Abstract
Hurricanes have the potential to alter the structures of coastal ecosystems and generate pathogen-laden floodwaters that threaten public health. To examine the impact of hurricanes on urban systems, we compared microbial community structures in samples collected after Hurricane Katrina and before and after Hurricane Rita. We extracted environmental DNA and sequenced small-subunit ribosomal RNA (SSU rRNA) gene clone libraries to survey microbial communities in floodwater, water and sediment samples collected from Lake Charles, Lake Pontchartrain, the 17th Street and Industrial Canals in New Orleans and raw sewage. Correspondence Analysis showed that microbial communities associated with sediments formed one cluster while communities associated with lake and Industrial Canal water formed a second. Communities associated with water from the 17th Street Canal and floodwaters collected in New Orleans showed similarity to communities in raw sewage and contained a number of sequences associated with possible pathogenic microbes. This suggests that a distinct microbial community developed in floodwaters following Hurricane Katrina and that microbial community structures as a whole might be sensitive indicators of ecosystem health and serve as “sentinels” of water quality in the environment.
Introduction
Hurricane Katrina, a Category 4 storm, and Hurricane Rita, a Category 3 storm, hit the shores of the US Gulf Coast on August 29th, and September 24th, 2005 respectively. These storms resulted in massive flooding and subsequent destruction of property and human lives. Hurricanes may have a limited impact on aquatic ecosystems relative to land systems (1), but Hurricane Katrina passed through a major city and generated floodwaters which contained a complex mixture of organic and inorganic chemicals associated with urban runoff, sediments resuspended from coastal areas, and human sewage (2).
The volume and composition of floodwaters created a potential public health crisis, and, when pumped from the city, threatened water quality in Lake Pontchartrain. Floodwaters contained high numbers of Aeromonas spp. and pathogenic Vibrio species; these microbes and organic pollutants were also detected in soils and sediments from flooded areas (3). Fecal-associated bacteria were observed for at least a month after the storm in the waters of Lake Pontchartrain within 5 km of the pump discharge site (4). Sediments deposited from floodwaters may still result in adverse effects on human health (5). Despite water quality returning to pre-hurricane conditions shortly afterwards, the ecological effects of these hurricanes are likely still an issue (5–7). Since many microbial communities show temporal and spatial patterns discernable with molecular techniques that are driven largely by environmental factors (8), microbial community analysis can provide a sensitive indication of the health of ecosystems (9).
This study builds on our previous study that found relatively high potential pathogen concentrations and sewage “signatures” in the 17th Street Canal, which received floodwater pumped from New Orleans (5). This prompted a more extensive survey of samples, including samples of floodwaters collected from New Orleans and Lake Charles, along with the application of more in-depth analyses that we present in this manuscript. In this study we present the first microbial community analysis with sufficient coverage (> 5,000 sequences from 28 samples) to explore the overall microbial community structure in systems disturbed by Hurricanes Katrina and Rita. We applied univariate and multivariate methods to compare community structure across the samples collected. We used ordination methods to summarize our multivariate data. This analysis revealed floodwaters contained a cluster of Operational Taxonomic Units (OTUs) that includes microbes associated with human waste.
Experimental Section
Sample Collection and DNA Extraction
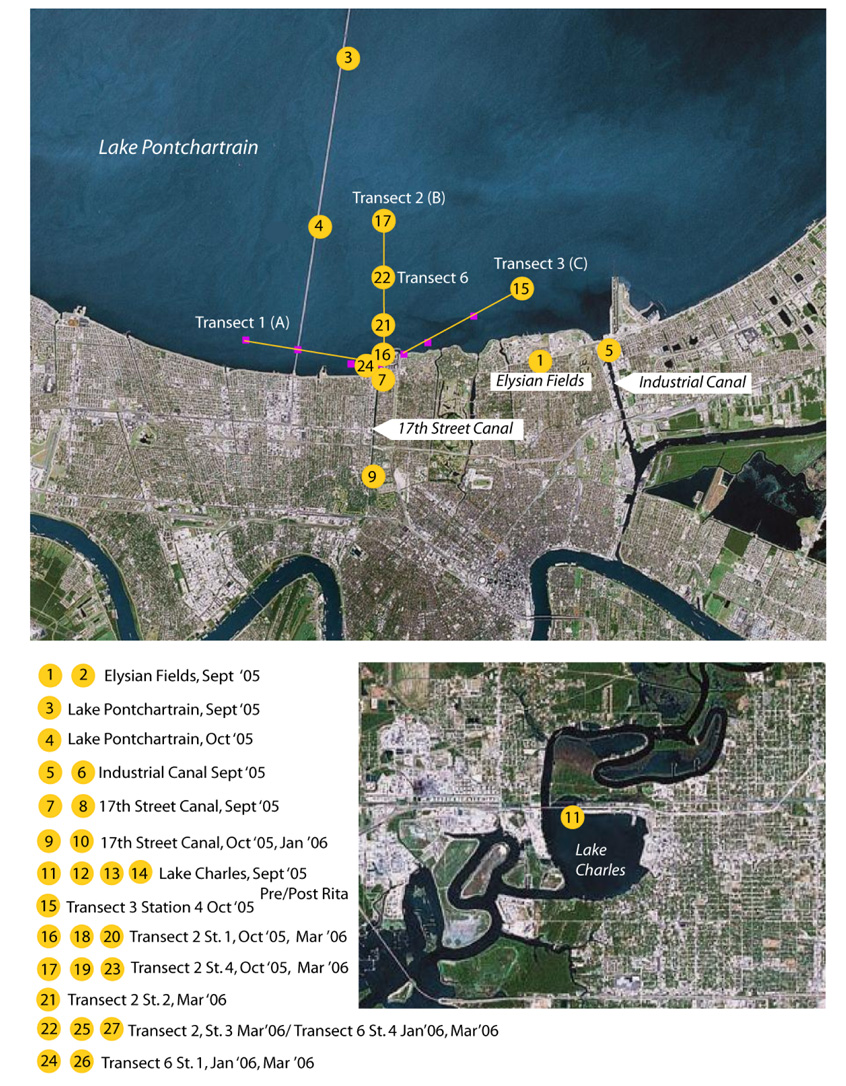
Sampling in the New Orleans and Lake Charles areas began on September 16th, 2005 (Figure 1, Table 1). The September samples were collected approximately 2 weeks after the levees broke and a week before the floodwaters were pumped out of the city. Water samples included two floodwater sites (Elysian Fields and Lake Charles after Rita), two canals (17th Street and Industrial Canals), transects radiating out from the 17th Street Canal into Lake Pontchartrain, and samples from Lake Pontchartrain and Lake Charles proper. The Lake Charles samples collected before Rita were collected off a dock at the Lake Charles Yacht Club at the end of the I–10 service road, while those collected after Rita were collected from floodwaters in the same area. Sediments were collected in March, 2006.
Figure 1.
Map overview of the sampling locations at Lake Pontchartrain and Lake Charles in Louisiana. Only the first sample number in a series of samples collected over time at the same location (see groupings below the map) is indicated on the map. Refer to the sampling codes in Table 1 and Table S1 for additional sample information. Pink squares indicate original transects sampled in Sinigalliano et al. (2007). This study focused on Transect 2 (Transect B in Sinigalliano et al. (2007)). Note that Transect 2, Station 3 and Transect 6, Station 4 refer to approximately the same location. Image modified from http://earth.google.com.
Table 1.
Sample codes, descriptions and geospatial coordinates.
| Sample | Sample | ||||||
|---|---|---|---|---|---|---|---|
| Code | Abbreviation | Location Name | Sampling Date | Sample Type | Size Fraction | Latitude | Longitude |
| 1 | A4S16P_ML | Elysian Fields | September 16, 2005 | Floodwater | particulate* | 30.0255 | −90.0610 |
| 2 | A4S16S_ML | Elysian Fields | September 16, 2005 | Floodwater | free** | 30.0255 | −90.0610 |
| 3 | C6S16_ML | Lake Pontchartrain | September 16, 2005 | Water | free | 30.1277 | −90.1353 |
| 4 | C7p3_ML | Lake Pontchartrain | October 5, 2005 | Water | particulate | 30.0710 | −90.1451 |
| 5 | I9S18D_ML | Industrial Canal | September 18, 2005 | Water | free | 30.0303 | −90.0331 |
| 6 | I9S18K_ML | Industrial Canal | September 18, 2005 | Water | particulate | 30.0303 | −90.0331 |
| 7 | L7S16_ML | 17th Street Canal | September 16, 2005 | Water | free | 30.0198 | −90.1219 |
| 8 | L73_ML | 17th Street Canal | September 16, 2005 | Water | particulate | 30.0198 | −90.1219 |
| 9 | 051024-S1-W-T | 17th Street Canal | October 24, 2005 | Water | Whole Water | 29.9868 | −90.1251 |
| 10 | 060127-S1-W-T | 17th Street Canal | January 27, 2006 | Water | Whole Water | 29.9868 | −90.1251 |
| 11 | Lc2Y10_ML | Lake Charles | September 22, 2005 | Water | free | 30.2363 | −93.2417 |
| 12 | Lc2yS22_ML | Lake Charles | September 22, 2005 | Water | particulate | 30.2363 | −93.2417 |
| 13 | Lc2z_ML | Lake Charles | September 26, 2005 | Floodwater | particulate | 30.2363 | −93.2417 |
| 14 | Lc2_ML | Lake Charles | September 26, 2005 | Floodwater | free | 30.2363 | −93.2417 |
| 15 | 051011-T3S4-W-T | Transect 3 Station 4 | October 11, 2005 | Water | Whole Water | 30.0500 | −90.0681 |
| 16 | 051011-T2S1-W-T | Transect 2 Station 1 | October 11, 2005 | Water | Whole Water | 30.0264 | −90.1210 |
| 17 | 051011-T2S4-W-T | Transect 2 Station 4 | October 11, 2005 | Water | Whole Water | 30.0719 | −90.1209 |
| 18 | 051025-T2S1-W-T | Transect 2 Station 1 | October 25, 2005 | Water | Whole Water | 30.0264 | −90.1210 |
| 19 | 051025-T2S4-W-T | Transect 2 Station 4 | October 25, 2005 | Water | Whole Water | 30.0719 | −90.1209 |
| 20 | 060329-T2S1-S-T | Transect 2 Station 1 | March 29, 2006 | Sediment | NA | 30.0264 | −90.1210 |
| 21 | 060329-T2S2-S-T | Transect 2 Station 2 | March 29, 2006 | Sediment | NA | 30.0355 | −90.1209 |
| 22 | 060329-T2S3-S-T | Transect 2 Station 3 | March 29, 2006 | Sediment | NA | 30.0543 | −90.1210 |
| 23 | 060329-T2S4-S-T | Transect 2 Station 4 | March 29, 2006 | Sediment | NA | 30.0719 | −90.1209 |
| 24 | 060127-T6S1-W-T | Transect 6 Station 1 | January 27, 2006 | Water | Whole Water | 30.0248 | −90.1227 |
| 25 | 060127-T6S4-W-T | Transect 6 Station 4 | January 27, 2006 | Water | Whole Water | 30.0543 | −90.1211 |
| 26 | 060329-T6S1-W-T | Transect 6 Station 1 | March 29, 2006 | Water | Whole Water | 30.0248 | −90.1227 |
| 27 | 060329-T6S4-W-T | Transect 6 Station 4 | March 29, 2006 | Water | Whole Water | 30.0543 | −90.1211 |
| 28 | Gctb_ML | Lake Charles Treatment Facility | December 21, 2005 | Raw Sewage | NA | NA | NA |
> 3 µm fraction
< 3 µm fraction
All surface water samples collected in the month of September and on October 5th were collected by hand with a bucket. These samples were size-fractionated by filtration to differentiate free-living and particle-associated communities as described previously (10) but preliminary results (LaMontagne et al., unpublished) showed that differences between sites were greater than differences between fractions, so we sampled whole water after September. A sewage sample was collected from the primary influent (raw, untreated) at the Lake Charles sewage treatment plant with a drop bucket on Dec 21st, 2005 and transported on ice to McNeese State University.
Raw effluent was shaken briefly and then 30 ml was centrifuged for 10 min at 10,000 g. The pellet was resuspended in 1 × Phosphate Buffered Saline (PBS), transferred to six microcentrifuge tubes and centrifuged again (2 min, 16,000 g). The sewage samples and all water samples collected in September were frozen and then extracted with a Gentra Puregene kit (Qiagen, Valencia, CA) followed by a CTAB/chloroform extraction. Samples collected on October 11th, 2005 through March 2006 followed protocols established in our laboratory and described in Sinigalliano et al. (5).
Microbial Assemblage Analysis
PCR amplifications and cloning reactions followed protocols described in Sinigalliano et al. (5) with the following additions. We added PrimeSafe™ (Smiths Detection, Boston, MA) to reactions to suppress mispriming, improve overall efficiency of the reactions, and minimize chimera formation. Between two and three separate PCR reactions were pooled prior to cloning and between 192 and 384 clones were sequenced from each library in the forward and reverse directions.
Our bioinformatics processing of sequence data followed protocols described in Sinigalliano et al. and Santelli et al. (5,11). We only retained and further processed clone sequences for which forward and reverse reads assembled to form a single contiguous consensus sequence. An initial BLAST search served to identify any vector sequences or non-rRNA sequences obtained. We then further screened for anomalous sequences in each of the clone libraries using the computer program MALLARD (URL: http://www.bioinformatics-toolkit.org/Mallard/index.html). The analysis of putative chimeras was restricted by the default cut-off line of 99.9% (12,13). All anomalous sequences identified by MALLARD were considered chimeric and excluded from further analysis. The percentage of chimeras in our clone libraries averaged 11% which is within the range reported in the literature of 0–46% (13) (Table S1).
We used BLAST and Ribosomal Database Project (RDP) II’s Classifier (14) to identify the closest relatives of the sequences and to obtain taxonomic information about our environmental sequence data. To assign domains to sequences, we aligned sequences within the ARB (15) program and further adjusted the alignment manually. We then used the “quick add sequences to tree” feature, to add all the sequences to the reference tree found in the SSU Reference 91 (07-18-07) version of the SILVA-ARB database (16).
We assigned OTUs, at a 99% cut-off criterion, to clusters of sequences using the program DOTUR (17). This level of similarity agreed with the placement of sequences in our ARB tree and also coincided well with RDP classifier taxonomic assignments. We employed customized Perl scripts to construct OTU abundance × sample matrices from DOTUR output. These abundance matrices served as the input for our multivariate and univariate analyses.
Multivariate and Univariate Analyses
We used ordination and scaling multivariate methods to explore patterns in our OTU abundance data. To decide between linear and unimodal response models for our data, we carried out a Detrended Correspondence Analysis (DCA) as implemented in the program CANOCO 4.5 (18). A length of the first DCA ordination axis greater than 4 S.D. indicated a unimodal species response. We conducted DCA analyses using a bacterial OTU abundance matrix (28 samples × 2,139 OTUs) created from DOTUR output as input for CANOCO 4.5 (see above). We ran the DCA analysis with default settings and log transformed the data. We chose not to run the analysis with the rare-species down-weighted option because we wanted to show the positioning of potentially pathogenic OTUs in relation to the samples on our ordination diagrams and most of the potentially pathogenic OTUs were rare. Due to the large numbers of OTUs it was not possible to display all names to provide their taxonomic affiliation but information regarding potentially pathogenic OTU clusters is available in the supplemental data.
We used CANOCO 4.5 to perform a Canonical Correspondence Analysis (CCA) on a subset of 9 samples (986 OTUs) for which corresponding environmental data was available (Green et al., unpublished). We performed CCA with scaling focused on inter-species distances so that OTUs and environmental variables formed a biplot. We first ran the program with automatic forward selection to limit the number of explanatory variables (environmental variables) to significant ones and then reran the analysis using manual selection to chose these variables as explanatory variables and retain the remaining variables as supplementary variables. To statistically evaluate the significance of the first canonical axis and of all canonical axes together we used a Monte Carlo reduced model test with 499 unrestricted permutations.
We used the program Primer v6 (19) to estimate species richness (Margalef’s d), species evenness (Pielou’s J’) and Simpson diversity indices (1-lambda’) for all our samples. We determined percent OTU overlap between sample pairs using the output from DOTUR as input for the SONS (Shared OTUs aNd Similarity) program that is a tool for making operational taxonomic unit-based comparisons of microbial community memberships and structures (20). We also used this program to report Theta-YC values with 95% confidence intervals for each pair of samples. Theta –YC is a non-parametric maximum likelihood estimator of similarity to compare community structures that accounts for similarities in the relative abundances among OTUs, not just community OTU overlap (21).
Results
Bacterial sequences dominated our 28 clone libraries, but we did recover 192 eukaryal and 437 archaeal clones out of the 32 libraries examined (28 considered for all three domains in this study and 4 for archaeal and eukaryal not considered in our previous study). DOTUR analysis identified 2,139 bacterial OTUs; 69 OTUs appeared related to potentially pathogenic species (Figure S1). These potential pathogens included members of the genus Aeromonas found in the 17th Street Canal and Elysian Fields samples. We also detected sequences related to Vibrio vulnificus in the sample collected at the 17th Street Canal in September and the sample collected after Hurricane Rita in Lake Charles. We refrain from calling our OTUs pathogenic “species” on the basis of an rRNA gene taxonomic assignment alone, but realize their potential for being pathogenic given their close relationship to known pathogens.
Microbial Community Structure in Raw Sewage as Compared to Environmental Samples
The most frequently occurring OTU in the library generated from sewage matched the genus Arcobacter, a genus commonly occurring in human feces and containing potential pathogens. We did not detect this OTU in any of our environmental libraries. Of the 37 OTUs occurring more than once in the library generated from sewage, 24 did not occur in libraries generated from environmental samples. Of the remaining 13 OTUs, the second most abundant overall in the Raw Sewage sample matched the genus Zoogloea (100%) and was also found in the September 17th Street Canal sample. Zoogloea species are associated with sewage sludge (22). The fourth most abundant OTU in the Raw Sewage sample matched the genus Aeromonas and was the same OTU shared by the 17th Street Canal samples and the Elysian Fields sample described above.
Species Richness and Diversity Index Measures
Species diversity, as assessed by richness and diversity indices, appeared higher in libraries generated from floodwater than from lake samples; libraries generated from sediment samples showed the highest diversity overall (Table S1). Rarefaction analyses (data not shown) performed using DOTUR confirmed the relative degrees of richness we observed in our Margalef’s d calculation of richness available through the Primer program. Simpson diversity indices covaried closely with evenness and Shannon indices (not shown). Richness estimates averaged (± S.D.) 26 ± 7 for the floodwater libraries and 17 ± 6 for the lake libraries (when classifying all canal samples as “lake” samples). When classifying the September 17th Street Canal samples as floodwater, these values change to flood 27 ± 6 and lake 15 ± 3. A two-factor ANOVA found a significant (P < 0.040) difference in richness, evenness and diversity indices between libraries generated from floodwater and lake water and no significant (P > 0. 538) difference in diversity between particle-associated and free-living fractions or interaction between sample type and fraction.
Detrended Correspondence Analyses and Percent OTU Overlap Between Samples
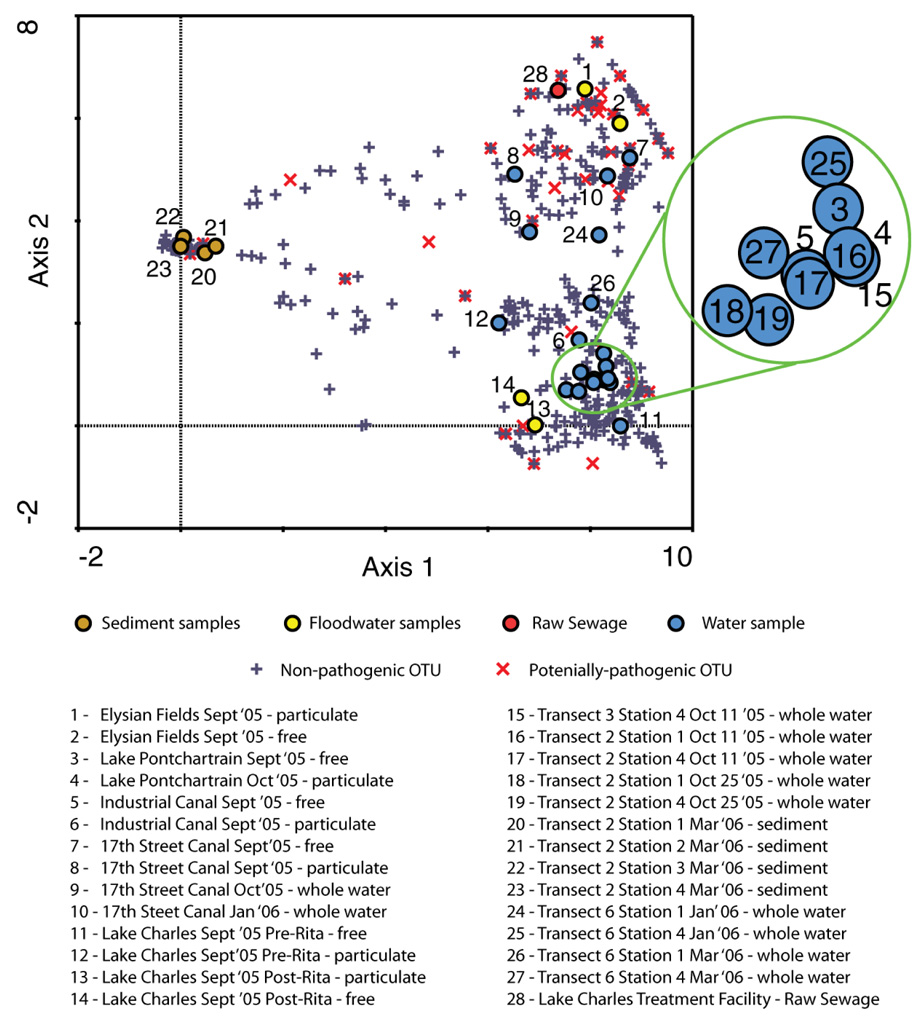
DCA revealed two distinct microbial community clusters: sediment samples and samples from lake and Industrial Canal waters (Figure 2). The remaining samples including raw sewage, floodwater, and 17th Street Canal water samples were more dispersed in the ordination plot. Samples collected from Lake Charles tended to cluster closer to samples from Lake Pontchartrain. Percent shared overlap between lake samples (Lake Charles (pre-Rita) and Lake Pontchartrain) ranged between 9–10% shared OTUs, floodwaters between 2–11% and sediments between 8–10% (Table S2).
Figure 2.
A Detrended Correspondence Analysis (DCA) ordination diagram showing 28 samples (filled circles) and bacterial OTUs at the 99% similarity cut-off value (crosses and x-marks) plotted on the first (horizontal) and second (vertical) DCA axes. The scalings on the axes are in multiples of standard deviation. The length of the gradient of our first axis (X-axis) was > 4 S.D. suggesting that some species show a unimodal response to the 1st DCA axis. This confirms a non-linear response of the species to their environment and thus justifies the use of a DCA approach. We can interpret our ordination diagram using the distance rule wherein a sample that is close to a species point is more likely to contain that species than one farther away. The first eigenvalue was 0.944 and the second was 0.787.
Elysian Fields floodwater samples clustered with the raw sewage sample and 17th Street Canal samples. Raw sewage shared the greatest percent overlap in OTUs with the Transect 2 Station 1 sediment sample (9%) followed by the 17th Street Canal sample free (<3 µm fraction) fraction (2%), although sediment samples shared very few species with all other samples in general. However, Theta-YC similarity indices, which take relative abundance into account, revealed the greater similarity of raw sewage to the September 17th Street Canal samples (Table S2).
The most abundant OTUs that clustered with the New Orleans floodwaters were classified as belonging to the genera Rhodobacter and Allochromatium (Figure S2). The Rhodobacter-related OTUs showed similarity to sequences previously generated from phototrophic sludge (23). The Allochromatium-related sequences showed similarity to sequences previously generated from landfill leachate (24). The abundance of these OTUs suggests that the floodwater contained bacteria typically associated with wastewater and highly polluted systems. Potentially pathogenic OTUs occur all over the ordination space but appear to be more concentrated in the upper right hand corner of the diagram coincident with raw sewage and floodwater samples.
Canonical Correspondence Analysis
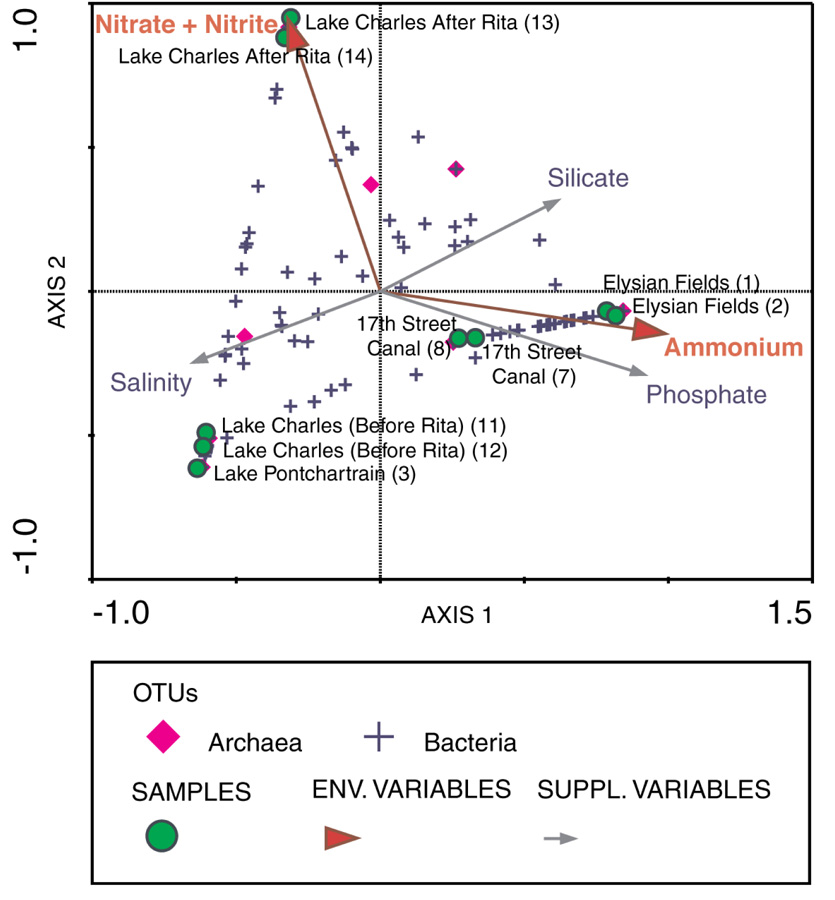
CCA suggests that microbial communities sampled from Elysian Fields and the 17th Street Canal are associated with high ammonium levels and that microbial communities sampled from Lake Charles after Hurricane Rita are associated with high nitrate and nitrite levels. The first two axes of the CCA triplot explained 30% of the variance (inertia) in the species (OTU) data, 100% of the variance in the fitted species data and the same percentage of the variance in the weighted averages and the class totals of the species with respect to the environmental variables (Figure 3). Axis one was highly correlated with ammonium (intra-set correlation coefficient = 0.9892); axis two was highly correlated with nitrate plus nitrite (intra-set correlation coefficient =0.9496). Monte Carlo tests for the first and all axes were highly significant (P=0.002) indicating that these environmental parameters may be important in explaining community diversity.
Figure 3.
Canonical Correspondence Analysis (CCA) triplot of samples and bacterial and archaeal OTUs at the 99% similarity cut-off value displaying 100% of the variance of the OTUs with respect to the environmental variables. The canonical eigenvalues for axes 1–2 of the sample analysis are 0.874 and 0.781 respectively. Environmental variables are indicated by red arrows while supplemental variables are shown using grey arrows. Samples are represented by green circles and sample numbers refer to sample codes further detailed in Table 1 and Table S1. Bacterial OTUs are represented by blue crosses while archaeal OTUs are shown as pink diamonds.
Eukaryal and Archaeal Diversity
Recovery of eukaryal and archaeal OTUs from our clone libraries varied greatly between samples. Some libraries contained only bacterial sequences (Table S1). Eukaryal sequences (Figure S3) ranged from 0 to 44% of the clone libraries and encompassed a phylogenetically broad range of taxa including representatives from the animals (such as calanoid and harpacticoid copepods, mites, mussels, oligochaete worms, bryozoans, and gastrotrichs), fungi, and protists related to lobose amoebae, unicellular green plants (such as chlamydomonad and chlorophyte relatives), cryptophytes, cercozoa, dinoflagellates, and a variety of ciliates dominated by stichotrichs. We did not detect any eukaryal sequences related to known pathogens.
Archaeal sequences comprised from 0 to 24% of our clone libraries (Figure S4) and included those branching with Methanosarcina (a methanogenic archaeon and obligate anaerobe found in a transect sediment sample from March), uncultured marine group II and III euryarchaeotes, and uncultured crenarchaeotes. The most abundant archaeal OTU branched among uncultured marine group I crenarchaeotes, including relatives of archaeal sequences derived from methane hydrates and those from Candidatus Nitrosopumilus maritimus, an ammonia-oxidizing autotrophic crenarchaeon. This cluster along with a second cluster of sequences related to archaeal sequences recently described from the Antarctic constituted the most abundant archaeal OTU class (25). Noteworthy is the fact that the former of these OTUs was almost exclusively associated with water samples while the latter predominantly with sediment samples.
Comparisons of Lake Pontchartrain Transects over Space and Time
In libraries generated from near-shore samples, the phylum Planctomycetes decreased over the sampling time period (Figure S5). In libraries generated from the offshore samples, the phylum Acidobacteria decreased over the same period. Between near-shore and offshore samples, January samples showed the largest differences. Cyanobacteria appeared relatively abundant offshore and Bacteroidetes appeared relatively abundant near-shore.
Discussion
Hurricanes Katrina and Rita appeared to change the overall microbial community of aquatic systems in flooded areas. Although we lack any pre-Hurricane Katrina samples, our comparisons of pre- and post- Hurricane Rita clone libraries revealed distinct differences in the underlying OTU composition of these samples. CCA suggested that these changes were associated with nutrient availability. Since we only had an opportunity to collect a limited number of environmental parameters, it is possible that other unmeasured parameters were equally important.
Samples originating from different kinds of sources (raw sewage vs. water column vs. sediments vs. floodwater) generally appeared to be quite distinct from one another – with some samples sharing no or few OTUs. The exceptions mostly occurred with the September and/or 17th Street Canal samples, which showed a community structure more closely associated with sewage than water and sediments. Since, when sampled in January 2006, the 17th Street Canal (#10) still plotted apart from the lake cluster, it seems that the OTUs associated with the 17th Street Canal are distinct from the lake/Industrial Canal samples despite the time of year or duration after the hurricanes. The 17th Street Canal appears chronically contaminated with fecal-associated microbes (4) and may contaminate Lake Pontchartrain. The influence of this contaminated canal decreased over the period of seven months; the microbial community observed along the transect sampled originating from the 17th Street Canal became progressively more similar to that observed in offshore waters of Lake Pontchartrain. The similarity between microbial communities in the floodwaters overlying New Orleans to sewage does not appear to be a universal feature of floodwaters generated by hurricanes, as the microbial communities in floodwaters generated by Rita appeared similar to those found in lake waters.
The high diversity in floodwater samples could reflect the contribution of sediments, soils and sewage to these communities or the effect of environmental factors on microbial diversity. Fractionation, by prefiltration, cannot explain the relatively high diversity of libraries generated from floodwaters as these samples had higher diversity than lake samples processed in a similar manner. Sediments also showed high diversity, which is consistent with the relatively high bacterial diversity of sediments (26, 27), but it does not appear likely that sediments contributed significantly to floodwater diversity. While we employed analytical methods that take into account variability in sampling depth, we acknowledge that it is very difficult to control for other biases such as extraction efficiencies of different sample types (sediments vs. water) and variations in biomass of a given domain in the starting material. DCA showed little similarity in libraries generated from sediments and those generated from floodwaters, even when only the particle-associated fraction of the floodwater samples was examined. We also ran a DCA with the four samples published in our collaborative paper (5) (data not shown) and found similar placement of 17th Street Canal samples with other 17th Street Canal samples analyzed in this study and likewise for the Industrial Canal sample. DCA revealed that the New Orleans Yard floodwater sediment sample was very different from all other sample types in the analyses (data not shown). The four DCA axes combined explained only ~20% of the variance in our data; however, non-metric multidimensional scaling (NMDS) analysis of the 28 samples also revealed distinct sediment and lake/Industrial Canal clusters with other samples falling in between (Figure S6). Because we were not able to sample through a full seasonal cycle, it is difficult to separate possible seasonal effects from hurricane-induced patterns of community structure. Sediment samples may have been very different than the initial sediments that were deposited in the city.
An alternative explanation for the high diversity is that the nutrients in the floodwater supported microbial growth. Floodwaters were laden with toxins (2), which tend to reduce microbial diversity (28); however, they also contained high concentrations of dissolved inorganic nitrogen and particulate organic carbon (Green et al., unpublished). These conditions could support a large microbial community, which following species-energy theory (29), should lead to high diversity.
Sediment clone libraries also had relatively more archaeal sequences (15–24%) than water (<10%) samples. The only exception was the Lake Pontchartrain sample collected on September 16th. This sample and the post-Rita Lake Charles sample also containing large numbers of archaeal sequences may have had a sediment archaeal signature because of the resuspension of sediments. Although we have no data on the relative efficiency of our primers in vitro using an artificial constructed microbial assemblage, we tested these primers in silico using the primer design and probe match features of the ARB software package (15) and have found them to be effective at recovering amplicons across all three domains of life. Because these primers are universal, we expect that they will amplify a greater diversity of bacterial targets often missed by other bacterial primers. While these primers themselves do not inherently favor the amplification of bacterial rRNA genes for reasons of specificity, length heterogeneities in the V4–V8 region in combination with lower eukaryotic and archaeal cell concentrations in the environment, likely contribute to a decreased number of eukaryal and archaeal amplicons overall.
Similarity between the microbial community in floodwaters generated by Hurricane Katrina and the 17th Street Canal suggests that this canal could significantly contribute to the microbial community in future flood events. The community observed in floodwaters does not appear closely associated with communities in sediments and lakes but appears to develop from the mixture of nutrients and pollution in the floodwater. Regular monitoring of the 17th Street Canal may serve as a mechanism for gauging future levels of pollution in the canal, Lake Pontchartrain, and floodwaters in the area. Microbial community analysis provides a sensitive method of monitoring the water quality in these systems.
Supplementary Material
Additional data are available on-line. Table S1 provides sample codes, clone library information, species richness and Simpson diversity indices. Table S2 provides pairwise percent OTU overlap between samples in the upper portion of the triangular matrix and Theta-YC community structure similarity values in the lower triangle. Figure S1 provides a list of 69 potentially pathogenic OTUs and their associated taxonomy. Figure S2 presents comprehensive pie charts showing a comparative bacterial taxonomic summary of the sequences in all 28 samples – clicking on the names allows one to view a taxonomic breakdown of the sequences down to genus when possible or some higher taxonomic level. Asterisks indicate taxonomic levels shared with the raw sewage sample. Figure S3 shows the placement of eukaryotic sequences within an ARB tree while Figure S4 shows the placement of archaeal sequences. Figure S5 shows pie charts indicating changes in relative abundances of bacterial phyla and classes of Proteobacteria over time at transect stations near-shore and offshore. Figure S6 is a NMDS 3-dimensional ordination of 28 samples based on standardization by total and Log(X+1) transformed OTU abundances and Bray-Curtis similarities. Refer to Table 1 for sample codes. This information is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This work was funded by the National Science Foundation (NSF)–National Institute of Environmental Health Sciences (NIEHS) Oceans and Human Health Program (NSF OCE-0430724 and NIEHS P50ES012742) and the NSF Small Grant for Exploratory Research Program (NSF OCE-0554850; NSF BES-0553138). We thank the Hou Laboratory at Louisiana State University and members of the Miami and Hawaii Oceans and Human Health Centers for logistical support. We thank Mitchell Sogin and Hilary Morrison for Woods Hole Oceans and Human Health Genomics facility support. We acknowledge Susan Huse for assistance with data matrix generation and Phillip Neal for assistance with pie chart constructions. Sequences have been deposited in GenBank under accession numbers FJ349630-FJ355433.
Footnotes
A microbial survey following Hurricanes Katrina and Rita suggests that microbial community structures might serve as “sentinels” of water quality in the environment.
Literature Cited
- 1.Valiela I, Peckol P, D'Avanzo C, Kremer J, Hersh D, Foreman K, Lajtha K, Seely B, Geyer WR, Isaji T, Crawford R. Ecological effects of major storms on coastal watersheds and coastal waters: Hurricane Bob on Cape Cod. J. Coastal Res. 1998;14:218–238. [Google Scholar]
- 2.Pardue JH, Moe WM, Mcinnis D, Thibodeaux LJ, Valsaraj KT, Maciasz E, Ven Heerden I, Korevec N, Yuan QZ. Chemical and microbiological parameters in New Orleans floodwater following hurricane Katrina. Environ. Sci. Technol. 2005;39:8591–8599. doi: 10.1021/es0518631. [DOI] [PubMed] [Google Scholar]
- 3.Presley SM, Rainwater T, Austin GP, Platt SG, Zak JC, Cobb IP, Marsland EJ, Kang T, Zhang B, Anderson TA, Cox SB, Abel MT, Leftwich B, Huddleston JR, Jeter RM, Kendall RJ. Assessment of pathogens and toxicants in New Orleans, LA following hurricane Katrina. Environ. Sci. Technol. 2006;40:468–474. doi: 10.1021/es052219p. [DOI] [PubMed] [Google Scholar]
- 4.Hou A, Laws EA, Gambrell RP, Bae H-S, Tan M, Delaune RD, Li Y, Roberts H. Pathogen indicator microbes and heavy metals in Lake Pontchartrain following hurricane Katrina. Environ. Sci. Technol. 2006;40:5904–5910. doi: 10.1021/es060946u. [DOI] [PubMed] [Google Scholar]
- 5.Sinigalliano CD, Gidley ML, Shibata T, Whitman D, Dixon TH, Laws E, Hou A, Bachoon D, Brand L, Amaral-Zettler LA, Gast RJ, Steward GF, Nigro OD, Fujioka R, Betancourt WQ, Vithanage G, Mathews J, Fleming LE, Solo-Gabriele HM. Impacts of hurricanes Katrina and Rita on the microbial landscape of the New Orleans area. Proc. Nat. Acad. Sci. 2007;104:9029–9034. doi: 10.1073/pnas.0610552104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwab KJ, Kristene G, Williams DAL, Kulbicki KM, Paullo C, Mihalic J, Breysse PN, Curriero FC, Geyh AS. Microbial and chemical assessment of regions within New Orleans, LA impacted by hurricane Katrina. Environ. Sci. Technol. 2006;41:2401–2406. doi: 10.1021/es062916x. [DOI] [PubMed] [Google Scholar]
- 7.U.S. Geological Survey. Stoeckel D, Bushon RN, Demcheck DK, Skrobialowski SC, Kephart CM, Bertke EE, Mailot BE, Mize SV, Fendick RBJ. Bacteriological water quality in the Lake Pontchartrain Basin, Louisiana, following hurricanes Katrina and Rita, September 2005. Data Series. 2005;143 [Google Scholar]
- 8.Hughes Martiny JB, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Øvreås L, Reysenbach A-L, Smith VH, Staley JT. Microbial biogeography: Putting microorganisms on the map. Nat. Rev. Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 9.Cao Y, Cherr GN, Cordova-Kreylos AL, Fan TW-M, Green PG, Higashi RM, LaMontagne MG, Scow KM, Vines CA, Yuan J, Holden PA. Relationships between sediment microbial communities and pollutants in two California salt marshes. Microbial Ecol. 2006;52:619–633. doi: 10.1007/s00248-006-9093-1. [DOI] [PubMed] [Google Scholar]
- 10.LaMontagne MG, Holden PA. Comparison of free-living and particle-associated bacterial communities in a coastal lagoon. Microbial Ecol. 2003;46:228–237. doi: 10.1007/s00248-001-1072-y. [DOI] [PubMed] [Google Scholar]
- 11.Santelli CM, Orcutt BN, Banning E, Bach W, Moyer CL, Sogin ML, Staudigel H, Edwards KJ. Abundance and diversity of microbial life in ocean crust. Nature. 2008;453:653–656. doi: 10.1038/nature06899. [DOI] [PubMed] [Google Scholar]
- 12.Ashelford KE, Chuzanova NA, Fry JC, Jones AJ, Weightman AJ. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashelford KE, Chuzanova NA, Fry JC, Jones AJ, Weightman AJ. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM. The Ribosomal Data Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucl. Acids. Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H. ARB: a software environment for sequence data. Nucl. Acids. Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruesse E, Quast C, Knittel K, Fuchs B, Ludwig W, Peplies J, Gloeckner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl. Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ter Braak CJF, Simlauer P. CANOCO Reference manual and CanoDraw for Window's User's guide: Software for Canonical Community Ordination (version 4.5) Ithaca: Microcomputer Power; 2002. [Google Scholar]
- 19.Clark KR, Gorely RN. PRIMER-E: Plymouth. 2006. [Google Scholar]
- 20.Schloss PD, Handelsman J. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 2006;72:6773–6779. doi: 10.1128/AEM.00474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yue JC, Clayton MK. A similarity measure based on species proportions. Commun. Stat. Theor. M. 2006;34:2123–2131. [Google Scholar]
- 22.Rosello-Mora R, Wagner M, Amann R, Schleifer K-H. The abundance of Zoogloea ramigera in sewage treatment plants. Appl. Environ. Microbiol. 1995;61:702–707. doi: 10.1128/aem.61.2.702-707.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang T, Liu H, Fang HHP. Microbial analysis of a phototrophic sludge producing hydrogen from acidified wastewater. Biotechnol. Lett. 2002;24:1833–1837. [Google Scholar]
- 24.Huang LN, Zhu S, Zhou H, Qu LH. Molecular phylogenetic diversity of bacteria associated with the leachate of a closed municipal solid waste landfill. FEMS Microbiol. Lett. 2005;242:297–303. doi: 10.1016/j.femsle.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Gillan DC, Danis B. The archaebacterial communities in Antarctic bathypelagic sediments. Deep Sea Res. Part II Top. Stud. Oceanogr. 2007;54:1682–1690. [Google Scholar]
- 26.Lozupone CA, Knight R. Global patterns of bacterial diversity. Proc. Nat. Acad. Sci. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of human intestinal flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atlas KE, Horowitz A, Krichevsky M, Bej AK. Response of microbial populations to environmental disturbance. Micro. Ecol. 1991;22:249–256. doi: 10.1007/BF02540227. [DOI] [PubMed] [Google Scholar]
- 29.Wright DH. Species-energy theory: an extension of species-area theory. Oikos. 1983;41:496–506. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional data are available on-line. Table S1 provides sample codes, clone library information, species richness and Simpson diversity indices. Table S2 provides pairwise percent OTU overlap between samples in the upper portion of the triangular matrix and Theta-YC community structure similarity values in the lower triangle. Figure S1 provides a list of 69 potentially pathogenic OTUs and their associated taxonomy. Figure S2 presents comprehensive pie charts showing a comparative bacterial taxonomic summary of the sequences in all 28 samples – clicking on the names allows one to view a taxonomic breakdown of the sequences down to genus when possible or some higher taxonomic level. Asterisks indicate taxonomic levels shared with the raw sewage sample. Figure S3 shows the placement of eukaryotic sequences within an ARB tree while Figure S4 shows the placement of archaeal sequences. Figure S5 shows pie charts indicating changes in relative abundances of bacterial phyla and classes of Proteobacteria over time at transect stations near-shore and offshore. Figure S6 is a NMDS 3-dimensional ordination of 28 samples based on standardization by total and Log(X+1) transformed OTU abundances and Bray-Curtis similarities. Refer to Table 1 for sample codes. This information is available free of charge via the Internet at http://pubs.acs.org.