Abstract
The checkpoint mediator protein Claspin facilitates the phosphorylation and activation of Chk1 by ATR and thus is required for efficient DNA replication. However, the physical association of Claspin homologues with replication factors and forks suggests that it might have additional functions in controlling DNA replication. DNA combing was used to examine the functions of Chk1 and Claspin at individual forks and to determine whether Claspin functions independently of Chk1. We find that Claspin, like Chk1, regulates fork stability and density in unperturbed cells. As expected, Chk1 regulates origin firing predominantly by controlling Cdk2-Cdc25 function. By contrast, Claspin functions independently of the Cdc25-Cdk2 pathway in mammalian cells. The findings support a model in which Claspin plays a role regulating replication fork stability that is independent of its function in mediating Chk1 phosphorylation.
Keywords: Chk1, Claspin, DNA replication
Introduction
A major challenge for all proliferating cells is the maintenance of genomic integrity during replication. The sheer size of the human genome requires that DNA synthesis be initiated at more than 50,000 individual sites called origins of replication. Accordingly, the accurate and efficient duplication of genetic material depends on the proper progression and coordinated activation of numerous independently functioning replication forks. When fork progression is halted it is necessary that all components of the replication apparatus remain associated with the fork if progression is to resume efficiently after removal or repair of the blockade. Thus, in response to replication stress, cells activate a checkpoint response known as the replication or intra S-phase checkpoint that stabilizes replication forks and suppresses further origin firing, facilitating the effective completion of replication. In mammalian cells the central players of this checkpoint pathway are the damage sensing kinase ATR and its down-stream effector kinase Chk1 [1]. The generation of RPA-coated regions of single-stranded DNA causes activation of the ATR-ATRIP complex [2] which in turn activates Chk1 by phosphorylation on two sites, Ser345 and Ser317 [3,4]. In addition to ATR, the activation and/or function of Chk1 requires the activity of several other sensor or mediator proteins including Claspin, TopBP1, the Rad9-Hus1-Rad1 (9-1-1) complex and the Rad17-RFC complex [5,6,7,8,9]. In addition to its function as a checkpoint mediator circumstantial evidence suggests that Claspin has a more direct role in the regulation of replication forks. For example, human Claspin is a ring-shaped molecule that binds branched DNA structures [10] and Xenopus Claspin binds replication forks and interacts with several components of the replisome [11,12]. The functional homologue of Claspin in budding yeast, Mrc1, travels with the replication fork and is required for normal rates of fork progression in an unperturbed S-phase [13,14,15]. Interestingly, these functions of Mrc1 are independent of its checkpoint functions [15]. The extent to which Claspin functions are dependent on Chk1 in mammalian cells is not clear [16,17].
Chk1 functions in the replication checkpoint to block cell cycle progression, suppress further origin firing, stabilize replication forks and facilitate the restart of collapsed forks [18]. The mechanism by which activated Chk1 prevents inappropriate cell cycle progression is well established. Phosphorylation of the protein phosphatase Cdc25A by Chk1 results in ubiquitin-mediated degradation of Cdc25A [19], thereby preventing Cdc25A from dephosphorylating and activating its cyclin-Cdk substrates [20]. In addition to the role that Chk1 plays in replication stress it is clear that it has key roles in regulating the timing and progression of replication in unperturbed cell cycles [21,22]. The mechanism by which Chk1 regulates origin firing is also thought to depend on phosphorylation of Cdc25A, inhibition of Cdk2 and the prevention of Cdc45 loading [23,24], but whether this pathway functions at the level of individual origins has not been tested. Labeling techniques that allow monitoring of replication in whole nuclei provided evidence that Chk1 regulates origin density, protects against replication-associated DNA breaks, and prevents the firing of late origins when early origins are stalled [21,24,25,26]. More recently, DNA combing technologies, a single molecule assay that allows the examination of events at individual replication forks, have provided evidence that Chk1 and Claspin regulate the rate of progression of individual forks [17,21,22]. We have employed DNA fiber labeling to assess the role of Claspin in replication fork firing and progression and to establish whether these functions are independent of Chk1 in human cells. We find that both Chk1 and Claspin are required to regulate origin firing and fork stability in HeLa cells. Furthermore, we demonstrate that although Chk1 regulates replication primarily via the Cdc25-Cdk2 pathway, Claspin regulates replication through a mechanism that is largely independent of Cdc25 and Cdk2 function.
Results
Claspin, like Chk1, regulates origin firing and fork stability in unperturbed cells
DNA fiber labeling and combing analysis has recently been use to demonstrate that Chk1 is required to maintain normal rates of DNA replication fork progression in vertebrate cells [21,22]. To determine whether Claspin plays a similar role, replication fork progression was examined in cells in which expression of either Claspin or Chk1 was reduced by siRNA (Figure 1A). Active replicons were labeled with bromodeoxyuridine (BrdU) for 10, 20 or 40 minutes and the mean lengths of labeled tracks were calculated for each time period. The mean replication rate in control siRNA-treated cells, 1.2kbp/min, is similar to previous reports for unperturbed HeLa cells [27]. Depletion of either Chk1 or Claspin severely impact replication fork progression such that the rate was < 25% of that seen in controls (Figure 1B). To determine which parameters of DNA replication are affected by Claspin or Chk1 depletion, a system in which replication fork progression is monitored by sequentially labeling tracks with two different nucleotide analogues was used [28,29]. Isolated DNA fibers were spread onto glass microscope slides and sites of replication were detected with antibodies that preferentially recognize the different halogenated nucleotides. Using this protocol three patterns of labeling can be distinguished (See Supplementary Figure 1 for examples). Tracks containing green staining next to a red region (BrdU followed by IdU) are classified as tracks of ongoing replication. Ongoing replication events include a single region of green followed by a single region of red (generated by a single fork), green flanked by red regions (generated by two diverging forks) and red flanked by green (terminal fusions). Origins that stalled during the first pulse and remain inactive in the second labeling period are green only and represent both natural terminations, due to completion of replication in that region, and forks that stalled, due to damage or fork instability. Red only tracks represent regions that initiated replication in the second labeling period only and are therefore classified as newly initiated origins. In addition, closely spaced short tracks of alternating green-red-green-red are occasionally seen in control cells (<1% of total events). This interspersed pattern represents the firing of several closely spaced origins and is thus included in the count of firing events [21]. HeLa cells were transfected with control siRNA, or siRNA against Chk1 or Claspin (Figure 1A). Forty-eight hours after transfection, cells were incubated with BrdU for 20 minutes; this was washed out and cells were then incubated with iododeoxyuridine (IdU) for 20 minutes and DNA spreads were prepared. This analysis showed significantly increased levels of origin firing and fork stalling both in Chk1- and Claspin-depleted cells (Figure 1C and 1D). The specificity of these effects on Chk1 and Claspin suppression was confirmed by repeating the analysis with independent siRNA oligonucleotides against Chk1 or Claspin (Supplementary Figure 2). These data show that both Claspin and Chk1 play critical roles in regulating origin firing and maintaining fork stability in untreated cells.
Figure 1. Chk1 and Claspin regulate origin firing and fork stability in unperturbed cells.
(A) HeLa cells were transfected with control (siCon), Chk1 (siChk1), or Claspin (siClasp) siRNA twice, 24 hours apart. Forty-eight hours following initial transfection, levels of Chk1 and Claspin protein were monitored using the relevant antibodies. Ku80 was used to verify equal loading. (B) siRNA transfected cells were labeled with 25μM BrdU for 10, 20 or 40 min before preparing DNA spreads. The mean length of at least 300 replication tracks, measured in two independent experiments, is plotted for each time point. Error bars indicate standard error of the mean. (C) Labeling protocol to determine replication origin firing: HeLa cells transfected with the indicated siRNA were pulsed with 25μM BrdU for 20 minutes followed by 250μM IdU for 20 minutes and DNA fiber spreads were prepared. Origin firing was calculated as a fraction of ongoing replication by dividing the number of red only and interspersed tracks by the total number of ongoing tracks. Graph represents fraction of origin firing measured from ten independent experiments; at least 100 replication tracks were counted in each experiment. Error bars represent standard error of the mean. (D) Fork stalling was calculated as a fraction of ongoing replication by dividing the number of green only tracks by the total of ongoing and stalled forks. Graph represents average fraction of fork stalling measured from ten independent experiments; at least 100 replication tracks were counted in each experiment. Error bars represent standard error of the mean.
Claspin and Chk1 regulate origin density
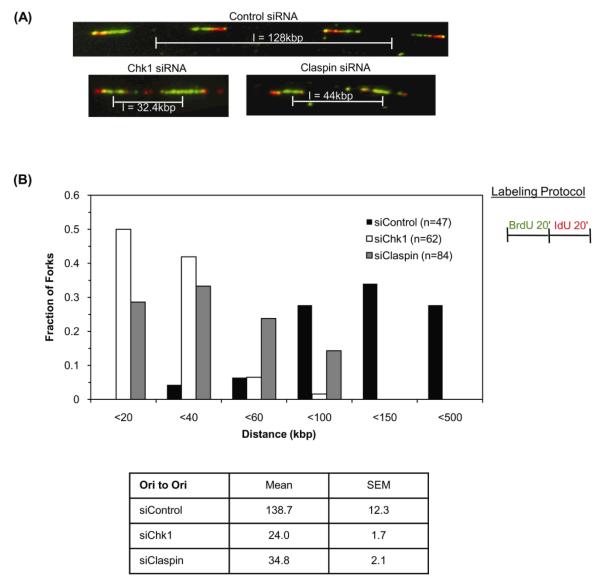
Dual labeling allows the direction that replication forks are traveling to be determined and from this the distance between adjacent replication origins can be calculated [28]. Disruption of Chk1 leads to a significant decrease in inter-origin distances in unperturbed cells [21]. Based on the similarity of Claspin and Chk1 knockdowns in fork stalling and firing assays we wished to determine if inter-origin distance is also reduced in Claspin-depleted cells. Cells expressing control, Chk1 or Claspin siRNA were labeled for 20 minutes with BrdU followed by IdU for 20 minutes and the mean inter-origin distance was measured (Figure 2A). In HeLa cells expressing control siRNA, adjacent origins were a distance of 138 ± 12kbp apart (Figure 2B). This is similar to the value of 145kbp measured by [27] for untreated HeLa cells. In contrast, the mean inter-origin distance in Chk1- and Claspin-depleted cells was 24kb and 35kb, respectively (p<0.001) (Figure 2B). The nature of the difference in inter-origin distances is more clearly seen when the distribution of distances is plotted as a histogram (Figure 2B). In control siRNA treated cells, less than 1% of origins are found within 20kb of each other, whereas in Chk1-siRNA cells 50% of origins were less than 20kb apart (Figure 2B). A similar, but less substantial shift to shorter inter-origin distances was seen in Claspin-depleted cells, in which nearly 30% of origins are less than 20kb apart. Inter-origin distances in Chk1 and Claspin depleted cells were significantly reduced. The increased incidence of origin firing in Chk1- and Claspin-depleted cells, together with the observed decrease in inter-origin distance suggests that, similar to Chk1 (Figure 1 and 2 and [21]), Claspin plays a key role in regulating origin density in an unperturbed S-phase.
Figure 2. Chk1 and Claspin regulate replication origin density.
(A) Representative images of DNA fibers prepared from Chk1, Claspin or control siRNA-transfected cells pulsed with 25μM BrdU for 20 minutes followed by 250μM IdU for 20 minutes. Pattern of red-green staining shows the direction of synthesis in adjacent replication forks and allows determination of initiation sites such that inter-origin distance (I) can be measured. (B) Graph represents distribution of inter-origin distances in cells expressing control, Chk1 or Claspin siRNA. Table summarizes mean and standard error of the mean for distances measured from five independent experiments.
Requirement for Chk1, but not Claspin, to suppress origin firing during replication stress
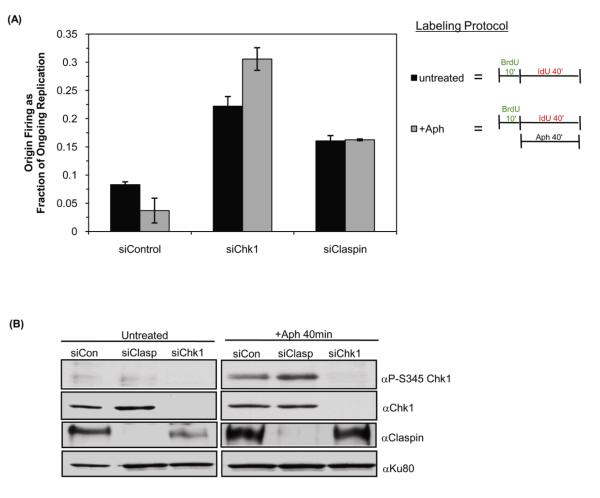
We sought to determine whether inhibition of late origin firing during replication stress was dependent on Chk1 and Claspin by examining the firing of individual origins during a replication block. Cells were transfected with siRNA and active replicons were labeled with BrdU for 10 minutes to establish the intrinsic rates of replication. To test the effect of a replication block on origin firing BrdU was removed and cells were incubated in aphidicolin and IdU for 40-minutes. As expected, aphidicolin significantly inhibited labeling of nascent tracks, it was therefore necessary to add IdU for 40 minutes to allow detection of new or continuing replication tracks (Supplementary Figure 3). Untreated samples were labeled with BrdU for 10 minutes followed by IdU labeling for 40 minutes in the absence of aphidicolin. Consistent with evidence that aphidicolin inhibits origin firing in a checkpoint-dependent manner [25,30,31,32], origin firing in control cells was reduced by the presence of aphidicolin (Figure 3A). In Chk1-siRNA treated cells, origin firing, which is already elevated, further increased in the presence of aphidicolin. Interestingly, in Claspin siRNA transfected cells the level of origin firing was elevated in the untreated sample but did not increase further upon addition of aphidicolin. Similarly, a Chk1-dependent, Claspin-independent response to aphidicolin and hydroxyurea was found using a different protocol in which cells were monitored for the ability to resume replication following exposure to replicational stress for 2 or 6 hours (Supplementary Figure 4).
Figure 3. Chk1, but not Claspin, is required to suppress origin firing during replication block.
(A) HeLa cells transfected with the indicated siRNA were labeled with 25μM BrdU for 10 minutes followed by 250μM IdU for 40 minutes in the presence or absence of aphidicolin. The level of origin firing measured in at least 100 replication tracks from three independent experiments is plotted. Error bars are standard error of the mean. (B) HeLa cells transfected with control (siCon), Chk1 (siChk1) or Claspin (siClasp) siRNA were treated with 15μM aphidicolin for 40 minutes or left untreated. The extent of Chk1 phosphorylation was monitored using anti-P-Ser345. Total levels of Chk1 and Claspin in cells were determined by probing with the relevant antibodies. Ku80 was used as a loading control.
To investigate the reason that Chk1- and Claspin-depleted cells behave differently in this assay we examined checkpoint activation in these cells. The phosphorylation of Chk1 on Ser345 in the presence and absence of aphidicolin was examined by western blotting (Figure 3B). As expected, the addition of aphidicolin to control-siRNA treated cells led to increased phosphorylation of Chk1. Similarly, when Claspin abundance was reduced by siRNA, Chk1 was efficiently phosphorylated upon addition of aphidicolin treatment (Figure 3B). The phosphorylation of Chk1 in Claspin-depleted cells suggests that at least some components of the replication stress checkpoint remain operational in these cells.
Chk1, but not Claspin, regulates origin firing by controlling Cdk activity
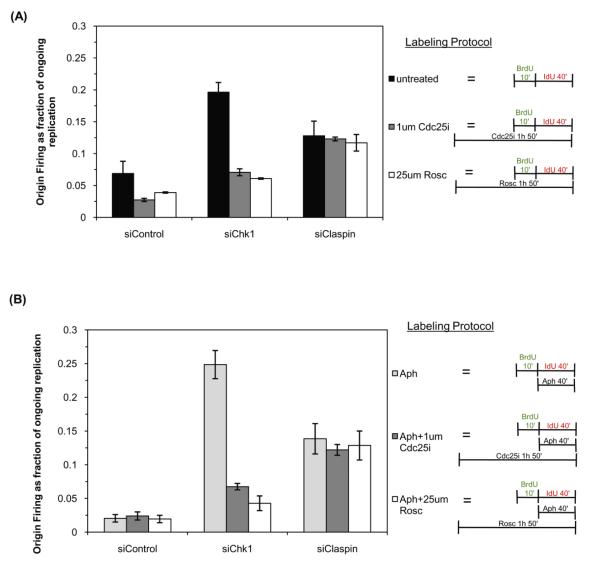
To determine whether the increased origin firing measured in Claspin and Chk1 knockdown cells is due to increased Cdk2 activity we used an inhibitor of Cdc25, (Cdc25i, NCS663284) [33] and roscovitine, a direct Cdk inhibitor [34]. Cells were transfected with siRNA against Chk1, Claspin or transfected with control siRNA. Forty-eight hours after transfection, cells were incubated with 25μM roscovitine or 1μM Cdc25i for one hour prior to and during labeling with BrdU for 10 minutes followed by IdU for 40 minutes. As shown in Figure 4A, origin firing in control siRNA cells was reduced by 40% and 55% by the presence of roscovitine and Cdc25i, respectively. Elevated levels of origin firing were seen in cells with reduced Chk1 expression and the incidence of origin firing was significantly decreased upon addition of either roscovitine, reduced by 70%, or Cdc25i, reduced by 60%. The data confirm that Cdc25-Cdk2 function is the major down-stream effector of Chk1 controlling the firing at individual replication forks. It is likely that the residual firing seen in Chk1-RNAi-roscovitine and Chk1-RNAi-Cdc25i treated cells, is due to incomplete suppression of Cdc25-Cdk2 function, although the possibility that other Chk1 targets also contribute to this signal cannot be formally excluded. Surprisingly, addition of either drug to Claspin-depleted cells did not significantly reduce origin firing (Figure 4A), suggesting that, unlike Chk1, Claspin does not function predominantly through the Cdc25-Cdk2 pathway in mammalian cells.
Figure 4. Claspin regulates origin firing by a Cdc25 and Cdk-independent mechanism.
(A) HeLa cells transfected with the indicated siRNA were left untreated (untreated) or pre-incubated with 1μM Cdc25 inhibitor (Cdc25i) or 25μM roscovitine (Rosc) for one hour. Cells were then labeled with 25μM BrdU for 10 minutes followed by 250μM IdU for 40 minutes in the continued presence or absence of Cdc25i or roscovitine. Origin firing was determined by measuring at least 100 replication tracks from three independent experiments. (B) HeLa cells transfected with the indicated siRNA were left untreated (aphidicolin samples) or pre-incubated with 1μM Cdc25 inhibitor (Cdc25i) or 25μM roscovitine (Rosc) for one hour. Cells were then labeled with 25μM BrdU for 10 minutes followed by 15μM aphidicolin for 40 minutes during the IdU label in the continued presence or absence of Cdc25i or roscovitine. DNA fiber spreads were prepared and origin firing was determined as a fraction of ongoing replication tracks from three independent experiments as above.
The next aim was to determine whether the elevated origin firing observed following addition of aphidicolin to Chk1-depleted cells is dependent on Cdk2 and Cdc25. Origin firing was monitored during an aphidicolin block in siRNA-transfected cells that had been incubated in the presence or absence of 25μM roscovitine or 1μM Cdc25i for 1 hour prior to labeling and during the labeling. As before, Chk1-depleted cells displayed a high incidence of firing upon aphidicolin-treatment (Figure 4B). Strikingly, this was significantly reduced in the presence of roscovitine or Cdc25 inhibitor, suggesting that under checkpoint-induced conditions Chk1 functions mainly to inhibit Cdc25-Cdk2 function. siRNA-mediated knockdown of Claspin also resulted in increased origin firing but this was not significantly altered by inhibition of Cdk2 or Cdc25. Together these data demonstrate that Chk1, but not Claspin, regulates origin firing by controlling Cdc25-Cdk2 function both in unperturbed S-phase and during replication blocks.
Discussion
Claspin was originally identified as an adapter protein that is essential for ATR-mediated phosphorylation of Chk1 in Xenopus extract [6]. However, several properties of the protein; its size, its ability to bind directly to branched DNA structures [10,11,12] and the fact that the Claspin homologue in yeast (Mrc1) has checkpoint independent functions [13,14,15] suggest that it might have functions that are independent of Chk1 activation. This study presents evidence that Claspin function does not depend exclusively on its ability to promote Chk1 phosphorylation in human cells.
DNA fiber analysis has been recently used to demonstrate that Chk1 and Claspin are required for normal rates of replication fork progression in human cells [17,21,22]. In agreement with these reports we found that depletion of Chk1 or Claspin by siRNA severely impacted replication fork progression. Reduced track lengths could be due to slower rates of progression along the same replication track, or they could result from increased rates of fork stalling causing an increased number of short tracks. Detailed assessment of the parameters of DNA replication by dual labeling analysis demonstrated that Chk1- and Claspin-depleted cells have an increased incidence of replication fork stalling. This increased rate of fork stalling suggests that the reduced rate of fork progression detected in single label experiments ([17,22] and Figure 1B) is likely due to increased rate of fork failure in Chk1- and Claspin-depleted cells.
The rate of replication fork progression is inversely correlated with the density of active replicons such that decreased rates of fork progression are associated with an increase in the number of active origins [28]. The adaptation to variable inter-origin distances in times of replication stress appears to be at least partly regulated by Chk1 [21,35]. However, the role of Claspin in regulating origin density was not known. Using dual labeling of Claspin-depleted cells we determined that Claspin, similar to Chk1, regulates the distance between active origins. These data are all consistent with models that place Claspin upstream in the activation of Chk1 in maintenance of replication fork function.
Mammalian cells execute a spatially and temporally ordered pattern of replication initiation that is visible as clusters of replication forks in labeled whole nuclei [36]. Drugs that directly inhibit replication also suppress the firing of late origins and this suppression of late origin firing depends on Chk1 function [25,30]. By examining the firing of single origins during an aphidicolin-induced replication block we determined that Chk1 is required to suppress the origin firing at individual forks during replication stress. Depletion of Claspin, however did not increase origin firing during replication stress and phosphorylation of Chk1 was seen. Claspin was first identified as a Chk1-binding protein that is required for ATR-mediated phosphorylation in Xenopus extracts, and initial reports suggested that it is essential for efficient Chk1 phosphorylation in human cells [5,16]. However, the presence of other mediator proteins such as TopBP1, Rad9 and Tim/Tipin that stimulate ATR-dependent phosphorylation of Chk1 suggest that phosphorylation is not uniquely dependent on Claspin [7,8,37,38]. More recent evidence confirms that Chk1 can be phosphorylated in a Claspin-independent manner [17].
The critical, proximal down-stream target of Chk1 in the regulation of DNA replication is thought to be the protein phosphatase Cdc25A that is phosphorylated and destabilized in a Chk1-dependent manner [39,40,41]. The inactivation of Cdc25A prevents the dephosphorylation and hence activation of the cyclin dependent kinases (Cdks) that in turn prevents the loading of Cdc45 which is needed to initiate replication [20]. Inhibition of Cdc25 or Cdk function in Chk1-depleted cells revealed that Chk1 regulates origin firing in a largely Cdc25-Cdk2 dependent manner. Interestingly and in striking contrast, the data show that Claspin does not function predominantly through the Cdc25-Cdk2 pathway in mammalian cells. An alternative or additional role for Claspin in replication fork regulation is consistent with evidence that it is a DNA binding protein and by its direct physical interaction with components of the replisome [5,10,42].
The mechanism by which Claspin regulates replication is not known. In addition to interacting with ATR and Chk1, human Claspin also interacts with Rad9, a component of the 9-1-1 complex which, like PCNA, is thought to encircle DNA and promote processive replication [43,44]. The physical interaction between Claspin and 9-1-1 or Claspin and PCNA may directly promote replication fork integrity. Interestingly, in budding yeast the Claspin homologue, Mrc1, maintains replication fork integrity by promoting the association of Cdc45 with replisomes in a checkpoint independent manner [13]. In addition, recent evidence suggests that Mrc1 physically interacts with the catalytic subunit of DNA polymerase epsilon and with components of the replicative helicase, the MCM complex [45]. It has therefore been proposed that in yeast, Mrc1 maintains replication fork stability by coupling polymerase and helicase activities [13,45]. Data presented here indicate that human Claspin has checkpoint-independent roles in replication and that it maintains replication fork integrity through a mechanism that is fundamentally distinct from the Chk1-Cdc25-Cdk2 pathway. The data presented here are consistent with a model in which human Claspin, like Mrc1 and Xenopus Claspin, plays a role both in the S-phase checkpoint and at the replication fork [11,12,13,45].
Materials and Methods
Cell lines and treatments
HeLa cells were grown in DMEM (Invitrogen) containing 10% bovine calf serum, penicillin and streptomycin, at 37°C and 5% CO2. Cells were treated with 2mM hydroxyurea, 15μM aphidicolin or 25 μM roscovitine (Sigma-Aldrich) where indicated. Cdc25 phosphatase inhibitor II (NSC 663284) was from Calbiochem. [33].
Oligonucleotides and transfections
siRNAs were transfected into cells using Oligofectamine (Invitrogen). HeLa cells were transfected twice, 24 hours apart and harvested 48 hours after initial transfection. The final concentration of the siRNA duplex in media was 75nM. All siRNA duplexes were from Dharmacon Research; Claspin: CCUUGCUUAGAGCUGAGUCdTdT, Chk1: GAAGCAGUCGCAGUGAAGAdTdT, Control: UUGGCCGAUAUCGCUCGACdTdT, Chk1-2: AACAGUAUUUCGGUAUAAUdTdT, Claspin-2: GCACAUACAUGAUAAAGAAdTdT.
Antibodies
Monoclonal Chk1 (G-4) antibodies were from Santa Cruz. Polyclonal phospho-Ser345 and phospho-Ser317 Chk1 antibodies were from Cell Signaling. Polyclonal antibodies to Claspin (ab3720) were from Abcam.
Western Blotting
Extracts were prepared by re-suspending an equal number of cells per sample in Laemmli sample buffer and briefly sonicating. Extracts were boiled and loaded on SDS-PAGE gels.
Replication Labeling and DNA Fiber Spreads
Replication tracks were labeled by adding 25μM BrdU (Sigma) to culture media for 10, 20 or 40 minutes. For dual labeling, cells were incubated with 25μM BrdU for 10 or 20 minutes, washed twice with PBS, twice with media and then incubated with 250μM IdU (Sigma) for 20 or 40 minutes. DNA spreads were prepared as described previously [21,44]. BrdU-containing tracks were detected with sheep anti-BrdU antibody (Biodesign; M20107S) and Cy3-conjugated donkey anti-sheep secondary (Jackson ImmunoResearch). In dual labeling experiments, BrdU labeled tracks were detected with rat anti-BrdU (Accurate; BU 1/75) and Alexa-fluor488-conjugated chicken anti-rat secondary (Invitrogen Molecular Probes), and IdU labeled track were detected using mouse anti-BrdU/IdU (Beckton Dickinson) and Cy3-conjugated goat anti-mouse (Jackson ImmunoResearch). Fibers were examined using a Nikon Eclipse E800 microscope using a 60X or 100X lens. Labeled track lengths were measured using ImageJ and converted to basepairs using the conversion 1μm = 2.59kb [27]. Measurements were made in randomly selected fields of discrete, untangled fibers.
Statistical Analysis
Statistical calculations were determined in Excel (Microsoft Office). Statistical difference was considered significant when p≤0.05 and was determined using an unpaired Student’s t-test.
Supplementary Material
Acknowledgements
The authors would like to thank D. Jackson for helpful technical advice pertaining to DNA combing assays. We thank E. Taylor for helpful suggestions and critical reading of the manuscript.
Abbreviations
- Chk1
checkpoint kinase 1
- Cdk2
cyclin-dependent kinase 2
- ATM
ataxia telangiectasia mutated related
- RPA
replication protein A
- BrdU
bromodeoxyuridine
- IdU
iododeoxyuridine
- siRNA
short-interfering RNA)
References
- 1.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 3.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chini CC, Chen J. Human claspin is required for replication checkpoint control. J Biol Chem. 2003;278:30057–30062. doi: 10.1074/jbc.M301136200. [DOI] [PubMed] [Google Scholar]
- 6.Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- 7.Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, et al. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol. 2006;26:6056–6064. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roos-Mattjus P, Hopkins KM, Oestreich AJ, Vroman BT, Johnson KL, et al. Phosphorylation of human Rad9 is required for genotoxin-activated checkpoint signaling. J Biol Chem. 2003;278:24428–24437. doi: 10.1074/jbc.M301544200. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Zou L, Lu T, Bao S, Hurov KE, et al. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol Cell. 2006;23:331–341. doi: 10.1016/j.molcel.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 10.Sar F, Lindsey-Boltz LA, Subramanian D, Croteau DL, Hutsell SQ, et al. Human claspin is a ring-shaped DNA-binding protein with high affinity to branched DNA structures. J Biol Chem. 2004;279:39289–39295. doi: 10.1074/jbc.M405793200. [DOI] [PubMed] [Google Scholar]
- 11.Lee J, Gold DA, Shevchenko A, Dunphy WG. Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response. Mol Biol Cell. 2005;16:5269–5282. doi: 10.1091/mbc.E05-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Kumagai A, Dunphy WG. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol Cell. 2003;11:329–340. doi: 10.1016/s1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 14.Szyjka SJ, Viggiani CJ, Aparicio OM. Mrc1 is required for normal progression of replication forks throughout chromatin in S. cerevisiae. Mol Cell. 2005;19:691–697. doi: 10.1016/j.molcel.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 15.Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 16.Lin SY, Li K, Stewart GS, Elledge SJ. Human Claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc Natl Acad Sci U S A. 2004;101:6484–6489. doi: 10.1073/pnas.0401847101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petermann E, Helleday T, Caldecott KW. Claspin Promotes Normal Replication Fork Rates in Human Cells. Mol Biol Cell. 2008 doi: 10.1091/mbc.E07-10-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 19.Mailand N, Falck J, Lukas C, Syljuasen RG, Welcker M, et al. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 20.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 21.Maya-Mendoza A, Petermann E, Gillespie DA, Caldecott KW, Jackson DA. Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO J. 2007;26:2719–2731. doi: 10.1038/sj.emboj.7601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petermann E, Maya-Mendoza A, Zachos G, Gillespie DA, Jackson DA, et al. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol. 2006;26:3319–3326. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorensen CS, Syljuasen RG, Lukas J, Bartek J. ATR, Claspin and the Rad9-Rad1-Hus1 complex regulate Chk1 and Cdc25A in the absence of DNA damage. Cell Cycle. 2004;3:941–945. [PubMed] [Google Scholar]
- 24.Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feijoo C, Hall-Jackson C, Wu R, Jenkins D, Leitch J, et al. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154:913–923. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heffernan TP, Simpson DA, Frank AR, Heinloth AN, Paules RS, et al. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol Cell Biol. 2002;22:8552–8561. doi: 10.1128/MCB.22.24.8552-8561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conti C, Sacca B, Herrick J, Lalou C, Pommier Y, et al. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol Biol Cell. 2007;18:3059–3067. doi: 10.1091/mbc.E06-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merrick CJ, Jackson D, Diffley JF. Visualization of altered replication dynamics after DNA damage in human cells. J Biol Chem. 2004;279:20067–20075. doi: 10.1074/jbc.M400022200. [DOI] [PubMed] [Google Scholar]
- 30.Dimitrova DS, Gilbert DM. Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat Cell Biol. 2000;2:686–694. doi: 10.1038/35036309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luciani MG, Oehlmann M, Blow JJ. Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J Cell Sci. 2004;117:6019–6030. doi: 10.1242/jcs.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zachos G, Rainey MD, Gillespie DA. Chk1-dependent S-M checkpoint delay in vertebrate cells is linked to maintenance of viable replication structures. Mol Cell Biol. 2005;25:563–574. doi: 10.1128/MCB.25.2.563-574.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazo JS, Aslan DC, Southwick EC, Cooley KA, Ducruet AP, et al. Discovery and biological evaluation of a new family of potent inhibitors of the dual specificity protein phosphatase Cdc25. J Med Chem. 2001;44:4042–4049. doi: 10.1021/jm0102046. [DOI] [PubMed] [Google Scholar]
- 34.Meijer L, Borgne A, Mulner O, Chong JP, Blow JJ, et al. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur J Biochem. 1997;243:527–536. doi: 10.1111/j.1432-1033.1997.t01-2-00527.x. [DOI] [PubMed] [Google Scholar]
- 35.Conti C, Seiler JA, Pommier Y. The mammalian DNA replication elongation checkpoint: implication of Chk1 and relationship with origin firing as determined by single DNA molecule and single cell analyses. Cell Cycle. 2007;6:2760–2767. doi: 10.4161/cc.6.22.4932. [DOI] [PubMed] [Google Scholar]
- 36.Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- 37.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25:3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshizawa-Sugata N, Masai H. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J Biol Chem. 2007;282:2729–2740. doi: 10.1074/jbc.M605596200. [DOI] [PubMed] [Google Scholar]
- 39.Mailand N, Bekker-Jensen S, Bartek J, Lukas J. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol Cell. 2006;23:307–318. doi: 10.1016/j.molcel.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, et al. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 41.Sorensen CS, Syljuasen RG, Falck J, Schroeder T, Ronnstrand L, et al. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003;3:247–258. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 42.Kim JM, Kakusho N, Yamada M, Kanoh Y, Takemoto N, et al. Cdc7 kinase mediates Claspin phosphorylation in DNA replication checkpoint. Oncogene. 2007 doi: 10.1038/sj.onc.1210994. [DOI] [PubMed] [Google Scholar]
- 43.Brondello JM, Ducommun B, Fernandez A, Lamb NJ. Linking PCNA-dependent replication and ATR by human Claspin. Biochem Biophys Res Commun. 2007;354:1028–1033. doi: 10.1016/j.bbrc.2007.01.091. [DOI] [PubMed] [Google Scholar]
- 44.Scorah J, Dong MQ, Yates JR, 3rd, Scott M, Gillespie D, et al. A conserved PCNA-interacting sequence in Chk1 is required for checkpoint function. J Biol Chem. 2008 doi: 10.1074/jbc.M800369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lou H, Komata M, Katou Y, Guan Z, Reis CC, Budd M, Shirahige K, Campbell JL. Mrc1 and DNA Polymerase e function together in linking DNA replication and the S phase Checkpoint. Cell. 2008;32:106–117. doi: 10.1016/j.molcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.