Abstract
The mitotic spindle is constructed from microtubules (MTs) nucleated from centrosomes, chromosome proximal regions, and preexisting spindle MTs. Augmin, a recently identified protein complex, is a critical factor in spindle MT-based MT generation in Drosophila S2 cells. Previously, we identified one subunit of human augmin. Here, by using mass spectrometry, we identified the full human augmin complex of 8 subunits and show that it interacts with the γ-tubulin ring complex (γ-TuRC). Unlike augmin-depleted S2 cells, in which the defect in spindle-mediated MT generation is mostly compensated by centrosomal MTs, augmin knockdown alone in HeLa cells triggers the spindle checkpoint, reduces tension on sister kinetochores, and severely impairs metaphase progression. Human augmin knockdown also reduces the number of central spindle MTs during anaphase and causes late-stage cytokinesis failure. A link between augmin and γ-TuRC is likely critical for these functions, because a γ-TuRC mutant that attenuates interaction with augmin does not restore function in vivo. These results demonstrate that MT generation mediated by augmin and γ-TuRC is critical for chromosome segregation and cytokinesis in human cells.
Keywords: centrosome, mitosis, RNAi, spindle checkpoint
Proper segregation of sister chromatids during cell division relies on the assembly of a bipolar spindle during mitosis. Sister kinetochores associate with microtubules (MTs) from opposite poles in metaphase. When all of the kinetochores are attached to MTs and under tension, the spindle checkpoint is satisfied and the anaphase segregation of sister chromatids takes place (1, 2). Beginning at anaphase, spindle MTs reorganize to form a bundled and antiparallel MT structure between the segregating chromatids, a structure referred to as the central spindle. Proper formation of the central spindle is required for contraction of the contractile ring and completion of cytokinesis (3).
On entry into mitosis, spindle MTs are generated at multiple locations in animal somatic cells. The dominant MT nucleation site is the centrosome, which binds the γ-tubulin ring complex (γ-TuRC), the major MT nucleator in cells. However, mitotic cells also have at least two centrosome-independent pathways for MT generation. The best studied is the chromatin-dependent pathway, in which Ran-GTP and the chromosome passenger complex enriched around chromosomes induce MT nucleation (4–6). In addition, an MT-based MT generation pathway, which was originally found in the cytoplasm of fission yeast (7) and plant cells (8), was recently uncovered within the mitotic spindle, in which MTs are generated throughout the spindle and not necessarily near the chromosomes (9–11). It has been suggested that γ-tubulin localized onto spindle MTs may play a significant role in MT generation within the spindle (9–12).
The protein complex augmin was recently identified in Drosophila S2 cells as a critical factor for centrosome-independent, spindle-based MT generation (10, 11). Originally, 5 augmin subunits (Dgt2–6) (Dgt stands for dim γ-tubulin) were identified in a genome-wide RNAi screen as genes required for γ-tubulin localization on the spindle but not at the centrosome (11). RNAi knockdown of Drosophila augmin led to a reduction in MT number within the spindle and consequently caused mild defects in spindle bipolarity and chromosome alignment. However, these defects became much more severe when centrosomes were simultaneously disrupted (note that functional spindles can be assembled without centrosomes in Drosophila) (10). Thus, the augmin-dependent MT generation pathway is used as a redundant mechanism with the centrosome-dependent pathway in Drosophila S2 cells. Previous work identified human Dgt6 based on sequence similarity and found that RNAi knockdown led to reduced MT and γ-tubulin intensities within the spindle, as in Drosophila (10). However, other components of augmin could not be found by sequence homology searches, and the mitotic defects of hDgt6 RNAi were not characterized in great detail.
In this study, we identify Drosophila and human augmin as 8-subunit complexes. We also show that augmin interacts with γ-TuRC and that attenuation of this interaction severely impairs spindle MT generation. Furthermore, we provide evidence that human augmin plays critical and nonredundant roles in the kinetochore–MT attachment and also central spindle formation during anaphase in human cells.
Results
The Augmin Complex Consists of 8 Subunits in Drosophila and Humans.
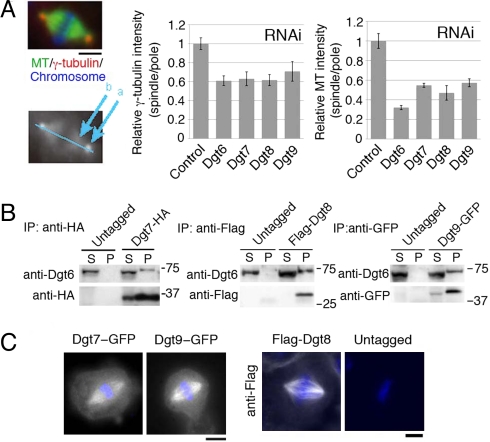
To search for additional components of Drosophila augmin, Dgt2-HA (HA indicating the hemagglutinin epitope) was immunoprecipitated from S2 cells, and the precipitated proteins were identified by liquid chromatography/tandem mass spectrometry (LC/MS/MS). Dgt2–6 appeared as top hits, confirming the fidelity of this assay. Among the other top hits were 7 proteins for which >5 peptides were uniquely detected in the Dgt2-HA sample (Table S1). We reevaluated their RNAi phenotypes in our whole-genome RNAi screen database (11) and repeated RNAi with newly designed dsRNA constructs (Table S2). The typical “dgt phenotype” (dim γ-tubulin and MT signals inside the spindle) were found after RNAi against CG2213, CG13879, and CG13914 (Fig. 1A). These phenotypes were present, but overlooked, in the previous high-throughput screen (11). The 3 proteins efficiently precipitated endogenous Dgt6 when immunoprecipitated (Fig. 1B) and were localized to spindle MTs (Fig. 1C), indicating that they are previously unrecognized subunits of augmin. We therefore designate them as Dgt7, Dgt8, and Dgt9. Drosophila augmin is predicted to be a 340-kDa protein complex on the basis of size-based fractionation (10). The individual molecular masses of proteins Dgt2–9 sum up to 327 kDa, which is very close to this value, suggesting that augmin consists of these 8 subunits. However, we cannot entirely exclude the possibility that smaller subunits are still missing.
Fig. 1.
Identification of Drosophila Dgt7, Dgt8, and Dgt9 as augmin subunits. (A) Dim γ-tubulin and MT intensities within the spindle after RNAi knockdown of Dgt7/CG2213, Dgt8/CG13879, and Dgt9/CG13914. The density of γ-tubulin or MTs inside the spindle decreased after RNAi of Dgt6–9 (n ≥ 5 each). The ratio of signal intensity in spindle (location at “b” in the lower image) to that in centrosome (“a”), which was normalized to control, is shown (±SEM). The normalized ratio for γ-tubulin and MT after Dgt7–9 knockdowns were significantly (P < 0.004) lower than in control. (B) Dgt7-HA, Flag-Dgt8, and Dgt9-GFP proteins coimmunoprecipitate with endogenous Dgt6. The supernatant (S) and precipitated (P: 40× volume of S) fractions after immunoprecipitation with anti-HA, anti-Flag, and anti-GFP antibodies were immunoblotted. (C) Dgt7-GFP, Dgt9-GFP, and Flag-Dgt8 were localized to spindle MTs during metaphase. Flag-Dgt8 was stained by anti-Flag antibody. Blue, DAPI. (Scale bars, 5 μm.)
To identify human augmin, a HeLa cell line stably expressing GFP-hDgt6 was generated (Fig. 2A), and the mitotic extracts were immunoprecipitated with anti-GFP antibody, followed by LC/MS/MS analysis. Ten proteins, including hDgt6 itself, were identified as strong hits (Table S3). Among them, 2 genes (RBM7 and CSDA) had clear orthologues in Drosophila, whose molecular functions and RNAi phenotypes (11) are not related apparently to augmin. We therefore considered them as unlikely candidates for augmin subunits and excluded them from further analyses. The remaining 7 genes (C4orf15, Hice1/NY-SAR48, KIAA0841, Ccdc5/Hei-C, Cep27, UCHL5IP, and C14orf94) were cloned, tagged with GFP and HA, and expressed in HeLa cells. All 7 proteins immunoprecipitated endogenous hDgt6, confirming the interactions detected in LC/MS/MS (Fig. 2 B and D).
Fig. 2.
Identification of human augmin as the 8-subunit complex. (A) Immunoblotting of hDgt6 for the control, siRNA-treated, and GFP-hDgt6-expressing cells. GFP-hDgt6 was expressed 2-fold more than endogenous hDgt6. (B) Coimmunoprecipitation of 7 HA-tagged hDgt6-associated proteins with endogenous hDgt6. The supernatant (S) and precipitated (P: 50× volume of S) fractions after anti-HA incubation were immunoblotted with ati-hDgt6 antibody. The asterisks in A and B indicate cross-reaction of anti-hDgt6 antibody to other proteins. (C) Localization of GFP-tagged C14orf94 and C4orf15 proteins (green) in metaphase (Left) and interphase (Right). The proteins were abundant at centrosomes during interphase, whereas spindle localization with slight enrichment near the poles was detected during metaphase. Chromosomes (blue) and γ-tubulin (red) were counterstained. (Scale bar, 5 μm.) (D) Two-hybrid interaction between full-length Hice1 and hDgt6. Colonies were visible on the 3-aminotriazole (3-AT)-containing plate only when both proteins were expressed. (E) Gel filtration analysis of human augmin subunits. A similar peak can be observed between hDgt6 and the hDgt6-interacting proteins at a Stokes radius of ≈9.5 nm, suggesting that they form a stable complex. The peak of KIAA0841 was shifted by one fraction to the left, probably because of GFP tagging of this protein (others were tagged with HA). The reason for the shift of Hice1 to higher molecular mass fractions is unclear but might be aggregation of a subpopulation of this protein in the extracts.
Observation of the GFP fusion proteins revealed that the 7 proteins, like GFP-hDgt6, localized to the metaphase spindle under a wide range of expression levels (Fig. 2C and Fig. S1). Signals were detected throughout spindle MTs, but in many cases a slight accumulation near the spindle pole regions was detected. These proteins accumulated at centrosomes during interphase. Gel filtration analysis indicated that they all migrated with a similar peak at a Stokes radius of ≈9.5 nm (Fig. 2E). On the basis of this value and the reported sedimentation coefficient of the Ccdc5-containing complex [≈10 S (13)], we estimated that the size of human augmin is 390 kDa. We suggest that these 8 subunits, which sum up to 442 kDa, compose human augmin.
Among the 8 genes, 3 (hDgt6, Ccdc5, and Hice1) were previously shown to localize to the mitotic spindle (10, 13, 14), 1 (Cep27) was reported as a centrosomal protein (15), and functions of the other 4 (UCHL5IP, C4orf15, C14orf94, and KIAA0841) were completely unknown. Our homology search suggested that C4orf15, Hice1, and KIAA0841 are the orthologues of Drosophila Dgt3, 4, and 5, respectively (Fig. S2).
Depletion of Human Augmin Arrests Cells in Metaphase and Reduces Tension on Bioriented Kinetochores.
We have previously shown that RNAi knockdown of hDgt6 (also called FAM29A) in HeLa cells resulted in the appearance of bipolar spindles with reduced γ-tubulin and MTs within the spindle and a modest (2- to 3-fold) increase in the mitotic index (10). Very recently, another study also reported the dgt phenotype and 2- to 4-fold increase of mitotic index after hDgt6 RNAi (16). These data suggested that MT attachment to kinetochores is modestly delayed without augmin-mediated MT generation and that the human augmin pathway acts redundantly with the centrosomal pathway for chromosome alignment, as is the case in Drosophila S2 cells (10, 16). However, the mitotic defects have not been characterized in great detail for each augmin subunit, and we also sought to obtain a more complete augmin knockdown by improving the RNAi efficiency in the current study [>95% depletion of the hDgt6 protein (Fig. S3A)] (see Methods).
Under the optimized RNAi condition, γ-tubulin and MT signals were greatly reduced inside the bipolar spindle for each hDgt gene RNAi (Fig. 3 A and B). In a severe case (hDgt6), the γ-tubulin signal intensity within the spindle was reduced to only 39% of control cells [mean of 2 independent experiments (n = 14 and 11)]. As is the case of Drosophila S2 cells (10), the observed reduction in γ-tubulin signals after augmin depletion was not simply the consequence of the spindle MT reduction, because a similar level of MT reduction by HURP RNAi [which reduces kinetochore MT numbers (17, 18)] or nocodazole treatment (which preferentially depolymerizes nonkinetochore MTs) did not dramatically affect γ-tubulin intensity compared with augmin RNAi (compare Fig. S4 and Fig. 3A). We conclude that human augmin is directly required for γ-tubulin localization to MTs and for spindle MT generation.
Fig. 3.
Dgt phenotype after RNAi knockdown of human augmin subunits. (A) Reduced spindle localization of γ-tubulin or MT after knockdowns of human augmin subunits. A hDgt3-knockdown cell is shown as a representative. (B) Quantification of spindle localization of γ-tubulin or MT. The ratio of spindle to centrosome signal intensity after knockdowns of human augmin subunits, which was normalized to control (±SEM), was significantly (P < 0.01) lower than that in the control (luciferase RNAi) (n ≥ 9). (C) Astral MTs (arrowheads) emanated from the centrosome (green) in metaphase-arrested cells depleted of hDgt6. GFP-tubulin was imaged in a living cell, and the region around one spindle pole is presented. (Scale bars, 5 μm.)
On hDgt6 RNAi, centrosomal γ-tubulin signals also were attenuated modestly (64% of the control cells), but astral MTs were clearly generated from centrosomes under this situation as revealed by GFP-tubulin in living cells (Fig. 3C). Thus, centrosomes preserve MT nucleating activity independent of augmin function, consistent with the result of an in vitro aster formation assay after Ccdc5 depletion (13). We also detected multipolar spindles in each RNAi sample, as was reported previously for Ccdc5 and Hice1 (13, 14). However, the frequency was only 10–35% of the mitotic cells, whereas almost all of the spindles, regardless of their polarity, exhibited the dim-γ-tubulin phenotype. Therefore, it is more likely that mislocalization of γ-tubulin is the primary defect of augmin knockdown.
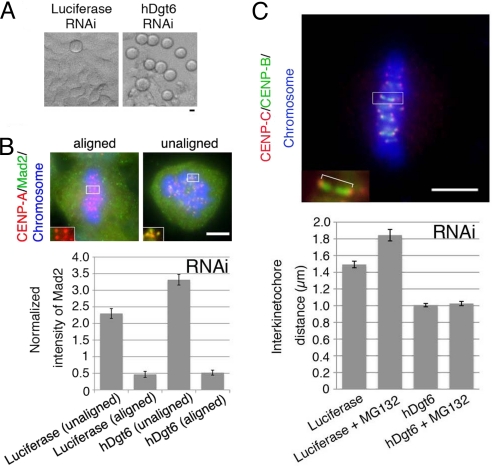
Surprisingly, the mitotic index was elevated much more dramatically for each hDgt siRNA treatment compared with previous reports for humans (10, 16) and Drosophila cells (10) (Fig. 4A). For RNAi of one subunit (hDgt6), the mitotic index even increased ≈9-fold [52% (n = 877) compared with 6.0% in control (n = 366)] (Fig. S3B). Unlike control cells, the majority of the mitotic cells were in prometaphase (53% versus 31% in the control) or metaphase (45% versus 26% in the control), and anaphase cells were scarce (0.7% versus 41% in the control; n > 241), suggesting that the spindle checkpoint is activated. Consistent with this interpretation, the mitotic index was restored when the spindle checkpoint component Mad2 or BubR1 was simultaneously deleted (Fig. S3B).
Fig. 4.
Reduced metaphase kinetochore tension in the absence of human augmin. (A) Mitotic cells (rounded cells in the phase-contrast microscopy image) were accumulated after hDgt6 RNAi. (B) The average intensity of the kinetochore-localized Mad2 signals normalized to that of CENP-A (n ≥ 41 kinetochores from ≥3 cells each). The Mad2 signals diminished on the aligned kinetochores in both the control and hDgt6-depleted cells. The brightest kinetochore Mad2 signal in each cell was also comparable between RNAi and control samples (Fig. S3C). (C) Reduced tension on sister kinetochores after hDgt6 RNAi. (Lower) The interkinetochore distance of the congressed chromosomes (blue) was quantified based on immunostaining of CENP-B (green) and CENP-C (red) (≥20 pairs of kinetochores from ≥6 cells each). (Upper) An enlarged image of a pair of sister kinetochores. Treatment with the proteasome inhibitor MG132 (10 μM, 90 min), which prolongs metaphase, did not alter the interkinetochore distance of hDgt6-depleted cells. (Scale bars, 5 μm.)
Mad2 accumulates on unattached, misaligned kinetochores, and MT attachment to the kinetochores triggers the loss of Mad2 from the kinetochores, which results in the progression into anaphase (19–21). In the absence of augmin, Mad2 was enriched on misaligned kinetochores (Fig. 4B). However, there was also an increase in the proportion of cells that had all of their chromosomes aligned at the spindle equator [23% of total cell (control; 1.6%)], indicating that augmin depletion also arrested cells even after their chromosomes had congressed in the metaphase plate. We found that Mad2 does not accumulate on those aligned kinetochores, similar to control cells (Fig. 4B). This result differs from the data reported by Zhu et al. (16), who showed elevated Mad2 signals on aligned kinetochores after hDgt6 RNAi (see Discussion). However, consistent with Zhu et al. (16), the sister kinetochore distance of aligned chromatids, which correlates with tension across kinetochores, was significantly shorter in the absence of hDgt6 than in control metaphase cells, even after further prolongation of metaphase by treatment with the proteasome inhibitor MG132 (Fig. 4C). These results suggest that, in the absence of spindle MTs generated by augmin, the spindle checkpoint is not satisfied, perhaps because of the lack of sufficient tension on kinetochores, even after all of the kinetochores attach to MTs and release Mad2.
Human Augmin Is Important for Central Spindle Formation in Anaphase and Cytokinesis.
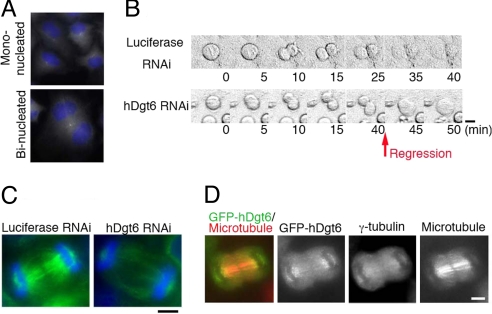
In addition to the dramatic accumulation of mitotic cells, we also noticed that the percentage of binucleate interphase cells increased after RNAi knockdowns of multiple augmin subunits [e.g., hDgt6, 24%; Ccdc5, 8%; control, 2% (n ≥ 394)], which is indicative of cytokinesis failure (Fig. 5A). This defect was rescued by ectopic expression of the augmin subunit insensitive to siRNA, excluding the possible off-target effect of the siRNA constructs (Fig. 6E). Live-cell imaging of hDgt6 RNAi cells revealed that the cleavage furrow forms and invaginates but later regresses in 14 of 18 cells (Fig. 5B). This finding suggests that augmin is required for the completion of cytokinesis. We further found that the MT density in the central spindle between the segregating chromosomes was decreased in hDgt6-depleted cells (Fig. 5C) (lagging chromosomes were scarcely observed). Consistent with this functional role in cytokinesis, GFP-tagged augmin subunits and γ-tubulin were localized to the central spindle, in particular in the region near the segregating chromosomes compared with the midzone (GFP-hDgt6 is displayed in Fig. 5D).
Fig. 5.
Cytokinesis failure in the absence of human augmin. (A) Binucleated cells were increased after hDgt6 RNAi. (B) Cleavage furrow regression was observed after hDgt6 RNAi (40–45 min) (phase-contrast image). Images were acquired every 5 min. (C) Lowered central spindle MT density (green) after hDgt6 RNAi in late anaphase. Blue, DNA. (D) Localization of GFP-hDgt6 and γ-tubulin on the central spindle MTs during late anaphase. (Scale bars, 5 μm.)
Fig. 6.
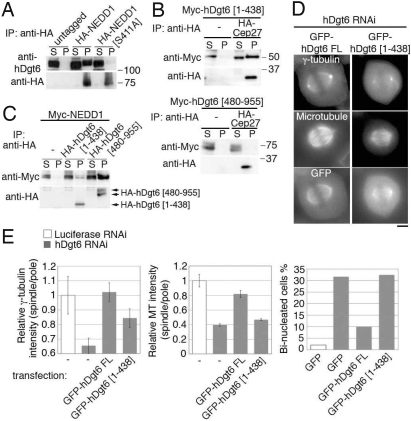
Augmin–γ-TuRC interaction is necessary for spindle MT generation. (A) hDgt6 was efficiently coimmunoprecipitated with NEDD1 but not with a point-mutant NEDD1 (S411A). HA-tagged NEDD1 or NEDD1 (S411A) expressed in HeLa mitotic cells was immunoprecipitated. (B) NEDD1 was efficiently coimmunoprecipitated with N-terminal truncated hDgt6(480–955) but not with C-terminal truncated hDgt6(1–438). Mitotic HeLa cells expressing Myc-NEDD1 with or without HA-hDgt6(1–438) or HA-hDgt6(480–955) mutant were used. (C) hDgt6(1–438), but not hDgt6(480–955), associates with augmin subunit Cep27. Myc-hDgt6(1–438) or Myc-hDgt6(480–955) with or without HA-Cep27 was immunoprecipitated. A 20-fold higher amount of the sample was loaded in the pellet lane in A–C. (D) The dgt phenotype was rescued by full-length GFP-hDgt6 but not by GFP-hDgt6(1–438). (Scale bar, 5 μm.) (E) Quantification of γ-tubulin (Left) or MT (Center) intensity within the spindle. The normalized ratio of spindle to centrosome signal intensity (±SEM) after knockdowns of hDgt6 was significantly (P < 0.0007) restored when siRNA-insensitive, full-length GFP-hDgt6 was expressed, but less efficiently by GFP-hDgt6(1–438) (n ≥ 7). (Right) Percentage of binucleated cells after depletion of endogenous hDgt6 was lowered by transfection of siRNA-insensitive full-length GFP-hDgt6, but not by a truncated GFP-hDgt6(1–438) or GFP alone (n ≥ 77).
The Interaction Between Augmin and γ-TuRC Is Necessary for Spindle-Based MT Generation.
γ-TuRC was recently shown to be coimmunoprecipitated with hDgt6 in HeLa cells (16). We further found coprecipitation of HA-tagged NEDD1 [also called GCP-WD, a component of γ-TuRC (12, 22)] with endogenous hDgt6 (Fig. 6A). A similar interaction was also found between their Drosophila orthologues (Fig. S5).
To investigate whether the augmin–γ-TuRC interaction is important for spindle MT generation, we analyzed the NEDD1 (S411A) mutant, which was previously shown to interfere with γ-TuRC localization to spindle MTs but not to centrosomes (12). NEDD1 (S411A) was much less efficiently coimmunoprecipitated with endogenous hDgt6 when expressed in mitosis (Fig. 6A), suggesting that physical interaction with augmin is important for γ-TuRC's localization to spindle MTs. We next sought to identify the γ-TuRC-interacting domain of augmin by generating several truncated forms of augmin subunits. Of those, a C-terminal hDgt6 fragment [hDgt6(480–955)] was efficiently coimmunoprecipitated with NEDD1 but failed to associate with another augmin subunit or spindle MTs (Fig. 6 B and C). In contrast, GFP-tagged hDgt6(1–438) retained an ability to associate with other augmin subunits and thereby localized to centrosomes in interphase and spindle MTs in mitosis, but was much less efficiently coprecipitated with wild-type NEDD1 (Fig. 6 B–D). This truncated gene could not fully rescue the dgt phenotype or the cytokinesis failure (Fig. 6E) [partial rescue might be due to weak interaction of hDgt6(1–438) with NEDD1 (Fig. 6B)]. These results suggest that the interaction between augmin and γ-TuRC is critical for the recruitment of γ-TuRC to spindle MTs and for augmin/γ-TuRC-mediated spindle MT generation.
Discussion
In this study, we identified the 8 subunits belonging to the human and Drosophila augmin complexes through mass spectrometry. Analogous to Drosophila augmin, human augmin bound to γ-TuRC and localized it to spindle MTs to generate noncentrosomal MTs within the mitotic spindle. However, unexpected from the previous fly augmin or human Dgt6 analysis (10, 16), depletion of human augmin caused severe checkpoint-dependent prometaphase/metaphase delay, likely because of a defect in kinetochore–MT interaction. We also uncovered cytokinesis failure with reduced MTs in the central spindle. Thus, the augmin-dependent MT generation pathway plays an indispensable role in cell division in human tissue culture cells. Augmin may play similarly critical roles in other cell types in humans and therefore might constitute a possible target for small-molecule inhibition in cancer chemotherapy.
Human Augmin Is Needed for Establishing Kinetochore Tension.
A significant population of the human augmin-depleted cells arrested in metaphase with a reduced level of Mad2 signals on aligned kinetochores. Thus, sister kinetochores appear to be attached to MTs (perhaps centrosomal MTs) without augmin in these cells. In contrast, Zhu et al. (16) recently showed that Mad2 intensity on metaphase chromosomes is 2.5-fold higher in hDgt6-knockdown cells than control, which suggests that MT attachment does not take place for some aligned chromosomes. The reason for this data discrepancy is uncertain. One possibility is that more efficient knockdown of hDgt6 (>95%) in the present work induced more severe metaphase delay, so that dynamic centrosomal MTs might establish attachment to all of the kinetochores during this prolonged period.
Our study (10) and that of Zhu et al. (16) both showed that augmin RNAi leads to a reduction in tension across sister kinetochores of aligned chromosomes. The reason for the reduced tension in augmin-depleted cells may be that chromosomes are never stably attached to MTs, possibly because of fewer kinetochore MTs. Alternatively, augmin may impact chromosome tension by producing cross-linked, short antiparallel MTs that act as substrates for motor-dependent sliding forces, as suggested for meiotic metaphase spindles reconstituted in Xenopus extracts (23). A reduction in sister kinetochore tension might be responsible for checkpoint activation after augmin depletion. However, it was recently shown that checkpoint can be satisfied in the absence of interkinetochore tension in some experimental conditions (24). Thus, defining the mechanism of checkpoint arrest after augmin depletion remains an important topic for future investigation.
The mitotic progression defect observed in human cells is considerably different from that in Drosophila cells, where severe chromosome alignment defect and mitotic delay were observed only when augmin and centrosomes were simultaneously depleted (10, 25). Thus, the augmin-dependent MT amplification pathway works as a redundant mechanism with the centrosomal pathway in Drosophila. However, our results indicate that the augmin-dependent pathway plays critical and nonredundant roles in chromosome segregation in human cells. A possible explanation for the discrepancy between fly and human cells is that human cells may need more MTs for building a functional spindle, in particular for stable kinetochore–MT attachment; the number of human kinetochore MTs (≈17) as well as chromosomes (46) is larger than that of Drosophila (≈11 and 8, respectively) (26).
Human Augmin Is Needed for Completing Cytokinesis.
We observed a reduction in central spindle MTs during anaphase and a subsequent cytokinesis failure in the absence of human augmin. A prevailing model postulates that bundling of interpolar MTs leads to central spindle formation during anaphase. According to this model, reduced MT number in the metaphase spindle might consequently lead to the defect in robust central spindle formation in the augmin-depleted cells. However, it has also been reported that MTs required for cytokinesis can be formed in the absence of preanaphase MTs under certain experimental conditions (27). Our localization data in anaphase imply that augmin-dependent de novo generation of MTs at the central spindle might also play an important role in mature central spindle formation.
Mechanism of Augmin- and γ-TuRC-Mediated Spindle MT Amplification.
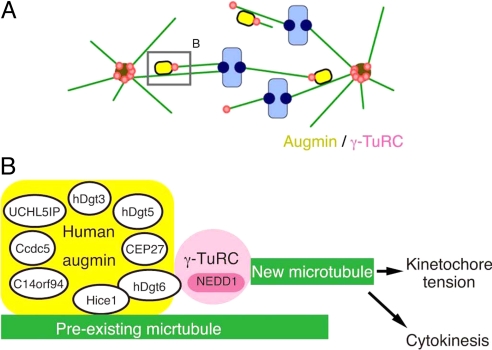
Hice1 protein was shown to bind directly to MTs in vitro via its N-terminal region (14). It is therefore likely that augmin directly binds to spindle MTs. Interestingly, hDgt6 directly interacts with the Hice1 (Fig. 2D), and the C terminus of hDgt6 coprecipitates with NEDD1-γ-TuRC. An intriguing possibility therefore is that γ-tubulin recruitment to spindle MTs is achieved through direct protein–protein interaction of a preexisting MT to Hice1 and then hDgt6 binding to γ-TuRC (Fig. 7).
Fig. 7.
Model for the augmin- and γ-TuRC-dependent MT amplification in the mitotic spindle in human cells. (A) The augmin/γ-TuRC machinery generates MTs within the spindle. The new MTs contribute to kinetochore MT formation and net kinetochore capture by the search-and-capture mechanism. This model was originally proposed in ref. 10. A similar mechanism may work within the central spindle during anaphase. (B) A speculative molecular model of the augmin/γ-TuRC-dependent MT generation. Human and Drosophila augmin were identified as 8-subunit complexes (human augmin subunits are described in this diagram). Hice1 is known to bind to MTs directly in vitro (14). Hice1-hDgt6 interaction was detected in the yeast 2-hybrid assay, whereas NEDD1-containing γ-TuRC was efficiently coprecipitated with the C-terminal fragment of hDgt6. Attenuation of this interaction caused defects in spindle MT generation. Thus, we suggest that augmin recruits γ-TuRC to spindle MTs, where γ-TuRC nucleates new MTs. This MT amplification process is critical for generating kinetochore tension and also completion of cytokinesis in human tissue culture cells.
Extensive phenotypic analyses (10, 16) alone could not elucidate the exact role of augmin and γ-TuRC in this process (discussed in ref. 10). For example, although it was tempting to speculate that γ-TuRC nucleates MTs by recruiting of augmin to preexisting spindle MTs, the observed phenotypes could also be explained by a model in which augmin nucleates MTs independently of γ-TuRC function and γ-TuRC stabilizes new MT minus-ends by capping. However, our results showing critical interaction of augmin and γ-TuRC for spindle MT generation make the former model more likely (Fig. 7). However, in this scenario, γ-TuRC must be specifically activated on recruitment to spindle MTs (note that γ-TuRC is abundant in the cytoplasm but apparently does not nucleate MTs). One possibility therefore is that augmin subunits other than Hice1 or hDgt6 might play a role in activating γ-TuRC within the spindle. Reconstitution of augmin and γ-TuRC-dependent MT generation in vitro would serve as the most decisive method of elucidating the molecular basis of this process.
Methods
Cell Culture and RNAi.
Cell culture, plasmid transfection, and RNAi were performed according to the method of Goshima et al. (10). Several different HeLa cell lines and transfection reagents were assessed, and the cell line used in ref. 28 was found to produce the most severe phenotype when siRNA was transfected by using RNAiMax (Invitrogen) or DreamFect (OZ Biosciences). The dsRNA (S2) and siRNAs (HeLa) sequences used in this study are listed in Table S2 and Table S4, respectively. The siRNA mixture for KIAA0841 was made by using ColdShock-Dicer (Takara).
Microscopy.
At the end of the RNAi treatment (day 4), S2 cells were resuspended, transferred to glass-bottom Con A-coated plates, and allowed to adhere for 2.5 h before fixation (10). Immunofluorescence microscopy for the spindle and kinetochore proteins was performed as described in refs. 10 and 28, respectively. siRNA-treated HeLa cells were observed at day 3 (Fig. 3B) or day 2 (others). Most of the images were acquired by wide-field microscopy. GFP-tubulin image was acquired by FV1000 confocal microscopy (Olympus).
Biochemistry and LC/MS/MS.
Rabbit polyclonal antibodies against hDgt6 were generated and affinity-purified by using mixtures of 2 synthesized peptides as antigens (CPAKKSDPFQKEQDHLVE and CNKSLDAKEPPSDLTR). Immunoprecipitation was performed according the method of Goshima et al. (10), the details of which are described in SI Methods. LC/MS/MS was performed as described in ref. 29. In brief, samples were separated with SDS/PAGE, and each lane was cut into 9 pieces. Tandem mass spectra of the trypsinized peptides from the each gel section obtained by LXQ (Thermo Finnigan) were obtained by using Mascot (Matrix Science) and searched against the International Protein Index human database or National Center for Biotechnology Information database (Drosophila). The MS/MS-based peptide and protein identifications were validated by Scaffold (Proteome Software).
Supplementary Material
Acknowledgments.
We are grateful to Drs. H. Hanafusa, T. Kamura, S. Kojima, Y. Maeda, H. Masumoto, T. Watanabe, and K. Yoda (Nagoya University, Nagoya, Japan), and T. Kiyomitsu and M. Yanagida (Kyoto University, Kyoto, Japan) for reagents and technical assistance, and H. Ohkura (University of Edinburgh, Edinburgh, UK) for communicating results. This work is supported by the Special Coordination Funds for Promoting Science and Technology, Grant-in-Aid for Scientific Research [Ministry of Education, Culture, Sports, Science and Technology (Japan)], Nakajima Foundation, Uehara Foundation, Cell Science Foundation, and Human Frontier Science Program (G.G.). R.U. is supported by the Global Centers of Excellence program. S.P. is a recipient of European Molecular Biology Organization postdoctoral fellowship. R.D.V. is supported by National Institutes of Health Grant 38499.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901587106/DCSupplemental.
References
- 1.Skoufias DA, Andreassen PR, Lacroix FB, Wilson L, Margolis RL. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc Natl Acad Sci USA. 2001;98:4492–4497. doi: 10.1073/pnas.081076898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McIntosh JR, Grishchuk EL, West RR. Chromosome-microtubule interactions during mitosis. Annu Rev Cell Dev Biol. 2002;18:193–219. doi: 10.1146/annurev.cellbio.18.032002.132412. [DOI] [PubMed] [Google Scholar]
- 3.Wheatley SP, Wang Y. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J Cell Biol. 1996;135:981–989. doi: 10.1083/jcb.135.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gadde S, Heald R. Mechanisms and molecules of the mitotic spindle. Curr Biol. 2004;14:R797–R805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 5.Gruss OJ, Vernos I. The mechanism of spindle assembly: Functions of Ran and its target TPX2. J Cell Biol. 2004;166:949–955. doi: 10.1083/jcb.200312112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly AE, et al. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev Cell. 2007;12:31–43. doi: 10.1016/j.devcel.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janson ME, Setty TG, Paoletti A, Tran PT. Efficient formation of bipolar microtubule bundles requires microtubule-bound gamma-tubulin complexes. J Cell Biol. 2005;169:297–308. doi: 10.1083/jcb.200410119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murata T, et al. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat Cell Biol. 2005;7:961–968. doi: 10.1038/ncb1306. [DOI] [PubMed] [Google Scholar]
- 9.Mahoney NM, Goshima G, Douglass AD, Vale RD. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr Biol. 2006;16:564–569. doi: 10.1016/j.cub.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 10.Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: A protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goshima G, et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luders J, Patel UK, Stearns T. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat Cell Biol. 2006;8:137–147. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- 13.Einarson MB, Cukierman E, Compton DA, Golemis EA. Human enhancer of invasion-cluster, a coiled-coil protein required for passage through mitosis. Mol Cell Biol. 2004;24:3957–3971. doi: 10.1128/MCB.24.9.3957-3971.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu G, et al. Hice1, a novel microtubule-associated protein required for maintenance of spindle integrity and chromosomal stability in human cells. Mol Cell Biol. 2008;28:3652–3662. doi: 10.1128/MCB.01923-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen JS, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Coppinger JA, Jang CY, Yates JR, III, Fang G. FAM29A promotes microtubule amplification via recruitment of the NEDD1-γ-tubulin complex to the mitotic spindle. J Cell Biol. 2008;183:835–848. doi: 10.1083/jcb.200807046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silljé HH, Nagel S, Körner R, Nigg EA. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol. 2006;16:731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 18.Wong J, Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol. 2006;173:879–891. doi: 10.1083/jcb.200511132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen RH, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 21.Waters JC, Chen RH, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haren L, et al. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J Cell Biol. 2006;172:505–515. doi: 10.1083/jcb.200510028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang G, et al. Architectural dynamics of the meiotic spindle revealed by single-fluorophore imaging. Nat Cell Biol. 2007;9:1233–1242. doi: 10.1038/ncb1643. [DOI] [PubMed] [Google Scholar]
- 24.O'Connell CB, et al. The spindle assembly checkpoint is satisfied in the absence of interkinetochore tension during mitosis with unreplicated genomes. J Cell Biol. 2008;183:29–36. doi: 10.1083/jcb.200801038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meireles AM, et al. Wac: a new Augmin subunit required for chromosome alignment, but not for acentrosomal microtubule assembly in female meiosis. J Cell Biol. 2009;184:777–784. doi: 10.1083/jcb.200811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maiato H, et al. The ultrastructure of the kinetochore and kinetochore fiber in Drosophila somatic cells. Chromosoma. 2006;115:469–480. doi: 10.1007/s00412-006-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canman JC, Hoffman DB, Salmon ED. The role of pre- and post-anaphase microtubules in the cytokinesis phase of the cell cycle. Curr Biol. 2000;10:611–614. doi: 10.1016/s0960-9822(00)00490-5. [DOI] [PubMed] [Google Scholar]
- 28.Goshima G, Kiyomitsu T, Yoda K, Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obuse C, et al. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.