Abstract
Viral strain differences influence the oncogenic potential of polyomavirus simian virus 40 (SV40). We hypothesized that viral strain differences might also affect vertical transmission of SV40 in susceptible hosts. Pregnant Syrian golden hamsters were inoculated intraperitoneally with 107 plaque-forming units of SV40 and offspring were sacrificed post-delivery (1-21 days, 6 mo). Organ extracts were analyzed for SV40 DNA by polymerase chain reaction assay. Transmission of SV40 from mother to offspring was detected in over half of litters. Most placentas were virus-positive. Mothers inoculated with SV40 strains containing complex regulatory regions transmitted virus more frequently than those infected with simple enhancer viruses (p<0.001). Virus was detected more often in progeny brain than in spleen (p<0.05). Several progeny were virus-positive at 6 months of age, suggesting viral persistence. Maternal animals retained virus in several tissues through day 21 and developed T-antigen antibodies. These results indicate that SV40 replicates in hamsters, vertical transmission of SV40 can occur, and the viral regulatory region influences transmission.
Keywords: SV40, polyomavirus, vertical transmission, hamster, vimentin gene, viral regulatory region
Introduction
Vertical transmission of various DNA and RNA viruses among humans (Blanche et al., 1989; Candotti et al., 2006; Demmler, 1991; Paisley et al., 2006; Paryani and Arvin, 1986; Pereira et al., 2005; Stevens et al., 1975) and animals (Amedee et al., 1995; McCance and Mims, 1977; Oldstone and Dixon, 1972; Zhang et al., 2005) is well documented. Murine polyoma virus has been implicated in intrauterine infections in mice (McCance and Mims, 1977; Zhang et al., 2005). However, the potential for this mode of transmission among other members of the family Polyomaviridae is less clear. Transplacental transmission of BK virus (BKV) has been suggested based on detection of BKV DNA in fetal tissues (Pietropaolo et al., 1998), but others failed to detect evidence of transmission (Boldorini et al., 2008). Studies based on assays for IgM class antibody to BKV and JC virus (JCV) in umbilical cord blood samples found no evidence for congenital transmission (Andrews et al., 1983; Brown et al., 1984). Simian virus 40 (SV40), a polyomavirus of macaque origin, was introduced into humans presumably as a result of the widespread use of poliovaccines inadvertently contaminated with this DNA virus (Butel and Lednicky, 1999; Cutrone et al., 2005; Stratton et al., 2003; Vilchez and Butel, 2004). The contamination occurred because vaccines were produced in cultures of kidney cells derived from rhesus macaques, which are frequently infected with SV40. As infectious SV40 survived the vaccine inactivation treatments in early killed (Salk) vaccines and was present in live (Sabin) vaccines, millions of people were exposed to live SV40 (Butel and Lednicky, 1999; Cutrone et al., 2005; Proceedings of the Second International Conference on Live Poliovirus Vaccines, 1960; Stratton et al., 2003; Vilchez et al., 2003; Vilchez and Butel, 2004).
SV40 infections have been detected in different human populations today (Butel, 2008; Vilchez and Butel, 2004). Significantly, some of the subjects found with SV40 markers were not exposed to contaminated poliovaccines, suggesting infections by other pathways (Butel et al., 1999a; Stratton et al., 2003; Vilchez and Butel, 2004). Maternal-infant transmission has been reported as a possible route of polyomavirus SV40 pathogenesis in the hamster model (Rachlin et al., 1988). This might also represent a pathway for occasional transmission of SV40 in humans, as SV40 large tumor antigen (T-ag) DNA or protein has been detected in primary brain and bone cancers in infants and young children (Bergsagel et al., 1992; Lednicky et al., 1995a; Malkin et al., 2001; Martini et al., 1996; Stewart et al., 1998; Suzuki et al., 1997; Weggen et al., 2000; Zhen et al., 1999). In addition, SV40 has been isolated (Brandner et al., 1977; Lednicky et al., 1995a) and detected in urine (Vanchiere et al., 2005b) and stool samples (Vanchiere et al., 2005a) from young children.
Different natural strains of SV40 have been recognized (Forsman et al., 2004) and appear to be distributed in the human population (Butel and Lednicky, 1999; Forsman et al., 2004; Stewart et al., 1998). Strains of SV40 are known to diverge in the structure of their regulatory region and some strains have variants based on the number of enhancer elements in this region (Lednicky and Butel, 2001; Stewart et al., 1998). SV40 variants containing two 72-base-pair enhancer elements or other sequence rearrangements or duplications are said to have complex regulatory region structures; those with one enhancer and no rearrangement have a simple regulatory region structure (Lednicky and Butel, 2001; Stewart et al., 1998). The number of enhancer elements in the regulatory region of SV40 influences the replication of the virus in cell cultures (Lednicky et al., 1995b; Lednicky and Butel, 2001). This report describes investigations that quantify vertical transmission of polyomavirus SV40 in the hamster model, identify infected tissues, reveal the potential contribution of the structure of the SV40 regulatory region on transmission of virus from mothers to offspring, and suggest that persistent infections may occur.
Results
Absolute quantification of hamster vimentin gene in real-time quantitative polymerase chain reaction (RQ-PCR) assays
The vimentin gene is a proven hamster single copy gene. The amplification of this gene serves as a control for the quality of cellular DNA isolated from hamster tissues. The quantitative analysis of the vimentin gene allows SV40 copy numbers to be normalized to cell numbers. The standard curve method of analysis was used for absolute quantification of the vimentin gene in RQ-PCR assays. A representative amplification plot of serial 10-fold dilutions of the vimentin standard plasmid is shown in Fig. 1A. The lower limit of reproducible detection of the vimentin standard in multiple assays was 101 copies of the target gene; in a few assays, 100 copy was detected. Standard curves were generated to allow calculation of amounts of the vimentin gene in experimental samples (Fig. 1B).
Fig. 1.
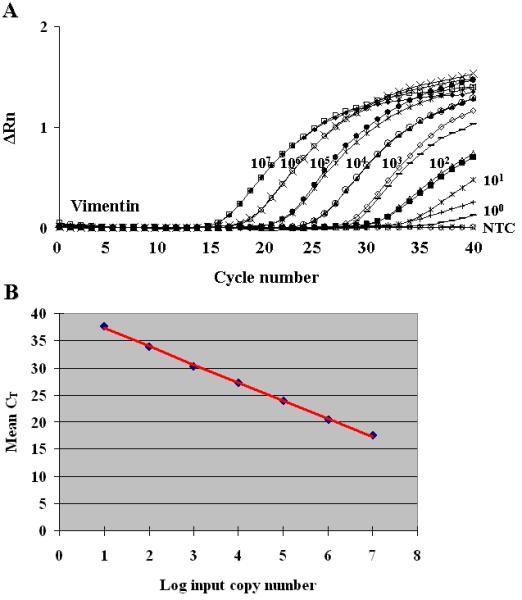
Quantitative assay for hamster vimentin gene. (A) A representative hamster vimentin gene amplification plot of normalized reporter fluorescence (ΔRn) against the cycle number. The log10 of the input copy number of each plasmid standard is indicated. NTC, Non-template control. (B) Typical standard curve for the hamster vimentin gene created by regression analysis of the observed CT value against the log10 of the input copy number of standard plasmid DNA encoding the vimentin gene sequences. R2 = 0.9991; y = 3.3295x + 40.579.
The precision of amplification of the vimentin target gene was assessed by measuring known quantities of two hamster lymphoma cell lines (McNees et al., 2008). The RQ-PCR assay was sensitive and reproducible up to 5 × 105 cell equivalents/reaction. Generally, 250-500 ng of input cellular DNA (representing approximately 37,000-75,000 cells) was used per reaction in assays of experimental samples, well within the limits of the assay.
Transmission of SV40 from mothers to offspring hamsters
A total of 14 pregnant Syrian golden hamsters were inoculated with SV40 strains and 6 with uninfected cell lysate as controls. Mothers and offspring were sacrificed 1, 3, 7, 14, and 21 days post-parturition (Table 1). Multiple organs were harvested from mothers and offspring from the inoculated and control groups and total DNA was extracted. A total of 127 progeny yielding 403 organs were produced from 14 virus-inoculated pregnant hamsters; 73 progeny (259 organs) were produced from the control animals. One tissue slice per organ was analyzed by RQ-PCR. Vimentin gene sequences were amplified from all samples tested. SV40 DNA was detected in tissues (spleen, kidney, lung, brain) from some offspring (Table 1). The mean SV40 viral loads in progeny animal tissues were variable, ranging from 376 to 23,966 copies/106 cells in positive tissues. A total of 8 of 14 (57%) litters, 18 of 127 (14%) progeny, and 21 of 403 (5%) organs were positive for SV40 from progeny in the inoculated group, compared to none (0 of 6 litters, 0 of 73 progeny, and 0 of 259 organs) in the control group (Table 2). From all offspring tested from 1 to 21 days of age, positive tissues detected were spleen (n=5), kidney (n=3), lung (n=7), and brain (n=6) (Table 1).
Table 1.
Vertical transmission of SV40 from maternal hamsters inoculated during pregnancy to progeny animalsa
| Days after birth | Viral strain (No. positive litters)b | No. progeny tested | Tissue | No. tissues positive/No. tested (%)c | Viral loads (copies/106 cells) Range (mean)d |
|---|---|---|---|---|---|
| 1, 3 | Baylor-2E (2) | 30 | Kidney Lung |
1/29 (3) 3/30 (10) |
649 (649) 234-1,944 (1,192) |
| 7 | Baylor-1E (1) SVCPC (2) |
23 | Spleen Kidney Lung Brain |
4/21 (19) 1/20 (5) 4/19 (21) 0/17 (0) |
817-2,406 (1,452) 23,966 (23,966) 488-2,532 (1,623) <5 (<5) |
| 14 | SVCPC (1) SVPML-1 (1) |
22 | Spleen Kidney Lung Brain |
1/22 (5) 0/22 (0) 0/20 (0) 4/22 (18) |
1,020 (1,020) <5 (<5) <5 (<5) 477-908 (719) |
| 21 | SVPML-1 (1) | 12 | Spleen Kidney Lung Brain |
0/12 (0) 1/10 (10) 0/12 (0) 2/12 (17) |
<5 (<5) 376 (376) <5 (<5) 477-893 (685) |
Pregnant hamsters were inoculated intraperitoneally with 107 PFU of SV40.
Vertical transmission was not detected in litters from 6 additional animals inoculated with SV40 strains Baylor-1E, Baylor-2E, SVCPC, and SVPML-1 and harvested at 3, 7/14/21, 21, and 7 days after birth, respectively (data not shown).
One tissue slice per organ was analyzed for SV40 DNA.
Viral loads among virus-positive samples.
Table 2.
Dynamics of detection of SV40 vertical transmissiona
| Strain/variant | No. litters positive/No. tested (%) | No. progeny positive/No. testedb (SV40 copies/106 cells)c |
Total (%) | |||
|---|---|---|---|---|---|---|
| 1, 3 days after birth | 7 days after birth | 14 days after birth | 21 days after birth | |||
| Baylor-1E | 1/2 (50) | 0/7 (<5) | 1/12 (488) | NDd | ND | 1/19 (5) |
| Baylor-2E | 2/5 (40) | 4/23 (303) | 0/14 (<5) | 0/6 (<5) | 0/5 (<5) | 4/48 (8) |
| SVCPC | 3/4 (75) | ND | 5/11 (4,473) | 1/10 (908) | 0/9 (<5) | 6/30 (20) |
| SVPML-1 | 2/3 (67) | ND | 0/6 (<5) | 4/12 (747) | 3/12 (582) | 7/30 (23) |
| Total (%): | 8/14 (57) | 4/30 (303) | 6/43 (2,883) | 5/28 (799) | 3/26 (582) | 18/127 (14) |
| Control | 0/6 (0) | 0/8 (<5) | 0/22 (<5) | 0/24 (<5) | 0/19 (<5) | 0/73 (0) |
Pregnant hamsters were inoculated intraperitoneally with 107 PFU of SV40. From Table 1.
One tissue slice per organ (spleen, kidney, lung, brain) was analyzed for SV40 DNA for each animal.
Mean viral loads among virus-positive samples.
ND, not done.
The dynamics of infection in progeny animals with different strains of SV40 are shown (Table 2). However, comparable harvests were not available for all viral strains at some time points. A litter was considered to be positive if SV40 DNA was detected in one littermate. Overall, 8 of 14 (57%) litters from mothers in the SV40-inoculated group were infected, as compared to none in the control group (p=0.02). Positive litters ranged from a single animal that contained detectable viral DNA to five littermates that were SV40-positive. The difference in mean viral loads observed among time points was not statistically significant because of small sample sizes.
Structure of the SV40 regulatory region and vertical transmission
In vitro studies have shown that SV40 isolates with a complex regulatory region produced higher virus yields and larger plaque sizes in cultured cells than did viruses with a simple regulatory region (Lednicky et al., 1995b; Lednicky and Butel, 2001). Hamster experiments demonstrated that SV40 strains with simple (1E) regulatory region structures were more oncogenic than strains with complex (2E) regulatory regions (Sroller et al., 2008). However, significantly more of the tumor-free hamsters exposed to 2E viruses developed antibodies to T-ag than did the non-tumor-bearing animals inoculated with 1E strains, suggesting more abundant replication of SV40 strains with complex regulatory regions in the animals (Sroller et al., 2008). These observations provided the rationale for examining whether SV40 strains with different regulatory region structures diverged in their frequency of transmission. All SV40 strains tested had resulted in vertical transmission (Table 1). To explore possible regulatory region effects on transmission in more detail, 4 litters harvested at 7 days after birth were analyzed further (Table 3). In this analysis, multiple tissue sections (3 tissue slices per organ per progeny) were assessed for the presence of SV40 DNA. Of mothers inoculated with SV40 strains containing simple regulatory regions (Baylor-1E and SVCPC), transmission occurred in 1 of 2 (50%) litters, and virus was detected in 2 of 20 (10%) progeny and 3 of 50 (6%) tissues. In contrast, in offspring of mothers inoculated with SV40 strains containing complex regulatory regions, 2 of 2 (100%) litters, 19 of 20 (95%) progeny, and 39 of 57 (68%) tissues were virus-positive. This difference between SV40 1E and 2E variants was statistically significant (p<0.001), with the transmission rate for 2E viruses being approximately 65% greater than for 1E variants. SV40 DNA was preferentially detected in brain tissue of progeny compared to spleen (p<0.05), but there was no difference between spleen and kidney (p>0.60) for virus detection.
Table 3.
Frequency of SV40 vertical transmission to progeny animals is higher from mothers inoculated with SV40 variants with complex regulatory region structuresa
| Type of regulatory region | Strain/variant | No. litters positive/No. tested (%) | No. progeny positive/No. tested (%)b | No. tissues positive/No. tested (%)b |
|---|---|---|---|---|
| Simple | Baylor-1E SVCPC |
0/1 (0) 1/1 (100) |
0/12 (0) 2/8 (25) |
0/36 (0) 3/14 (21) |
| Total: | 1/2 (50) | 2/20 (10)c | 3/50 (6)d | |
| Complex | Baylor-2E SVPML-1 |
1/1 (100) 1/1 (100) |
13/14 (93) 6/6 (100) |
23/39 (59) 16/18 (89) |
| Total: | 2/2 (100) | 19/20 (95)c | 39/57 (68)d | |
Analysis of litters harvested 7 days after birth.
Three tissue slices for each of three different organs (spleen, kidney, brain) were analyzed for SV40 DNA for each animal.
p<0.001
p<0.001.
A long-term experiment was conducted to examine the possibility of persistent infection by SV40 in offspring of inoculated mothers. Three litters of progeny hamsters were removed from their mothers at day 21 and were held for 6 months after birth (Table 4). The animals from given litters were caged together. Results showed that 6 of 14 (43%) animals were SV40-positive, with SV40 viral loads ranging from 518 to 3,539 copies/106 cells in positive tissues. Viral DNA was detected in lung and spleen tissues in 7 of 36 (19%) tissues of progeny from SV40-injected mothers. Tumors did not develop in any of the progeny animals in the short- or long-term experiments.
Table 4.
Evidence of SV40 vertical transmission and persistence of virus in progeny animals 6 months after birth
| Strain/variant (regulatory region) | No of litters positive/No. tested (%) | No. progeny positive/No. tested (%) | Tissue | No. tissues positive/No. tested (%) | Viral load (copies/106 cells) Range (mean)a |
|---|---|---|---|---|---|
| SVCPC (simple) |
1/1 (100) | 1/5 (20) | Spleen Kidney Lung |
1/1 (100) 0/5 (0) 0/5 (0) |
2,343 (2,343) <5 <5 |
| 776-2E (complex) |
1/2 (50) | 5/9 (56) | Spleen Kidney Lung |
1/7 (14) 0/9 (0) 5/9 (56) |
615 (615) <5 518-3,539 (1,800) |
| Total: | 2/3 (67) | 6/14 (43) | 7/36 (19) | 518-3,539 (1,721) | |
Viral loads among virus-positive samples.
Distribution of SV40 in maternal hamster tissues
SV40 was detected in tissues from all virus-inoculated mother hamsters that were tested except two; those two animals from the Baylor-2E group (and another animal from the SVCPC group that was not tested) are not shown (Table 5). Results from eight animals injected with virus variants containing simple regulatory regions and eight animals with variants having complex regulatory regions are reported (Table 5). Maternal animals were sacrificed at the same times as their litters, ranging from 2 to 24 days postinoculation (p.i.) (either before delivery or from 3 to 21 days after delivery), maternal tissues were collected, and total DNAs were extracted. Viral loads were assayed in three organs (spleen, kidney, lung). Viral loads were low at 2 days p.i. (3,270 copies/106 cells or less), were highest in tissues from animals sacrificed 4 and 6 days p.i., reaching over 600,000 copies/106 cells (animals 4, 9, 10), and declined by 10 to 24 days p.i., but remained detectable in spleen and kidney. In contrast, infections often had been cleared from the lungs at the later times with detectable levels of SV40 remaining in only 3 of 9 animals. Blood was available from 9 animals. Virus was detected in white blood cells (WBCs) soon after infection and persisted, at least in two animals, up to 17 and 24 days p.i. Virus was also detected in plasma, but at very low levels (usually less than 50 genome copies per 10 μl). The amount of virus present in blood samples did not appear to correlate with SV40 viral loads detected in the maternal tissues of the same animals, suggesting that tissue-associated virus did not reflect simple presence of circulating blood-associated virus. No virus was found among hamsters in the control group of 8 animals (data not shown).
Table 5.
Viral loads in maternal tissues and blood following inoculation of pregnant hamsters with SV40 strains with different regulatory region structures
| Animal number | Type of regulatory region | Strain/variant | Days post-inoculation (days post-delivery) | SV40 copies/106 cells |
Blood |
|||
|---|---|---|---|---|---|---|---|---|
| Spleen | Kidney | Lung | WBC (SV40 copies/106 cells) | Plasma (SV40 copies/10 μ1) | ||||
| 1 | Simple | Baylor-1E | 2a | 255 | 1,770 | 869 | NDb | 41 |
| 2 | 2a | 1,590 | 3,270 | 3,020 | 10,400 | 22 | ||
| 3 | 2a | <5 | 360 | 891 | 962,000 | 12 | ||
| 4 | 6 (3) | 11,300 | 8,020 | 609,000 | <5 | <5 | ||
| 5c | 10 (7) | 72 | 559 | <5 | ND | ND | ||
| 6c | SVCPC | 10 (7) | 2,460 | 25,000 | <5 | ND | ND | |
| 7c | 17 (14) | 8,680 | 17,800 | <5 | ND | ND | ||
| 8 | 24 (21) | 3,670 | 32,900 | <5 | ND | ND | ||
| 9c | Complex | Baylor-2Ed | 4 (1) | 233,000 | 789,000 | 442,000 | ND | 580 |
| 10c | 6 (3) | 45,400 | 883,000 | 72,200 | ND | 17 | ||
| 11c | 10 (7) | 17,500 | 34,400 | 1,780 | <5 | 16 | ||
| 12 | 17 (14) | 26,100 | 44,400 | 1,470 | 20,600 | 14 | ||
| 13 | 24 (21) | 409 | 3,890 | <5 | 383,000 | <5 | ||
| 14c | SVPML-1 | 10 (7) | 7,040 | 36,900 | 769 | ND | ND | |
| 15c | 17 (14) | 3,180 | 11,900 | <5 | ND | ND | ||
| 16c | 24 (21) | 249 | 810 | <5 | ND | ND | ||
Animals from transplacental transmission experiments.
ND, not done.
Animals with virus-positive offspring.
Two animals inoculated with SV40 strain Baylor-2E and harvested 2 days p.i. were negative for SV40 DNA in all tissues tested; one animal inoculated with SV40 strain SVCPC and harvested at 10 days p.i. was not tested (data not shown).
Potential for transplacental transmission of SV40
As vertical transmission of SV40 in progeny hamsters could have occurred either before, during, or after birth, the possibility of transplacental transmission was examined. Two groups of pregnant hamsters were inoculated with SV40 strains Baylor (variant 1E) and 776-2E; a control animal was inoculated with uninfected cell lysate. Pregnant hamsters were sacrificed prior to delivery (48 h p.i. to 6 days) and their tissues, fetuses, and placentas were harvested and processed as described in Materials and methods. Placentas were frequently virus-positive (23 of 25, 92%) and were found only among the SV40-inoculated group (Table 6). Mean SV40 viral loads in virus-positive placentas were 11,000 copies/106 cells. Viral loads were higher in placentas from animals exposed to strain 776-2E, as compared to those inoculated with Baylor-1E. SV40 DNA was detected in 4 of 47 (8.5%) fetuses from 1 of 5 litters tested from SV40-infected mothers, suggesting the possibility of transplacental transmission. The positive litter was from an animal inoculated with strain 776-2E. No virus was detected in fetuses of the uninfected animals.
Table 6.
Infection of placental tissue in hamsters by SV40
| SV40 strain/variant | No. litters positive/No. tested (%) | Placentas |
|
|---|---|---|---|
| No. positive/No. tested (%) | Viral loads (copies/106 cells) Range (mean)a | ||
| Baylor-1Eb | 0/3 (0) | 21/23 (91) | 233-8,010 (3,670) |
| 776-2Ec | 1/2 (50) | 2/2 (100) | 27,400-148,000 (87,700) |
| Total: | 1/5 (20) | 23/25 (92) | 233-148,000 (11,000) |
| Control | 0/1 (0) | 0/12 (0) | <5 (<5) |
Viral loads among virus-positive samples.
Harvested 2 days p.i., prior to delivery.
Harvested 6 days p.i., prior to delivery.
SV40 antibody responses in mothers and offspring
Serologic responses to SV40 were determined for mothers and offspring in the inoculated and control groups. Maternal hamsters from SV40-inoculated groups that were sacrificed between 10 and 24 days p.i. were tested for virus neutralizing antibodies and all were positive. Median neutralizing titers for animals inoculated with SV40 strains SVCPC (n=3), SVPML-1 (n=3) and Baylor-2E (n=3) were 1:100, 1:1000, and 1:1000, respectively. Some of the tested mothers from the virus-inoculated groups also had detectable T-ag antibody responses. Positivity rates and median T-ag antibody titers for the different groups of maternal hamsters were the following: SVCPC, 3 of 4 positive (1:100); SVPML-1, 2 of 3 positive (1:5); Baylor-2E, 2 of 3 positive (1:5); and 776-2E, 1 of 1 positive (1:10). None of the maternal animals from the control group (n=5) had serologic evidence of SV40 infection. None of 12 progeny animals harvested at the 21-day timepoint had detectable virus neutralizing antibodies.
Discussion
Vertical transmission of a virus to the fetus may occur during the birth process, in the period soon after birth, or across the placenta prior to birth (Virgin, 2007). The studies described here involving Syrian golden hamsters indicate that vertical transmission is a possible mode of spread of polyomavirus SV40. These are intriguing findings as SV40 DNA has been detected in infants (Bergsagel et al., 1992) and recent reports have indicated that the virus is shed in the urine (Vanchiere et al., 2005b) and stool (Vanchiere et al., 2005a) of young children. The overall rates of SV40 transmission from mothers to individual offspring in the hamster model under our experimental conditions averaged 14%, a value comparable to those of varicella-zoster virus and human immunodeficiency virus vertical transmission in humans (2-20% and 13-30%, respectively) (Blanche et al., 1989; Paryani and Arvin, 1986), but higher than those reported for cytomegalovirus and West Nile virus in humans (0.2% to 4%, respectively) (Demmler, 1991; Paisley et al., 2006). The frequency of SV40-positive litters following inoculation of pregnant hamsters was high at 57% (8 of 14 litters with one or more positive progeny).
This study was not designed to study transplacental transmission in detail, but we did detect virus transmission in 1 of 5 litters. This was lower than reported for murine polyoma virus in mice (6 of 7 litters, 86%) (Zhang et al., 2005), perhaps reflecting differences in experimental conditions or in maternal or viral factors that affect the establishment of infection. One difference is that the Syrian golden hamsters are outbred, making it likely that they exhibit broader individual variations in susceptibilities to infection than inbred mice.
Although it was assumed in the past that hamsters are nonpermissive hosts for SV40, recent evidence indicates that SV40 can infect and undergo replication in some hamsters. In a previous study, infectious SV40 was rescued from 39% of tumor cell lines established from primary tumors induced by SV40 (Sroller et al., 2008). Tumors from which the cell lines were established had arisen 5-8 months p.i., making it highly unlikely that residual input SV40 was resident in the tumors in vivo and survived during subculture of the cell lines in vitro. This observation was compatible with earlier reports of recovery of infectious virus and/or infectious viral DNA from SV40 tumor cells (Black and Rowe, 1964; Boyd and Butel, 1972; Gerber, 1964; Sabin and Koch, 1963). Many virus-inoculated, tumor-free animals developed antibodies to T-ag that persisted for one year following either intraperitoneal or intravenous exposure (McNees et al., 2008; Sroller et al., 2008). The percentage of T-antibody responders among tumor-free hamsters was higher for those inoculated with viruses having complex regulatory regions as compared to those with simple enhancers. We believe induction of durable T-antibodies in the absence of tumor formation reflects synthesis of T-ag during virus infection and that viruses with complex enhancers replicate more abundantly in vivo than those with simple enhancers. SV40 regulatory region rearrangements were detected in several SV40-induced tumors (Sroller et al., 2008), and it has been proposed that such rearrangements reflect recombination during viral DNA replication (Cubitt, 2006; Yogo and Sugimoto, 2001). In this study, SV40 strains with complex regulatory regions were vertically transmitted significantly more frequently than strains with simple enhancers. Our interpretation is that the 2E viruses replicated to higher titers, increasing the likelihood of transmission. Maternal animals exhibited a rise in viral load from day 2 p.i. followed by viral persistence in maternal spleen, kidney and WBCs through day 24 (the latest time point examined), plus the production of anti-T-ag antibodies. These analyses provided evidence of SV40 replication in hamsters and identified tissues that can be virus positive following IP inoculation.
We postulate that higher viral loads in the mother increase the potential for vertical transmission of polyomavirus. There was a statistically significant higher frequency of transmission of virus to progeny from mothers infected with virus with complex enhancer structures than with simple enhancers (Table 3). The frequent detection of SV40 DNA in brain tissue of progeny is compatible with other studies suggesting that SV40 can be neurotropic (Bergsagel et al., 1992; Lednicky et al., 1995a; Lednicky and Butel, 2001).
The SV40-infected maternal hamsters had detectable virus in tissues that might serve as sources of virus for transmission to their young after birth, including the kidney (urine) and lung (respiratory droplets). It is of interest that lung tissue was sometimes positive in progeny animals, raising the possibility that some polyomavirus infections might occur via the respiratory route. It is expected that blood would be the route of fetal infection in utero. Compatible with that premise, SV40 was detected in the blood of maternal hamsters. It is noteworthy that in the case in which transplacental transmission was documented, there had been a longer duration p.i. prior to fetal harvest than in the other litters tested. In addition, the placentas from that litter had significantly higher viral loads than those from litters in which transmission was not detected, suggesting that placental viral loads may impact transmission to the fetus or may reflect overall viral loads in those maternal animals.
Any long-term effects of SV40 vertical transmission in progeny animals were not addressed in this study. However, none of 43 animals held for 6 months developed neoplasms. This finding contrasts with a previous report that 43-54% of progeny from SV40-inoculated pregnant hamsters developed tumors (Rachlin et al., 1988). There were several differences between the two studies that may explain the results. The hamsters were obtained from different sources, the pregnant animals were inoculated earlier during gestation in the Rachlin study than in ours, and they observed the animals for 10 months as compared to only 6 months in our study. SV40 tumor latency and tumor frequency are virus-dose dependent (Gerber and Kirschstein, 1962; Girardi et al., 1963). It is possible that earlier gestational inoculation led to higher amounts of virus being transmitted to the fetuses in the Rachlin study, increasing their effective exposure dose; that exposure, coupled with the longer period of observation, perhaps made it more likely that tumors could be observed.
Our findings of SV40 vertical transmission in the hamster model differ from cross-sectional serological surveys in captive rhesus macaques (Minor et al., 2003) and humans (Engels et al., 2004) that suggested that vertical transmission had not occurred. Serologic analyses of BKV in humans led to the conclusion that transplacental transmission was not a mode of spread for this polyomavirus (Andrews et al., 1983; Brown et al., 1984). Recent molecular-based studies have differed in evidence for transplacental transmission of human polyomaviruses (Boldorini et al., 2008; Pietropaolo et al., 1998). Additional studies are warranted to clarify the possible vertical transmission of polyomavirus in humans.
It is known that SV40 strains may have different growth properties in vitro (Lednicky et al., 1995b; Lednicky and Butel, 2001) and recent observations have proven variation in oncogenic potential in vivo (Sroller et al., 2008; Vilchez et al., 2004). This study shows that viral strains can differ in the biologic property of frequency of vertical transmission, as well. All three examples reflected differences in the regulatory region structures of the viral variants. This suggests that the viral regulatory region can be considered a determinant that may affect pathogenesis of infection and disease in vivo.
Animal models have been powerful experimental tools for studies of human disease pathogenesis, including cancer (Mizgerd and Skerrett, 2008; Virgin, 2007). Syrian golden hamsters are uniquely susceptible to a variety of intracellular pathogens. They serve as infection models that mimic human disease for several parasites, including Leishmania (Melby et al., 2001) and Opistorchis (Jittimanee et al., 2007). Hamsters are permissive immunocompetent animal models for infections by numerous viruses able to infect humans, such as herpesviruses (van Ekdom et al., 1987), vaccinia virus (Nelles et al., 1981), adenovirus (Thomas et al., 2006), rubella virus (Rayfield et al., 1986), measles virus (Vanchiere et al., 1995), Nipah virus (Wong et al., 2003), arenaviruses (Gowen et al., 2005), flaviviruses (Siirin et al., 2007; Tesh et al., 2005), and hantaviruses (Campen et al., 2006). As described above, it appears that SV40 can replicate in hamsters. SV40 has long been recognized for its ability to induce tumors in hamsters (Butel et al., 1972; Butel, 2000; Butel and Lednicky, 1999; Cicala et al., 1992; Diamandopoulos, 1973) and the types of tumors that develop include the same spectrum of malignancies as the human cancers associated with SV40 (Butel, 2008; Gazdar et al., 2002). The hamster model has shown that asbestos and SV40 can cooperate in induction of mesotheliomas (Kroczynska et al., 2006). Similarities have been found between humans and hamsters with respect to tissues that can harbor SV40 and the spectrum of malignancies associated with the virus. Hamsters are especially well-suited for future comparative studies of the effects of viral strain genetic differences on pathogenesis of SV40 infections and disease.
In conclusion, the results described here show that vertical transmission by polyomavirus SV40 can occur in susceptible hosts, that the viral regulatory region is a determinant of transmission, and that SV40 appears to replicate in hamsters. The possibility of vertical transmission of polyomaviruses in humans should be considered.
Materials and methods
Experimental animals
Outbred female and male Syrian golden hamsters (Mesocricetus auratus) were obtained from Harlan Sprague Dawley, Indianapolis, IN. Pregnant hamsters were inoculated intraperitoneally at late gestation (day 10-13 of a gestation period of 16 days, estimated by subtraction from the date of delivery) with 1 × 107 plaque-forming units (PFU) of SV40. Control animals of similar gestation were inoculated with 1.0 ml of uninfected TC-7 cell lysate. Animals were housed in the biohazard facility of the Center for Comparative Medicine at Baylor College of Medicine. Animal protocols were approved by the Institutional Animal Care and Use Committee and all care was in accordance with established national guidelines as outlined in DHEW Publication No. (NIH) 78-23.
Viruses
SV40 strains from different phylogenetic groups included: (i) SVCPC (simple regulatory region), isolated from a brain tumor from a 4-year-old child (Lednicky et al., 1995a) and detected in various human cancers (Arrington et al., 2004; Krieg and Scherer, 1984; Lednicky et al., 1995a; Vilchez et al., 2002) and also found in Russian oral poliovaccine used until the late 1970’s (Cutrone et al., 2005); (ii) SVPML-1 (complex regulatory region), a virus isolated from a patient with progressive multifocal leukoencephalopathy (Weiner et al., 1972); (iii) Baylor, an isolate recovered from a 1956 type 2 Sabin poliovaccine (Melnick and Stinebaugh, 1962), with one variant having a complex (Baylor-2E) and another a simple (Baylor-1E) regulatory region (Lednicky and Butel, 1997, 2001); and (iv) reference strain SV40-776 variant 2E (complex regulatory region), isolated from an adenovirus type 1 vaccine seed stock (Sweet and Hilleman, 1960). The structures of the regulatory regions of these strains are shown (Fig. 2). Virus stocks were prepared at 37 °C in TC-7 cells and were assayed using a plaque assay in TC-7 cells (Butel et al., 1999b).
Fig. 2.
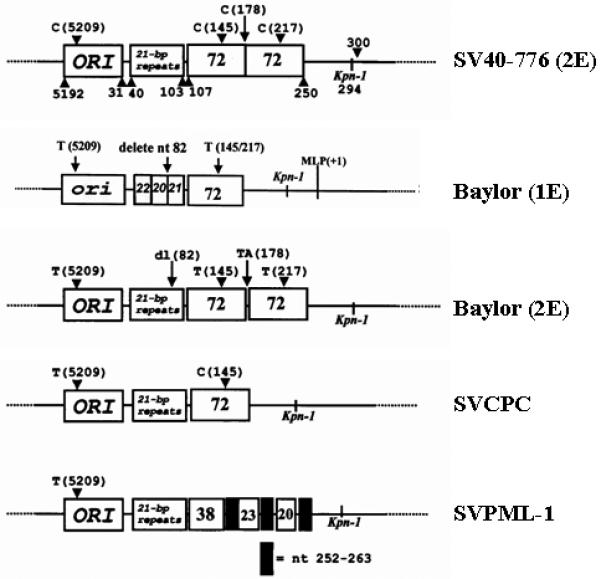
Regulatory regions of SV40 viral isolates. The viral origin of DNA replication spanning nucleotides 5192 to 31 is represented by the box labeled ORI; the 21-bp repeat region between nucleotides 40 and 103 is shown; the boxed number 72 is the 72-bp sequence within the enhancer region that is duplicated in some laboratory-adapted strains, with the rearranged enhancer region of SVPML-1 represented with boxes labeled 38, 23, 20 and a shaded box referring to nucleotides 252-263. Nucleotide numbers are based on that of SV40-776. Modified from Lednicky and Butel (2001) and Stewart et al. (1998).
Sample preparation and DNA
Litters delivered by SV40-inoculated hamsters were sacrificed between 1 and 21 days after birth and at 6 months post-parturition (litters contained from 6 to 12 progeny). Organs harvested from progeny hamsters included kidney, brain, spleen, and lung. Pregnant hamsters were sacrificed from 2 to 6 days p.i., prior to delivery, and placentas and fetuses were harvested. Maternal tissues collected included kidney, spleen, lung, and blood.
Small pieces (∼100 mg, one to three separate slices) of harvested tissues were minced, weighed, and incubated at 55 °C overnight with proteinase K (Roche Diagnostics Corp., Indianapolis, IN) and nuclei lysate solution (Promega Corp., Madison, WI). Protein was removed and DNA was precipitated by isopropanol, resuspended in TE buffer (pH 8.0), and stored at -20 °C. DNA from WBCs and plasma was recovered using a proteinase K and phenol/chloroform protocol (Lednicky and Butel, 1998).
Oligonucleotides and RQ-PCR analysis
SV40 was detected by RQ-PCR with primers and TaqMan probe designed to detect sequences from the conserved N-terminal region of the SV40 T-ag gene (McNees et al., 2005). An RQ-PCR assay to measure the single copy hamster vimentin gene (Quax et al., 1983) was developed in order to normalize SV40 gene copy numbers to cell numbers. The standard curve method of analysis was used for absolute quantification of SV40 and vimentin genes. Hamster tissues were analyzed for SV40 and vimentin gene in separate RQ-PCR assays. The standard curves were generated from 10-fold dilutions of plasmid standards containing known input gene copy numbers in each RQ-PCR assay. The amounts of SV40 and vimentin genes in hamster tissues were determined from the appropriate standard curve. The SV40 genome copies per hamster cell were calculated as the SV40 amount divided by one-half of the vimentin amount, as the diploid hamster cell contains two copies of the vimentin gene.
The plasmid pSV40-B2E was used as the SV40 standard in the RQ-PCR assay (McNees et al., 2005). The vimentin plasmid standard was constructed by PCR amplification of the vimentin gene from hamster tissue using primers VIM3 5′-ATA GAA TTC CGA CAA GGT GCG CTT CCT GG-3′ and VIM4 5′-ATA GAA TTC CTC GCA GCC GCA TGA TGT CC-3′. The PCR product was cloned using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA) and digested with EcoRI enzyme. The 206-bp fragment of the hamster vimentin gene was inserted into a vector pUC19, and the identity of the insert was confirmed by restriction digestion and sequence analysis. Plasmid stocks were prepared using the Endofree Plasmid Maxi Kit (Qiagen, Valencia, CA) and concentrations were determined by average A260 readings of multiple dilutions.
Primer and probes for the hamster vimentin gene RQ-PCR assay were designed using Primer Express software from Applied Biosystems. The forward and reverse primer sequences were 5′-TTC CTG GAG CAG CAG AAC AA-3′ and 5′-TGC CCT GAC CCT TGA GTT G-3′. The probe sequence was VIC-ATC CTG CTA GCC GAG CT-MGB. The target sequences for the RQ-PCR primers and probe were within the 206-bp fragment of the hamster vimentin gene in the plasmid standard.
RQ-PCR analyses were performed using the PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). 50-μl PCR reactions were prepared using 900 nM of each primer, 100 nM of TaqMan VIC-MGB-probe, and 25 μl of the 2× TaqMan Universal PCR Master Mix (Applied Biosystems). Reactions were prepared using the TaqMan PCR kit (Applied Biosystems) in PCR clean rooms. Test DNA samples (10 μl) were added outside the PCR clean room facility after tubes containing master mix and negative controls were sealed. After test DNA tubes were sealed, 10 μl of standard plasmid dilutions or positive control samples were added in duplicate. RQ-PCR reaction conditions for amplification of SV40 and vimentin target genes were as follows: 50 °C for 2 min, denaturing at 95 °C for 10 min, and 40 cycles of denaturing at 95 °C for 15 sec followed by annealing and extension at 60 °C for 1 min. Amplification data measured as an increase in reporter fluorescence were collected in real time and analyzed by the Sequence Detection System software.
Detection of SV40 T-ag and neutralizing antibodies
Antibody directed against SV40 T-ag in hamster sera was assayed by indirect immunofluorescence staining of T-ag in SV40-transformed cells (Butel and Ozbun, 1994). A plaque-reduction assay was used to measure SV40 neutralizing antibodies (Butel et al., 1999b).
Statistical analysis
Categorical data are presented as percentages. The standard chi square test was used to test differences between percentages. The Fisher’s exact test was used when one or more values was less than 5. Generalized estimating equations (SPSS 16) were used to assess the difference between simple and complex regulatory regions, tissue type, and the interaction of these with respect to SV40 vertical transmission rates. This model allows for the clustering of litter and littermates within region and the repeated measures of tissue types within offspring. This method assumed a binary logistic model and exchangeable correlation structure within offspring. A p-value less than 0.05 was considered statistically significant.
Acknowledgments
This work was supported by the National Institutes of Health (K12 RR17665 Mentored Clinical Investigator Award to N.C.P.; research grants R21 CA96951 and R01 CA104818 from the National Cancer Institute to J.S.B.; and training grant T32 CA09197 support to A.S.A.). The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Cancer Institute, the National Center for Research Resources, or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amedee AM, Lacour N, Gierman JL, Martin LN, Clements JE, Bohm R, Jr., Harrison RM, Murphey-Corb M. Genotypic selection of simian immunodeficiency virus in macaque infants infected transplacentally. J. Virol. 1995;69:7982–7990. doi: 10.1128/jvi.69.12.7982-7990.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews CA, Daniel RW, Shah KV. Serologic studies of papovavirus infections in pregnant women and renal transplant recipients. Prog. Clin. Biol. Res. 1983;105:133–141. [PubMed] [Google Scholar]
- Arrington AS, Moore MS, Butel JS. SV40-positive brain tumor in scientist with risk of laboratory exposure to the virus. Oncogene. 2004;23:2231–2235. doi: 10.1038/sj.onc.1207341. [DOI] [PubMed] [Google Scholar]
- Bergsagel DJ, Finegold MJ, Butel JS, Kupsky WJ, Garcea RL. DNA sequences similar to those of simian virus 40 in ependymomas and choroid plexus tumors of childhood. N. Engl. J. Med. 1992;326:988–993. doi: 10.1056/NEJM199204093261504. [DOI] [PubMed] [Google Scholar]
- Black PH, Rowe WP. Viral studies of SV40 tumorigenesis in hamsters. J. Natl. Cancer Inst. 1964;32:253–265. [PubMed] [Google Scholar]
- Blanche S, Rouzioux C, Moscato ML, Veber F, Mayaux MJ, Jacomet C, Tricoire J, Deville A, Vial M, Firtion G, HIV Infection in Newborns French Collaborative Study Group A prospective study of infants born to women seropositive for human immunodeficiency virus type 1. HIV Infection in Newborns French Collaborative Study Group. N. Engl. J. Med. 1989;320:1643–1648. doi: 10.1056/NEJM198906223202502. [DOI] [PubMed] [Google Scholar]
- Boldorini R, Veggiani C, Amoruso E, Allegrini S, Miglio U, Paganotti A, Ribaldone R, Monga G. Latent human polyomavirus infection in pregnancy: investigation of possible transplacental transmission. Pathology. 2008;40:72–77. doi: 10.1080/00313020701716458. [DOI] [PubMed] [Google Scholar]
- Boyd VAL, Butel JS. Demonstration of infectious deoxyribonucleic acid in transformed cells. I. Recovery of simian virus 40 from yielder and nonyielder transformed cells. J. Virol. 1972;10:399–409. doi: 10.1128/jvi.10.3.399-409.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner G, Burger A, Neumann-Haefelin D, Reinke C, Helwig H. Isolation of simian virus 40 from a newborn child. J. Clin. Microbiol. 1977;5:250–252. doi: 10.1128/jcm.5.2.250-252.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DW, Gardner SD, Gibson PE, Field AM. BK virus specific IgM responses in cord sera, young children and healthy adults detected by RIA. Arch. Virol. 1984;82:149–160. doi: 10.1007/BF01311159. [DOI] [PubMed] [Google Scholar]
- Butel JS. Viral carcinogenesis: revelation of molecular mechanisms and etiology of human disease. Carcinogenesis. 2000;21:405–426. doi: 10.1093/carcin/21.3.405. [DOI] [PubMed] [Google Scholar]
- Butel JS. SV40, human infections, and cancer: emerging concepts and causality considerations. In: Khalili K, Jeang KT, editors. Viral Oncology: Basic Science and Clinical Applications. Wiley-Blackwell; 2008. in press. [Google Scholar]
- Butel JS, Arrington AS, Wong C, Lednicky JA, Finegold MJ. Molecular evidence of simian virus 40 infections in children. J. Infect. Dis. 1999a;180:884–887. doi: 10.1086/314915. [DOI] [PubMed] [Google Scholar]
- Butel JS, Jafar S, Wong C, Arrington AS, Opekun AR, Finegold MJ, Adam E. Evidence of SV40 infections in hospitalized children. Hum. Pathol. 1999b;30:1496–1502. doi: 10.1016/s0046-8177(99)90173-9. [DOI] [PubMed] [Google Scholar]
- Butel JS, Lednicky JA. Cell and molecular biology of simian virus 40: implications for human infections and disease. J. Natl. Cancer Inst. 1999;91:119–134. doi: 10.1093/jnci/91.2.119. [DOI] [PubMed] [Google Scholar]
- Butel JS, Ozbun MA. Viral oncoprotein interactions with cellular proteins: Simian virus 40 large T-antigen and p53 as models. In: Adolph KW, editor. Methods in Molecular Genetics. Vol. 4: Molecular Virology Techniques, Part A. Academic Press; San Diego: 1994. pp. 282–309. [Google Scholar]
- Butel JS, Tevethia SS, Melnick JL. Oncogenicity and cell transformation by papovavirus SV40: The role of the viral genome. Adv. Cancer Res. 1972;15:1–55. doi: 10.1016/s0065-230x(08)60371-1. [DOI] [PubMed] [Google Scholar]
- Campen MJ, Milazzo ML, Fulhorst CF, Akata CJO, Koster F. Characterization of shock in a hamster model of hantavirus infection. Virology. 2006;356:45–49. doi: 10.1016/j.virol.2006.07.044. [DOI] [PubMed] [Google Scholar]
- Candotti D, Danso K, Parsyan A, Dompreh A, Allain JP. Maternal-fetal transmission of human parvovirus B19 genotype 3. J. Infect. Dis. 2006;194:608–611. doi: 10.1086/506450. [DOI] [PubMed] [Google Scholar]
- Cicala C, Pompetti F, Nguyen P, Dixon K, Levine AS, Carbone M. SV40 small t deletion mutants preferentially transform mononuclear phagocytes and B lymphocytes in vivo. Virology. 1992;190:475–479. doi: 10.1016/0042-6822(92)91237-o. [DOI] [PubMed] [Google Scholar]
- Cubitt CL. Molecular genetics of the BK virus. In: Ahsan N, editor. Polyomaviruses and human diseases. Springer Science; Georgetown, TX: 2006. pp. 85–95. [Google Scholar]
- Cutrone R, Lednicky J, Dunn G, Rizzo P, Bocchetta M, Chumakov K, Minor P, Carbone M. Some oral poliovirus vaccines were contaminated with infectious SV40 after 1961. Cancer Res. 2005;65:10273–10279. doi: 10.1158/0008-5472.CAN-05-2028. [DOI] [PubMed] [Google Scholar]
- Demmler GJ, Infectious Diseases Society of America and Centers for Disease Control Summary of a workshop on surveillance for congenital cytomegalovirus disease. Rev. Infect. Dis. 1991;13:315–329. doi: 10.1093/clinids/13.2.315. [DOI] [PubMed] [Google Scholar]
- Diamandopoulos GT. Induction of lymphocytic leukemia, lymphosarcoma, reticulum cell sarcoma, and osteogenic sarcoma in the Syrian golden hamster by oncogenic DNA simian virus 40. J. Natl. Cancer Inst. 1973;50:1347–1365. doi: 10.1093/jnci/50.5.1347. [DOI] [PubMed] [Google Scholar]
- Engels EA, Chen J, Viscidi RP, Shah KV, Daniel RW, Chatterjee N, Klebanoff MA. Poliovirus vaccination during pregnancy, maternal seroconversion to simian virus 40, and risk of childhood cancer. Am. J. Epidemiol. 2004;160:306–316. doi: 10.1093/aje/kwh219. [DOI] [PubMed] [Google Scholar]
- Forsman ZH, Lednicky JA, Fox GE, Willson RC, White ZS, Halvorson SJ, Wong C, Lewis AM, Jr., Butel JS. Phylogenetic analysis of polyomavirus simian virus 40 from monkeys and humans reveals genetic variation. J. Virol. 2004;78:9306–9316. doi: 10.1128/JVI.78.17.9306-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Butel JS, Carbone M. SV40 and human tumours: Myth, association or causality? Nat. Rev. Cancer. 2002;2:957–964. doi: 10.1038/nrc947. [DOI] [PubMed] [Google Scholar]
- Gerber P. Virogenic hamster tumor cells: induction of virus synthesis. Science. 1964;145:833. doi: 10.1126/science.145.3634.833. [DOI] [PubMed] [Google Scholar]
- Gerber P, Kirschstein RL. SV40-induced ependymomas in newborn hamsters. I. Virus-tumor relationships. Virology. 1962;18:582–588. doi: 10.1016/0042-6822(62)90061-2. [DOI] [PubMed] [Google Scholar]
- Girardi AJ, Sweet BH, Hilleman MR. Factors influencing tumor induction in hamsters by vacuolating virus, SV40. Proc. Soc. Exp. Biol. Med. 1963;112:662–667. doi: 10.3181/00379727-112-28133. [DOI] [PubMed] [Google Scholar]
- Gowen BB, Barnard DL, Smee DF, Wong MH, Pace AM, Jung KH, Winslow SG, Bailey KW, Blatt LM, Sidwell RW. Interferon alfacon-1 protects hamsters from lethal Pichinde virus infection. Antimicrob. Agents Chemother. 2005;49:2378–2386. doi: 10.1128/AAC.49.6.2378-2386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jittimanee J, Sermswan RW, Puapairoj A, Maleewong W, Wongratanacheewin S. Cytokine expression in hamsters experimentally infected with Opisthorchis viverrini. Parasite Immunol. 2007;29:159–167. doi: 10.1111/j.1365-3024.2006.00929.x. [DOI] [PubMed] [Google Scholar]
- Krieg P, Scherer G. Cloning of SV40 genomes from human brain tumors. Virology. 1984;138:336–340. doi: 10.1016/0042-6822(84)90357-x. [DOI] [PubMed] [Google Scholar]
- Kroczynska B, Cutrone R, Bocchetta M, Yang H, Elmishad AG, Vacek P, Ramos-Nino M, Mossman BT, Pass HI, Carbone M. Crocidolite asbestos and SV40 are cocarcinogens in human mesothelial cells and in causing mesothelioma in hamsters. Proc. Natl. Acad. Sci. USA. 2006;103:14128–14133. doi: 10.1073/pnas.0604544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky JA, Butel JS. Tissue culture adaptation of natural isolates of simian virus 40: changes occur in viral regulatory region but not in carboxy-terminal domain of large T-antigen. J. Gen. Virol. 1997;78:1697–1705. doi: 10.1099/0022-1317-78-7-1697. [DOI] [PubMed] [Google Scholar]
- Lednicky JA, Butel JS. Consideration of PCR methods for the detection of SV40 in tissue and DNA specimens. Dev. Biol. Stand. 1998;94:155–164. [PubMed] [Google Scholar]
- Lednicky JA, Butel JS. Simian virus 40 regulatory region structural diversity and the association of viral archetypal regulatory regions with human brain tumors. Semin. Cancer Biol. 2001;11:39–47. doi: 10.1006/scbi.2000.0345. [DOI] [PubMed] [Google Scholar]
- Lednicky JA, Garcea RL, Bergsagel DJ, Butel JS. Natural simian virus 40 strains are present in human choroid plexus and ependymoma tumors. Virology. 1995a;212:710–717. doi: 10.1006/viro.1995.1529. [DOI] [PubMed] [Google Scholar]
- Lednicky JA, Wong C, Butel JS. Artificial modification of the viral regulatory region improves tissue culture growth of SV40 strain 776. Virus Res. 1995b;35:143–153. doi: 10.1016/0168-1702(94)00093-r. [DOI] [PubMed] [Google Scholar]
- Malkin D, Chilton-MacNeill S, Meister LA, Sexsmith E, Diller L, Garcea RL. Tissue-specific expression of SV40 in tumors associated with the Li-Fraumeni syndrome. Oncogene. 2001;20:4441–4449. doi: 10.1038/sj.onc.1204583. [DOI] [PubMed] [Google Scholar]
- Martini F, Iaccheri L, Lazzarin L, Carinci P, Corallini A, Gerosa M, Iuzzolino P, Barbanti-Brodano G, Tognon M. SV40 early region and large T antigen in human brain tumors, peripheral blood cells, and sperm fluids from healthy individuals. Cancer Res. 1996;56:4820–4825. [PubMed] [Google Scholar]
- McCance DJ, Mims CA. Transplacental transmission of polyoma virus in mice. Infect. Immun. 1977;18:196–202. doi: 10.1128/iai.18.1.196-202.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNees AL, Vilchez RA, Heard TC, Sroller V, Wong C, Herron AJ, Hamilton MJ, Davis WC, Butel JS. SV40 lymphomagenesis in Syrian golden hamsters. Virology. 2008 doi: 10.1016/j.virol.2008.10.035. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNees AL, White ZS, Zanwar P, Vilchez RA, Butel JS. Specific and quantitative detection of human polyomaviruses BKV, JCV, and SV40 by real time PCR. J. Clin. Virol. 2005;34:52–62. doi: 10.1016/j.jcv.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Melby PC, Chandrasekar B, Zhao W, Coe JE. The hamster as a model of human visceral leishmaniasis: Progressive disease and impaired generation of nitric oxide in the face of a prominent Th1-like cytokine response. J. Immunol. 2001;166:1912–1920. doi: 10.4049/jimmunol.166.3.1912. [DOI] [PubMed] [Google Scholar]
- Melnick JL, Stinebaugh S. Excretion of vacuolating SV-40 virus (papova virus group) after ingestion as a contaminant of oral poliovaccine. Proc. Soc. Exp. Biol. Med. 1962;109:965–968. doi: 10.3181/00379727-109-27392. [DOI] [PubMed] [Google Scholar]
- Minor P, Pipkin PA, Cutler K, Dunn G. Natural infection and transmission of SV40. Virology. 2003;314:403–409. doi: 10.1016/s0042-6822(03)00435-5. [DOI] [PubMed] [Google Scholar]
- Mizgerd JP, Skerrett SJ. Animal models of human pneumonia. Am. J. Physiol. Lung Cell. Mol. Physiol. 2008;294:L387–L398. doi: 10.1152/ajplung.00330.2007. [DOI] [PubMed] [Google Scholar]
- Nelles MJ, Duncan WR, Streilein JW. Immune response to acute virus infection in the Syrian hamster. II. Studies on the identity of virus-induced cytotoxic effector cells. J. Immunol. 1981;126:214–218. [PubMed] [Google Scholar]
- Oldstone MB, Dixon FJ. Disease accompanying in utero viral infection. The role of maternal antibody in tissue injury after transplacental infection with lymphocytic choriomeningitis virus. J. Exp. Med. 1972;135:827–838. doi: 10.1084/jem.135.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paisley JE, Hinckley AF, O’Leary DR, Kramer WC, Lanciotti RS, Campbell GL, Hayes EB. West Nile virus infection among pregnant women in a northern Colorado community, 2003 to 2004. Pediatrics. 2006;117:814–820. doi: 10.1542/peds.2005-1187. [DOI] [PubMed] [Google Scholar]
- Paryani SG, Arvin AM. Intrauterine infection with varicella-zoster virus after maternal varicella. N. Engl. J. Med. 1986;314:1542–1546. doi: 10.1056/NEJM198606123142403. [DOI] [PubMed] [Google Scholar]
- Pereira L, Maidji E, McDonagh S, Tabata T. Insights into viral transmission at the uterine-placental interface. Trends Microbiol. 2005;13:164–174. doi: 10.1016/j.tim.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Pietropaolo V, Di Taranto C, Degener AM, Jin L, Sinibaldi L, Baiocchini A, Melis M, Orsi N. Transplacental transmission of human polyomavirus BK. J. Med. Virol. 1998;56:372–376. doi: 10.1002/(sici)1096-9071(199812)56:4<372::aid-jmv14>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Proceedings of the Second International Conference on Live Poliovirus Vaccines; Washington, DC, Pan American Health Organization. 1960; Papers and discussions held. Scientific Publication No. 50. [Google Scholar]
- Quax W, Egberts WV, Hendriks W, Quax-Jeuken Y, Bloemendal H. The structure of the vimentin gene. Cell. 1983;35:215–223. doi: 10.1016/0092-8674(83)90224-6. [DOI] [PubMed] [Google Scholar]
- Rachlin J, Wollmann R, Dohrmann G. Inoculation of simian virus40 into pregnant hamsters can induce tumors in offspring. Lab. Invest. 1988;58:26–30. [PubMed] [Google Scholar]
- Rayfield EJ, Kelly KJ, Yoon JW. Rubella virus-induced diabetes in the hamster. Diabetes. 1986;35:1278–1281. doi: 10.2337/diab.35.11.1278. [DOI] [PubMed] [Google Scholar]
- Sabin AB, Koch MA. Behavior of noninfectious SV 40 viral genome in hamster tumor cells: induction of synthesis of infectious virus. Proc. Natl. Acad. Sci. USA. 1963;50:407–417. doi: 10.1073/pnas.50.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siirin MT, Duan T, Lei H, Guzman H, Travassos da Rosa APA, Watts DM, Xiao SY, Tesh RB. Chronic St. Louis encephalitis virus infection in the golden hamster (Mesocricetus auratus) Am. J. Trop. Med. Hyg. 2007;76:200–306. [PubMed] [Google Scholar]
- Sroller V, Vilchez RA, Stewart AR, Wong C, Butel JS. Influence of the viral regulatory region on tumor induction by simian virus 40 in hamsters. J. Virol. 2008;82:871–879. doi: 10.1128/JVI.01626-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CE, Beasley RP, Tsui J, Lee WC. Vertical transmission of hepatitis B antigen in Taiwan. N. Engl. J. Med. 1975;292:771–774. doi: 10.1056/NEJM197504102921503. [DOI] [PubMed] [Google Scholar]
- Stewart AR, Lednicky JA, Butel JS. Sequence analyses of human tumor-associated SV40 DNAs and SV40 viral isolates from monkeys and humans. J. Neurovirol. 1998;4:182–193. doi: 10.3109/13550289809114518. [DOI] [PubMed] [Google Scholar]
- Stratton K, Almario DA, McCormick MC. Immunization Safety Review: SV40 Contamination of Polio Vaccine and Cancer. The National Academies Press; Washington, DC: 2003. [PubMed] [Google Scholar]
- Suzuki SO, Mizoguchi M, Iwaki T. Detection of SV40 T antigen genome in human gliomas. Brain Tumor Pathol. 1997;14:125–129. doi: 10.1007/BF02478881. [DOI] [PubMed] [Google Scholar]
- Sweet BH, Hilleman MR. The vacuolating virus, S.V.40. Proc. Soc. Exp. Biol. Med. 1960;105:420–427. doi: 10.3181/00379727-105-26128. [DOI] [PubMed] [Google Scholar]
- Tesh RB, Siirin M, Guzman H, Travassos da Rosa APA, Wu X, Duan T, Lei H, Nunes MR, Xiao SY. Persistent West Nile virus infection in the golden hamster: Studies on its mechanism and possible implications for other flavivirus infections. J. Infect. Dis. 2005;192:287–295. doi: 10.1086/431153. [DOI] [PubMed] [Google Scholar]
- Thomas MA, Spencer JF, La Regina MC, Dhar D, Tollefson AE, Toth K, Wold WSM. Syrian hamster as a permissive immunocompetent animal model for the study of oncolytic adenovirus vectors. Cancer Res. 2006;66:1270–1276. doi: 10.1158/0008-5472.CAN-05-3497. [DOI] [PubMed] [Google Scholar]
- van Ekdom LT, Herbrink P, Meddens MJ. Hamster model for herpes simplex virus infection of the central nervous system. Infection. 1987;15:125–127. doi: 10.1007/BF01650214. [DOI] [PubMed] [Google Scholar]
- Vanchiere JA, Bellini WJ, Moyer SA. Hypermutation of the phosphoprotein and altered mRNA editing in the hamster neurotropic strain of measles virus. Virology. 1995;207:555–561. doi: 10.1006/viro.1995.1116. [DOI] [PubMed] [Google Scholar]
- Vanchiere JA, Nicome RK, Greer JM, Demmler GJ, Butel JS. Frequent detection of polyomaviruses in stool samples from hospitalized children. J. Infect. Dis. 2005a;192:658–664. doi: 10.1086/432076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanchiere JA, White ZS, Butel JS. Detection of BK virus and simian virus 40 in the urine of healthy children. J. Med. Virol. 2005b;75:447–454. doi: 10.1002/jmv.20287. [DOI] [PubMed] [Google Scholar]
- Vilchez RA, Brayton CF, Wong C, Zanwar P, Killen DE, Jorgensen JL, Butel JS. Differential ability of two simian virus 40 strains to induce malignancies in weanling hamsters. Virology. 2004;330:168–177. doi: 10.1016/j.virol.2004.09.024. [DOI] [PubMed] [Google Scholar]
- Vilchez RA, Butel JS. Emergent human pathogen simian virus 40 and its role in cancer. Clin. Microbiol. Rev. 2004;17:495–508. doi: 10.1128/CMR.17.3.495-508.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez RA, Kozinetz CA, Butel JS. Conventional epidemiology and the link between SV40 and human cancers. Lancet Oncol. 2003;4:188–191. doi: 10.1016/s1470-2045(03)01024-6. [DOI] [PubMed] [Google Scholar]
- Vilchez RA, Madden CR, Kozinetz CA, Halvorson SJ, White ZS, Jorgensen JL, Finch CJ, Butel JS. Association between simian virus 40 and non-Hodgkin lymphoma. Lancet. 2002;359:817–823. doi: 10.1016/S0140-6736(02)07950-3. [DOI] [PubMed] [Google Scholar]
- Virgin S. Pathogenesis of viral infection. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Straus SE, Martin MA, Roizman B, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 328–388. [Google Scholar]
- Weggen S, Bayer TA, Von Deimling A, Reifenberger G, Von Schweinitz D, Wiestler OD, Pietsch T. Low frequency of SV40, JC and BK polyomavirus sequences in human medulloblastomas, meningiomas and ependymomas. Brain Pathol. 2000;10:85–92. doi: 10.1111/j.1750-3639.2000.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner LP, Herndon RM, Narayan O, Johnson RT, Shah K, Rubinstein LJ, Preziosi TJ, Conley FK. Isolation of virus related to SV40 from patients with progressive multifocal leukoencephalopathy. N. Engl. J. Med. 1972;286:385–390. doi: 10.1056/NEJM197202242860801. [DOI] [PubMed] [Google Scholar]
- Wong KT, Grosjean I, Brisson C, Blanquier B, Fevre-Montange M, Bernard A, Loth P, Georges-Courbot MC, Chevallier M, Akaoka H, Marianneau P, Lam SK, Wild TF, Deubel V. A golden hamster model for human acute Nipah virus infection. Am. J. Pathol. 2003;163:2127–2137. doi: 10.1016/S0002-9440(10)63569-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogo Y, Sugimoto C. The archetype concept and regulatory region rearrangement. In: Khalili K, Stoner GL, editors. Human polyomaviruses: Molecular and clinical perspectives. Wiley-Liss, Inc.; New York: 2001. pp. 127–148. [Google Scholar]
- Zhang S, McNees AL, Butel JS. Quantification of vertical transmission of Murine polyoma virus by real-time quantitative PCR. J. Gen. Virol. 2005;86:2721–2729. doi: 10.1099/vir.0.81168-0. [DOI] [PubMed] [Google Scholar]
- Zhen HN, Zhang X, Bu XY, Zhang ZW, Huang WJ, Zhang P, Liang JW, Wang XL. Expression of the simian virus 40 large tumor antigen (Tag) and formation of Tag-p53 and Tag-pRb complexes in human brain tumors. Cancer. 1999;86:2124–2132. [PubMed] [Google Scholar]


