3,5,10,11,21) and insulin sensitivity (2,5,11), whereas others found no associations (6,12,20). Thus, the quantitative trait results are less consistent and the statistical significance is less when compared with case-control studies.
Our study is the first population-based cohort study that tested for associations with precursor proteins of insulin. The associations between the TCF7L2 polymorphisms and intact proinsulin levels, and to a lesser extent for 32,33 split proinsulin levels, are the strongest thus far reported for quantitative traits.
The strong associations with proinsulin suggest that the TCF7L2 gene is associated with defects in insulin synthesis, processing, and/or secretion of insulin precursors. This is consistent with findings in previous studies and with the theory that TCF7L2 variants might impair β-cell function and insulin secretion (2,3,5,6,10,11). Proinsulin levels predict the risk of type 2 diabetes independent of insulin levels (22). This supports the hypothesis that a reduced efficiency in β-cell proinsulin processing might be the mediating factor in the TCF7L2–type 2 diabetes association.
That PC1, PC2, and CPE are key proteins in the processing of proinsulin to insulin (19,23) suggests that TCF7L2 variants affect proinsulin levels through transcriptional regulation of their genes. An impaired function of PC1 and PC2 would lead to an excess of intact proinsulin, whereas a defect in only PC1 or PC2 would result in accumulation of 65,66 split and 32,33 split proinsulin, respectively. A defect in CPE would result in accumulation of both split proinsulin forms. Support for our hypothesis comes from the identification of at least four conserved TCF-binding sites for PC1 and at least one for PC2. This observation needs further investigation and confirmation from experimental data. Given that PC1 and PC2 are involved in the processing of several other prohormones and neuropep-tide precursors, the potential effects of TCF7L2 could manifest itself in other phenotypes.
Furthermore, the minor allele was also associated with reduced insulin secretion (IGR) at 60 min, but not at 30 min, during an OGTT. These findings suggest that, upon glucose stimulation, the first-phase insulin release from the insulin-storing granules is normal. However, the insulin release at 60 min, which takes place once the insulin-storing granules are depleted and requires insulin synthesis, processing, and secretion, is impaired in the minor allele carriers. This corroborates the observed associations showing that the TCF7L2 gene variants are associated with increased intact proinsulin and 32,33 split proinsulin concentrations. In contrast, Saxena et al. (2) found association with insulin response at 30 min in the control subjects of a case-control study. No data at 60-min OGTT were available.
TCF7L2 activates many genes downstream of the Wnt signaling pathway, a complicated regulatory network implicated in many biological events such as the development of the pancreas and islets during embryonic growth (14,24). Therefore, our results might reflect impaired β-cell function due to reduced cell mass or to defects in cell differentiation and maturation.
It has been suggested that TCF7L2 could influence the susceptibility to type 2 diabetes by altering the expression of GLP-1 in enteroendocrine cells as TCF7L2 regulates the expression of its preprotein, proglucagon (13). Also, other pathways make GLP-1 a good candidate. GLP-1 is a β-cell mitogen, capable of promoting β-cell proliferation and inhibiting β-cell apoptosis in animal models (25). In addition, GLP-1 replenishes insulin stores via stimulation of proinsulin gene expression and translation (25). Interestingly, the processing of proglucagon to GLP-1 and other peptides also requires PC1 (25).
In summary, our findings confirm and provide further insight in the role of TCF7L2 in insulin secretion. The strong association with proinsulin concentrations and the association with insulin secretion at 60-min but not at 30-min OGTT suggest that TCF7L2 might be involved in insulin synthesis, processing, and secretion. This needs confirmation by experimental data, while other mechanisms cannot be excluded.
Supplementary Material
ACKNOWLEDGMENTS
The Ely Study was funded by the MRC and Diabetes UK. Additional support was provided by the Stroke Association, British Heart Foundation, Department of Health, Food Standards Agency, and the Wellcome Trust. I.B. is funded by the Wellcome Trust. P.W.F. was supported in part via grants from Novo Nordisk (370579201) and the Swedish Diabetes Association (DIA2006-013). S.O. and I.B. also acknowledge support from EU FP6 funding (contract no. LSHM-CT-2003-503041).
We are grateful to the volunteers in the Ely Study, who gave their time to take part in these studies.
Glossary
- GLP-1
glucagon-like peptide 1
- HOMA-S
homeostasis model assessment of insulin sensitivity
- IGR
insulin-to-glucose ratio
- OGTT
oral glucose tolerance test
- SNP
single nucleotide polymorphism
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Grant SFA, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thor-steinsdottir U, Gulcher JR, Kong A, Stefansson K. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 2.Saxena R, Gianniny L, Burtt NP, Lyssenko V, Giuducci C, Sjogren M, Florez JC, Almgren P, Isomaa B, Orho-Melander M, Lindblad U, Daly MJ, Tuomi T, Hirschhorn JN, Ardlie KG, Groop LC, Altshuler D. Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes. 2006;55:2890–2895. doi: 10.2337/db06-0381. [DOI] [PubMed] [Google Scholar]
- 3.Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PIW, Shuldiner AR, Knowler WC, Nathan DM, Altshuler D. The Diabetes Prevention Program Research Group: TCF7L2 Polymorphisms and progression to diabetes in the diabetes prevention program. N Engl J Med. 2006;355:241–250. doi: 10.1056/NEJMoa062418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Vliet-Ostaptchouk JV, Shiri-Sverdlov R, Zhernakova A, Strengman E, van Haeften TW, Hofker MH, Wijmenga C. Association of variants of transcription factor 7-like 2 (TCF7L2) with susceptibility to type 2 diabetes in the Dutch Breda cohort. Diabetologia. 2006;50:59–62. doi: 10.1007/s00125-006-0477-z. [DOI] [PubMed] [Google Scholar]
- 5.Damcott CM, Pollin TI, Reinhart LJ, Ott SH, Shen H, Silver KD, Mitchell BD, Shuldiner AR. Polymorphisms in the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in the Amish: replication and evidence for a role in both insulin secretion and insulin resistance. Diabetes. 2006;55:2654–2659. doi: 10.2337/db06-0338. [DOI] [PubMed] [Google Scholar]
- 6.Scott LJ, Bonnycastle LL, Willer CJ, Sprau AG, Jackson AU, Narisu N, Duren WL, Chines PS, Stringham HM, Erdos MR, Valle TT, Tuomilehto J, Bergman RN, Mohlke KL, Collins FS, Boehnke M. Association of transcription factor 7-like 2 (TCF7L2) variants with type 2 diabetes in a Finnish sample. Diabetes. 2006;55:2649–2653. doi: 10.2337/db06-0341. [DOI] [PubMed] [Google Scholar]
- 7.Humphries SE, Gable D, Cooper JA, Ireland H, Stephens JW, Hurel SJ, Li KW, Palmen J, Miller MA, Cappuccio FP, Elkeles R, Godsland I, Miller GJ, Talmud PJ. Common variants in the TCF7L2 gene and predisposition to type 2 diabetes in UK European Whites, Indian Asians and Afro-Caribbean men and women. Journal of Mol Med. 2006;84(Suppl. 12):1–10. doi: 10.1007/s00109-006-0108-7. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C, Qi L, Hunter DJ, Meigs JB, Manson JE, van Dam RM, Hu FB. Variant of transcription factor 7-like 2 (TCF7L2) gene and the risk of type 2 diabetes in large cohorts of U.S. women and men. Diabetes. 2006;55:2645–2648. doi: 10.2337/db06-0643. [DOI] [PubMed] [Google Scholar]
- 9.Groves CJ, Zeggini E, Minton J, Frayling TM, Weedon MN, Rayner NW, Hitman GA, Walker M, Wiltshire S, Hattersley AT, McCarthy MI. Association analysis of 6,736 U.K. subjects provides replication and confirms TCF7L2 as a type 2 diabetes susceptibility gene with a substantial effect on individual risk. Diabetes. 2006;55:2640–2644. doi: 10.2337/db06-0355. [DOI] [PubMed] [Google Scholar]
- 10.Munoz J, Lok KH, Gower BA, Fernandez JR, Hunter GR, Lara-Castro C, De Luca M, Garvey WT. Polymorphism in the transcription factor 7-like 2 (TCF7L2) gene is associated with reduced insulin secretion in nondiabetic women. Diabetes. 2006;55:3630–3634. doi: 10.2337/db06-0574. [DOI] [PubMed] [Google Scholar]
- 11.Chandak GR, Janipalli CS, Bhaskar S, Kulkarni SR, Mohankrishna P, Hattersley AT, Frayling TM, Yajnik CS. Common variants in the TCF7L2 gene are strongly associated with type 2 diabetes mellitus in the Indian population. Diabetologia. 2006;50:63–67. doi: 10.1007/s00125-006-0502-2. [DOI] [PubMed] [Google Scholar]
- 12.Cauchi S, Meyre D, Dina C, Choquet H, Samson C, Gallina S, Balkau B, Charpentier G, Pattou F, Stetsyuk V, Scharfmann R, Staels B, Fruhbeck G, Froguel P. Transcription factor TCF7L2 genetic study in the French population: expression in human β-cells and adipose tissue and strong association with type 2 diabetes. Diabetes. 2006;55:2903–2908. doi: 10.2337/db06-0474. [DOI] [PubMed] [Google Scholar]
- 13.Yi F, Brubaker PL, Jin T. TCF-4 mediates cell type-specific regulation of proglucagon gene expression by beta-catenin and glycogen synthase kinase-3beta. J Biol Chem. 2005;280:1457–1464. doi: 10.1074/jbc.M411487200. [DOI] [PubMed] [Google Scholar]
- 14.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mesa JL, Loos RJF, Franks PW, Ong KKL, Luan J, O'Rahilly S, Wareham NJ, Barroso I. Lamin A/C Polymorphisms, type 2 diabetes, and the metabolic syndrome: case-control and quantitative trait studies. Diabetes. 2007;56:884–889. doi: 10.2337/db06-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy JC, Matthews DR, Hermans MP. Correct homeostasis model assessment (HOMA) evaluation uses the computer program. Diabetes Care. 1998;21:2191–2192. doi: 10.2337/diacare.21.12.2191. [DOI] [PubMed] [Google Scholar]
- 17.The Diabetes Prevention Program Research Group Role of insulin secretion and sensitivity in the evolution of type 2 diabetes in the Diabetes Prevention Program: effects of lifestyle intervention and metformin. Diabetes. 2005;54:2404–2414. doi: 10.2337/diabetes.54.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu X, Orci L, Carroll R, Norrbom C, Ravazzola M, Steiner DF. Severe block in processing of proinsulin to insulin accompanied by elevation of des-64,65 proinsulin intermediates in islets of mice lacking prohormone convertase 1/3. Proc Natl Acad Sci U S A. 2002;99:10299–10304. doi: 10.1073/pnas.162352799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naggert JK, Fricker LD, Varlamov O, Nishima PM, Rouille Y, Steiner DF, Carroll RJ, Paigen BJ, Leiter EH. Hyperproinsulinaemia in obese fat/fat mice associated with a carboxypeptidase E mutation which reduces enzyme activity. Nat Genet. 1995;10:135–142. doi: 10.1038/ng0695-135. [DOI] [PubMed] [Google Scholar]
- 20.Cauchi S, Meyre D, Choquet H, Dina C, Born C, Marre M, Balkau B, Froguel P, the DESIR Study Group TCF7L2 variation predicts hyperglycemia incidence in a French general population: the Data From an Epidemiological Study on the Insulin Resistance Syndrome (DESIR) Study. Diabetes. 2006;55:3189–3192. doi: 10.2337/db06-0692. [DOI] [PubMed] [Google Scholar]
- 21.Melzer D, Murray A, Hurst AJ, Weedon MN, Bandinelli S, Corsi AM, Ferrucci L, Paulisso G, Guralnik JM, Frayling TM. Effects of the diabetes linked TCF7L2 polymorphism in a representative older population. BMC Med. 2006;4:34. doi: 10.1186/1741-7015-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zethelius B, Hales CN, Lithell HO, Berne C. Insulin resistance, impaired early insulin response, and insulin propeptides as predictors of the development of type 2 diabetes: a population-based, 7-year follow-up study in 70-year-old men. Diabetes Care. 2004;27:1433–1438. doi: 10.2337/diacare.27.6.1433. [DOI] [PubMed] [Google Scholar]
- 23.Jansen E, Ayoubi TAY, Meulemans SMP, de Ven WJMV. Neuroendocrinespecific expression of the human prohormone convertase 1 gene. J Biol Chem. 1995;270:15391–15397. doi: 10.1074/jbc.270.25.15391. [DOI] [PubMed] [Google Scholar]
- 24.Papadopoulou S, Edlund H. Attenuated Wnt signaling perturbs pancreatic growth but not pancreatic function. Diabetes. 2005;54:2844–2851. doi: 10.2337/diabetes.54.10.2844. [DOI] [PubMed] [Google Scholar]
- 25.Drucker DJ. The biology of incretin hormones. Cell Metab. 2006;3:153–165. doi: 10.1016/j.cmet.2006.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.