Abstract
The yeast [PSI+] prion is an epigenetic modifier of translation termination fidelity that causes nonsense suppression. The prion [PSI+] forms when the translation termination factor Sup35p adopts a self-propagating conformation. The presence of the [PSI+] prion modulates survivability in a variety of growth conditions. Nonsense suppression is essential for many [PSI+]-mediated phenotypes, but many do not appear to be due to read-through of a single stop codon, but instead are multigenic traits. We hypothesized that other global mechanisms act in concert with [PSI+] to influence [PSI+]-mediated phenotypes. We have identified one such global regulator, the Paf1 complex (Paf1C). Paf1C is conserved in eukaryotes and has been implicated in several aspects of transcriptional and posttranscriptional regulation. Mutations in Ctr9p and other Paf1C components reduced [PSI+]-mediated nonsense suppression. The CTR9 deletion also alters nonsense suppression afforded by other genetic mutations but not always to the same extent as the effects on [PSI+]-mediated read-through. Our data suggest that the Paf1 complex influences mRNA translatability but not solely through changes in transcript stability or abundance. Finally, we demonstrate that the CTR9 deletion alters several [PSI+]-dependent phenotypes. This provides one example of how [PSI+] and genetic modifiers can interact to uncover and regulate phenotypic variability.
INTRODUCTION
Termination of translation is signaled when a stop codon in the mRNA reaches the A site of the ribosome. In eukaryotes, the conserved proteins eRF1 and eRF3 recognize the stop codon and catalyze polypeptide release from the ribosome to terminate translation (Frolova et al., 1994; Stansfield et al., 1995b; Zhouravleva et al., 1995). Termination is a highly accurate process, and several factors that contribute to this fidelity have been identified (reviewed in von der Haar and Tuite, 2007). However, the molecular mechanism of how many of these factors contribute to faithful translation termination has not been fully elucidated. Mutant screens in Saccharomyces cerevisiae have identified genetic factors such as suppressor tRNAs, 18S rRNA, and ribosomal proteins that contribute to the accuracy of translation termination (reviewed in Beier and Grimm, 2001; Bertram et al., 2001; Valente and Kinzy, 2003). Factors that regulate polyA tail length and mRNA stability, such as Upf1p, Upf2p, Upf3p, Dcp1p, Pab1p, Pop2p, and Tpa1p, also influence faithful termination (Weng et al., 1996; Czaplinski et al., 1998; Cosson et al., 2002; Keeling et al., 2006; Kofuji et al., 2006). Overexpression of Mtt1p/Ecm32p, Itt1p, Sso1p, Stu2p, eEF1Bα, snR18, and Ssb1p, specific mutations in actin, up-regulation of tRNAs, and loss of SAL6/PPQ1 or DBP5 have all been shown to modify the efficiency of translation termination (Song and Liebman, 1987; Czaplinski et al., 2000; Urakov et al., 2001; Kandl et al., 2002; Kwapisz et al., 2002; Namy et al., 2002; Gross et al., 2007; Hatin et al., 2007); however, the mechanisms by which many of these factors affect termination have not been elucidated. Epigenetic factors that influence translation termination such as [PSI+] and [ISP+] have also been identified (Cox, 1965; Volkov et al., 2002).
In S. cerevisiae and other yeasts, eRF3 is encoded by SUP35 (Stansfield et al., 1995b). Interestingly, eRF3/Sup35p exists in two different states in yeast (Patino et al., 1996; Paushkin et al., 1996). In the soluble state, Sup35p functions with eRF1/Sup45p to terminate translation (Stansfield et al., 1995b). However, Sup35p can fold into an alternative conformation that is self-propagating, referred to as the prion [PSI+] (Patino et al., 1996; Paushkin et al., 1996). In the [PSI+] state, Sup35p is aggregated and the protein aggregates are heritable through mitosis and meiosis. [PSI+] cells exhibit increased nonsense suppression (or read-through of stop codons), presumably because aggregated Sup35p cannot participate effectively in translation termination. Multiple strain variants of the [PSI+] prion, differing in their level of nonsense suppression, can exist within different cells of the same genetic background (Derkatch et al., 1996). The level of nonsense suppression exhibited by strains of [PSI+] inversely correlates with the amount of soluble Sup35p (Zhou et al., 1999; Kochneva-Pervukhova et al., 2001; Uptain et al., 2001). Strong [PSI+] strain variants have high nonsense suppression, show little soluble Sup35p, and generally have high mitotic and meiotic stability (Derkatch et al., 1996; Zhou et al., 1999; Kochneva-Pervukhova et al., 2001; Uptain et al., 2001). In contrast, [PSI+] strain variants with lower nonsense suppression have more soluble Sup35p as well as lower mitotic and meiotic stability and are called weak [PSI+]. [PSI+] strain variants rarely interconvert and primarily arise by spontaneous or induced conversion from [psi−]. Prion strain variants are not due to genetic differences; rather they are due to different self-replicating Sup35p conformations (Derkatch et al., 1996; Krishnan and Lindquist, 2005; Tanaka et al., 2005; Toyama et al., 2007).
The domains of Sup35p that function in prion propagation and translation termination are separable (Ter-Avanesyan et al., 1993, 1994). The N-terminal region alone is capable of forming transmissible prion aggregates (Ter-Avanesyan et al., 1994; Li and Lindquist, 2000). Mutations in, or deletion of, the Sup35p N-terminal domain result in an inability to propagate [PSI+] but allow faithful translation termination (Doel et al., 1994; Ter-Avanesyan et al., 1994; DePace et al., 1998). Mutations in the essential C-terminal domain that reduce function result in less efficient translation termination and cause nonsense suppression (Ter-Avanesyan et al., 1993; Bradley et al., 2003). However, there is cross-talk between the domains. The N-terminal domain influences nonsense suppression in Sup35 C-terminal domain mutants (Volkov et al., 2007). Nonetheless, the N-terminal region is both necessary and sufficient for [PSI+] prion propagation, and the C-terminal domain alone is necessary and sufficient for translation termination.
Interestingly, we and others have previously shown that completely isogenic [PSI+] and [psi−] yeast strains differ in many different growth phenotypes, including changes in colony morphology (Eaglestone et al., 1999; True and Lindquist, 2000; Namy et al., 2008). Furthermore, the specific phenotypic effects were dependent on the genetic background of the strains used. [PSI+] permitted better growth in some conditions tested, whereas [psi−] enhanced growth in other conditions. In further studies, we demonstrated that many of the phenotypic changes could be attributed to [PSI+]-mediated nonsense suppression and not other properties of [PSI+], such as protein aggregation (True et al., 2004). Nonsense suppression can occur at stop codons at the 3′ ends of normal mRNAs or at premature stop codons. As a result, [PSI+]-mediated nonsense suppression has the potential to produce polypeptides with C-terminal extensions, to merge contiguous open reading frames into a single polypeptide, and to facilitate the translation of pseudogenes and other disabled open reading frames (Harrison et al., 2002; Namy et al., 2002, 2003, 2008; Giacomelli et al., 2007). The hidden variation revealed by [PSI+] in these normally untranslated regions has lead to the proposal that [PSI+] may generate adaptive phenotypic variants and increase the rate of evolvability (True and Lindquist, 2000; Masel and Bergman, 2003; Masel, 2006). In addition, [PSI+]-mediated read-through affects mRNA stability both by stabilizing mRNAs normally subjected to nonsense mediated decay and by triggering nonstop decay (Wilson et al., 2005). These may be additional sources of [PSI+]-mediated genetic variation (True et al., 2004; Wilson et al., 2005). Interestingly, although read-through was essential for the phenotypic effects, many did not appear to be due to nonsense suppression of a single transcript but instead were complex, polygenic traits. This suggests that many modifiers, and perhaps many other cellular pathways, can contribute to the effects of [PSI+] on phenotypic diversity. Furthermore, because many of the phenotypes are multigenic, modifiers may exist that broadly affect the level of read-through and have the potential to influence many phenotypes. In this study, we identify a genetic modifier of [PSI+]-mediated traits and show that [PSI+] and genetic modifiers can interact to regulate phenotypic variability.
MATERIALS AND METHODS
Yeast Strains and Plasmids
Yeast cells were cultured and manipulated using standard techniques. Yeast strains were grown in rich media (YPD: 1% yeast extract, 2% peptone, 2% dextrose) or synthetic dropout media (SD) lacking one or more components as needed to select for plasmids or assay for nonsense suppression. Solid media contained 2% agar. Additions of G418, guanidine hydrochloride (GdnHCl), hygromycin B, neomycin sulfate, and cycloheximide were at the final concentrations noted in the figure legends. Colony morphology was assayed on KOAc media (1% yeast extract, 2% peptone, 3% potassium acetate, 2% agar). Suppression of the lys1-1 allele in 10B-H49 was assayed on SD medium lacking lysine with 3% glycerol, as the use of glycerol as the carbon source enhances nonsense suppression in 10B-H49. Diploids were sporulated on 0.98% potassium acetate, 0.05% glucose, 0.1% yeast extract, 1.5% agar, 0.08% CSM powder (BIO-101, Carlsbad, CA) for ∼10 d before micromanipulation of asci.
All yeast strains used in this study except for those used in Figure 9 were derived from 74-D694 (MATa ade1-14 trp1-289 his3-Δ200 ura3-52 leu2-3112). 74-D694 containing strong [PSI+] was a gift of Y. Chernoff (Georgia Institute of Technology, Atlanta, GA) (Chernoff et al., 1995), as was the 74-D694 strain OT69 containing the sup45 mutation. The weak [PSI+] (Derkatch et al., 1996) and [psi−] sup35-Y351C (Bradley et al., 2003) strains were provided by S. Liebman (University of Illinois at Chicago, Chicago, IL). The [psi−] strain was generated in our laboratory by multiple passages of the strong [PSI+] strain on YPD supplemented with 3 mM GdnHCl. Strains used in Figure 9 were derivatives of 5V-H19 (MATa ade2-1 can1-100 leu2-3112 ura3-52 SUQ5; Ter-Avanesyan et al., 1993) or 10B-H49 (MATα ade2-1 SUQ5 lys1-1 his3-11,15 leu1 kar1-1; Ter-Avanesyan et al., 1994) and were obtained from M. Ter-Avanesyan (Institute of Experimental Cardiology, Moscow, Russia). In these strains the nonsense suppression depends on both [PSI+] and SUQ5. The coding regions of CTR9, PAF1, CDC73, LEO1, and RTF1 were replaced with HIS3MX6 or loxP-LEUMX-loxP using the method of Baudin et al. (1993) and pFA6a-HIS3MX6 (Wach et al., 1997) or pUG73 (Gueldener et al., 2002) as template. All knockouts were confirmed by colony PCR.
Figure 9.
Δctr9 modifies [PSI+]-dependent phenotypes. (A) CTR9 was deleted in [PSI+] and [psi−] derivatives of 10B-H49 and assayed for nonsense suppression of the lys1-1 allele on SD-lys + glycerol and for the alteration of [PSI+]-dependent cycloheximide resistance (YPD + 0.2 μg/ml cycloheximide). (B) Wild-type (WT) [psi−], wild-type [PSI+], and [PSI+] Δctr9 derivatives of 5V-H19 were grown on YPD (pre-wash) and assayed for invasive growth by washing the plate with distilled water (post-wash). (C) Colony morphology of the same strains as B grown on media containing potassium acetate as the sole carbon source. The puckered colony morphology was reduced in strong [PSI+] Δctr9 but not completely lost.
Plasmids were constructed using standard molecular biology techniques and maintained in DH5α. The shuttle vectors pRS315 (LEU2 CEN6 ARSH4) and pRS316 (URA3 CEN6 ARSH4) were described previously (Sikorski and Hieter, 1989). pRS316kanMX4 contains kanMX4 in the f1 origin of pRS316 and can be selected in yeast via URA3 or kanMX4. This plasmid was constructed by removing kanMX4 from pFA6a-kanMX4 (Wach et al., 1994) with BglII and SacI, making the fragment blunt with Klenow, and ligating it into the NgoMIV site (made blunt with Klenow) of pRS316 (Sikorski and Hieter, 1989). Wild-type CTR9 from 74-D694a and mutant ctr9 from SV16 were cloned into pRS316kanMX4 by PCR amplifying the gene from each strain with oligos (5′-CTACGGCCGCTTTTTTGAATTACACTATCCATC-3′ and 5′-CTACTCGAGAGGACACGAAAAGGTGGATG-3′), digesting the resulting PCR products with EagI and XhoI and ligating them into the same sites of pRS316kanMX4.
Screen for Factors that Reduce Nonsense Suppression
Strain 74-D694a containing strong [PSI+] was mutagenized with ethylmethane sulfonate as previously described (Lawrence, 1991) to 17% viability. Cells were plated onto YPD plates at ∼500–700 colonies per plate and incubated at 30°C for 6–7 d before visually screening for colonies that were darker pink than the original, unmutagenized light pink parental strain. Approximately 133,000 colonies were screened. Solid, darker pink colonies were restruck to YPD at least two additional times to assess the maintenance of the phenotype. Thirty-two mutants that consistently and uniformly produced solid, darker pink colonies were kept for further analysis.
Cloning SV16 by Complementation
The gene that is responsible for the reduced nonsense suppression in mutant SV16 was cloned by complementation of the poor growth phenotypes on SD-ade and YPD containing 3 mM GdnHCl. SV16 was independently transformed with two different libraries. S. cerevisiae genomic libraries in either p366 (LEU2 CEN; American Type Culture Collection, Manassas, VA; number 77162, deposited by Philip Hieter) or pRS316kanMX4 (URA3 kanMX4 CEN) made in our laboratory (unpublished data) were used. Approximately 108,000 and 17,400 transformants from the LEU2 CEN and URA3 kanMX4 CEN libraries, respectively, were screened. Library plasmids that complemented both poor growth phenotypes were rescued from transformants and retested. On retesting, eight isolates were identified that complemented both phenotypes. To evaluate specificity, each of the plasmids was tested in strong [PSI+], weak [PSI+], and [psi−] strains for an effect on nonsense suppression by color or growth on SD-ade. Six of the plasmids passed this secondary test. Both ends of the inserts in the complementing plasmids were sequenced (Protein Nucleic Acid Chemistry Laboratory, Washington University School of Medicine). All six library plasmids contained overlapping regions of the same genomic region. CTR9 was the only gene common to all six plasmids.
To compare the sequences of CTR9 from SV16 and wild-type, the sequence of the CTR9 locus from SV16 or 74-D694a was gap-repaired into plasmid A186 (isolated from the LEU2 CEN genomic library) digested with SwaI and MluI to remove most of the CTR9 coding region. CTR9 in the two gap-repaired plasmids was sequenced using ABI Big Dye v3.1 chemistry and the sequences from wild-type 74-D694 and SV16 were compared.
Growth Assays
[PSI+]-mediated phenotypes were assayed by growing cultures to mid-log phase in YPD or dropout media to select for a plasmid if necessary. All cultures within a single experiment were normalized to the same density by A600 before making fivefold serial dilutions. All phenotypes were assayed multiple times from independent cultures with identical results.
Biochemical Analysis
Sup35p aggregates were analyzed by semidenaturing detergent agarose gel electrophoresis (SDD-AGE) as previously described (Bagriantsev and Liebman, 2004) with the modifications of Tank et al. (2007). Unless indicated differently in the figure legend or text, samples were incubated for 7 min at room temperature before being loaded on the gel.
To examine Sup35p and Ctr9p expression levels, protein lysates were prepared as for SDD-AGE except denaturing loading buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 0.1% bromophenol blue, 10% glycerol, and 100 mM dithiothreitol) was added to cleared lysates, and samples were incubated at 95°C for 5 min. Equal amounts of protein, as determined by Bradford Assay, were loaded on polyacrylamide gels for SDS-PAGE. The protein was subsequently transferred to PVDF for Western blotting. Membranes were probed with antibodies recognizing Sup35p (Patino et al., 1996; diluted 1:2000), Ctr9p yN-18 (Santa Cruz Biotechnology, Santa Cruz, CA; diluted 1:400), or Pgk1p (Molecular Probes, Eugene, OR; diluted 1:10,000) followed by horseradish peroxidase-conjugated goat anti-rabbit IgG (Sigma-Aldrich, St. Louis, MO; diluted 1:10,000), rabbit anti-goat IgG (Jackson ImmunoResearch Laboratories, West Grove, PA; diluted 1:4000), or rabbit anti-mouse IgG (Sigma-Aldrich; diluted 1:10,000), respectively. Immunoblots were developed using enhanced chemiluminescence.
Fluorescence Microscopy
Yeast strains were transformed with the copper-inducible plasmid mCNMG containing a fusion between sequences coding for the Sup35p prion domain (NM) and green fluorescent protein (GFP; Patino et al., 1996). Expression of the fusion protein was induced with a final concentration of 60 μM copper sulfate for 6 h before visualization. Visualization was on an Olympus IX70 fluorescence microscope (Melville, NY) equipped with an enhanced GFP filter cube and a Photometrics Coolsnap HQ camera and QED in vivo software (Tucson, AZ).
Read-through Assay
ß-Galactosidase reporter plasmids pUKC817, pUKC818, and pUKC819 (Stansfield et al., 1995a) were provided by M. Tuite (University of Kent, Canterbury, United Kingdom). Transformants were grown to log-phase in liquid culture, harvested by centrifugation, lysed with glass beads, and assayed by the Galacto-Light Chemiluminescent Reporter Gene Assay System (Applied Biosystems, Bedford, MA). Detection was on a Sirius Single Tube Luminometer (Berthold Detection Systems, Pforzheim, Germany) with a delay time of 2.0 s and read time of 5.0 s. For each strain, read-through was calculated from three independent cultures as the average relative light units per microgram of protein assayed as determined by Bradford Assay. Read-through for the [psi−] culture was set to a value of 1.0 and read-through for each of the other strains was expressed as the fold-change from [psi−]. This method of normalization was used because our data suggest that each individual transcript may be influenced differently by the loss of Ctr9.
Northern Blot
Total RNA was isolated as described (Schmitt et al., 1990). Briefly, total RNA was extracted using three organic extractions: once in hot, acidic phenol-chloroform and twice in cold, acidic phenol-chloroform. The RNA was precipitated overnight with 3 M sodium acetate, pH 5.2, and 100% ethanol at −20°C. The RNA pellet was washed once in 70% ethanol and resuspended in diethylpyrocarbonate (DEPC)-treated water. Equal amounts (20 μg) of RNA was loaded onto a 0.8% denaturing formaldehyde agarose gel in 1× MOPS buffer. RNA was transferred onto a nylon membrane (Amersham, Piscataway, NJ; Hybond-N) by capillary action overnight. The membrane was stained with methylene blue to visualize the 18S and 25S rRNA. The RNA was UV-cross-linked onto the membrane. The membrane was prehybridized in Amersham Rapid-hyb Buffer at 55°C for 1 h. Radioactive probes for the Northern analysis were prepared by random primed labeling of ADE1 PCR product (Roche, Indianapolis, IN). The oligos used to amplify ADE1 product were 5′ ATGTCAATTACGAAGACT and 5′ TTAGTGAGACCATTTAGACCC. Hybridization was performed at 55°C for 2 h. The membrane was washed three times: once for 15 min at room temperature, followed by a 30 min wash at 60°C (1× SSPE, 0.1% SDS), and final wash for 45 min at 60°C (0.5× SSPE, 0.25% SDS). The membrane was exposed to film for visualization of bands.
3′ RACE and Quantitative RT-PCR Analysis
Total RNA was isolated as described above. Total RNA, 5 μg, was reverse-transcribed using Superscript III (Invitrogen, Carlsbad, CA) and 50 μM of an oligo adaptor-dT19: 5′ GTTTCCCAGTCAGATCTTTTTTTTTTTTTTTTTTT. The RNA was reverse transcribed at 50°C for 1 h, and the reverse transcriptase was inactivated at 70°C for 15 min. The cDNA was subsequently treated with RNase H at 37°C for 20 min. The cDNA was diluted 20-fold into a 50-μl PCR reaction. The oligos used in the PCR amplification were 5′ TACGGATCCTATGAAACATTGACAGGGTC and 5′ GTTTCCCAGTCAGATCT. The 3′ RACE reactions were resolved on a 1.5% agarose gel. Quantitative RT-PCR (qRT-PCR) was performed using Sybr-Green (Bio-Rad, Richmond, CA) according to the manufacturer's instructions. The PCR products for LacZ and ACT1 were generated using the following primer pairs: LacZ, 5′ GTTGCTGATTCGAGGCGTTAAC and 5′ GGCTTCATCCACCACATACAG and ACT1, 5′ CTACGTTTCCATCCAAGCCG and 5′ CCACGTTCACTCAAGATCTTC.
Invasive Growth Assay
Invasive growth was assayed as described previously (Roberts and Fink, 1994).
RESULTS
Identification of Mutants with Weakened Nonsense Suppression
Read-through of stop codons during translation (nonsense suppression) to produce elongated polypeptide chains is one mechanism that can give rise to phenotypic variability (True and Lindquist, 2000; True et al., 2004; Wilson et al., 2005). The [PSI+] prion reduces the fidelity of translation termination in a slightly metastable, yet heritable manner, and has been shown to influence the phenotype of numerous traits in yeast (True and Lindquist, 2000). Further, many of these traits are complex in nature (True et al., 2004). Although many of the phenotypes require nonsense suppression, other gene products appear to interact and contribute. With the goal of understanding how [PSI+] interacts with other cellular components to contribute to genetic variation and phenotypic diversity, we screened for factors that weakened the nonsense suppression of a [PSI+] yeast strain.
Nonsense suppression is commonly monitored using auxotrophic alleles of biosynthetic pathway genes that contain premature stop codons (Chernoff et al., 2002). For example, the ade1-14 allele contains a premature stop codon in the ADE1 gene that encodes a component of the adenine biosynthetic pathway. Because cells with the ade1-14 allele do not make enough functional Ade1 protein, they require adenine in the media to grow. Although they grow well on rich medium, [psi−] ade1-14 cells accumulate an intermediate in the adenine biosynthetic pathway that causes the colonies to be red in color. In [PSI+] cells, however, translation proceeds through the stop codon to produce some full-length Ade1 protein and thereby complete the adenine biosynthetic pathway. Thus, [PSI+] ade1-14 cells do not require adenine in the media, and the colonies are white or pale pink in color.
We utilized the ade1-14 allele in the 74-D694 yeast strain to screen for factors that weaken the nonsense suppression of a strong [PSI+] variant. Cells with weakened nonsense suppression should make less Ade1 protein and produce darker pink colonies than strong [PSI+]. Wild-type pale pink [PSI+] ade1-14 cells were mutagenized with ethylmethane sulfonate to 17% viability, and the resulting colonies were screened visually for a darker pink color. Of 133,000 colonies screened, 32 consistently produced darker pink colonies than wild-type strong [PSI+] after multiple restreaks.
ctr9 Weakens Nonsense Suppression
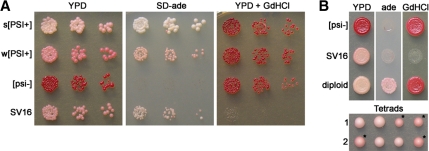
One mutant (SV16) will be described here. The SV16 mutant strain produces medium pink colonies on rich media (YPD) and grows slowly on media lacking adenine (SD-ade) compared with unmutagenized strong [PSI+] cells (Figure 1A). The [PSI+] prion can be cured by transient growth on media containing millimolar amounts of GdnHCl (Tuite et al., 1981). Interestingly, SV16 cells were inviable in the presence of 3 mM GdnHCl (Figure 1A). However, the SV16 mutant was viable on lower concentrations of GdnHCl and was able to be cured of the [PSI+] prion (data not shown).
Figure 1.
Mutant strain SV16 contains a recessive, Mendelian mutation that weakens the nonsense suppression of [PSI+]. (A) Fivefold serial dilutions of indicated yeast cultures at equal cell density were spotted onto YPD, SD-ade, and YPD containing 3 mM GdnHCl. s[PSI+] and w[PSI+], strong and weak [PSI+] strains, respectively. (B) Spotting of [psi−], SV16, and the wild-type [psi−] × SV16 diploid on YPD, SD-ade, and YPD containing 3 mM GdnHCl (top). Two representative tetrads from sporulation of the wild-type [psi−] × SV16 diploid are shown (bottom). Asterisks denote spores that inherited the mutant allele.
The SV16 mutant was backcrossed to a wild-type 74-D694 [psi−] strain, and the resulting diploid was analyzed. The diploid produced pale pink colonies on rich media and grew well on SD-ade (Figure 1B), indicating that the SV16 mutation was recessive and that the mutant still contained the [PSI+] prion. When the meiotic progeny were analyzed, two haploids were pale pink and two were medium pink in each tetrad. The 2:2 segregation pattern suggested that a single mutation was contributing to the phenotype. Furthermore, the recovery of strong [PSI+] haploids in each tetrad indicated that the mutation had to be present to obtain the reduced nonsense suppression and that the mutation had not permanently altered the structure to create a new prion strain variant.
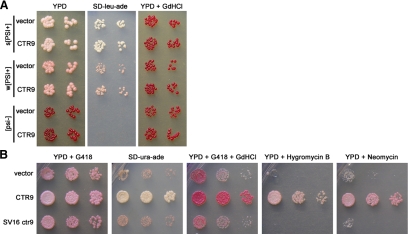
To identify the mutation in SV16, we complemented the weak nonsense suppression phenotype with two yeast genomic libraries. Complementation was assayed by robust growth on media lacking adenine and by resistance to 3 mM GdnHCl. Six plasmids were identified that complemented both phenotypes and passed all secondary tests (data not shown). The ends of each rescuing library plasmid were sequenced. All six plasmids contained overlapping genomic fragments. Only one gene, CTR9, was common to all six plasmids. Ctr9p is a component of the Paf1 protein complex (Paf1C) implicated in the transcription and transcriptional processing of some mRNAs (Shi et al., 1996; Koch et al., 1999; Krogan et al., 2002; Mueller and Jaehning, 2002; Pokholok et al., 2002; Squazzo et al., 2002; Mueller et al., 2004; Rondon et al., 2004; Penheiter et al., 2005). To test whether the increase in nonsense suppression conferred by the library plasmids to SV16 was specific to the SV16 mutant or due to a general increase in nonsense suppression, we transformed a representative library plasmid containing CTR9 into a wild-type [psi−] strain as well as strong and weak variants of [PSI+]. The library plasmid did not significantly affect the color of the wild-type [PSI+] or [psi−] strains on rich media or their growth on media lacking adenine (Figure 2A).
Figure 2.
CTR9 complements SV16 phenotypes. (A) Strong (s) [PSI+], weak (w) [PSI+], and [psi−] strains were transformed with pRS315 empty vector or the A3 library plasmid (LEU2 vector with a genomic region containing CTR9). Liquid cultures were normalized to equal cell density and fivefold serial dilutions were spotted onto YPD, SD-leu-ade, and YPD + 3 mM GdnHCl. (B) SV16 was transformed with pRS316kanMX4 empty vector, pRS316kanMX4-CTR9, or pRS316kanMX4-SV16ctr9. Liquid cultures of the resulting transformants were normalized and spotted as in A onto YPD + 200 mg/l G418 (to select for the plasmid), SD-ura-ade, YPD + 200 mg/l G418 + 3 mM GdnHCl, YPD + 25 μg/ml hygromycin B, and YPD + 20 mg/ml neomycin sulfate.
Because CTR9 was the only gene in common to all six library plasmids, we tested whether CTR9 was responsible for the complementation of SV16. CTR9 was cloned and tested for complementation of the reduced growth of SV16 on SD-ade and for rescue of other phenotypes associated with SV16. During our analysis we found that the SV16 mutant grows slower than wild-type, is slightly temperature sensitive at 37°C, and is hypersensitive to hygromycin B, neomycin sulfate, and hydroxyurea (Figure 2B and data not shown). The introduction of wild-type CTR9 on a plasmid permitted the rapid growth of SV16 on SD-ade compared with the empty vector (Figure 2B). In addition, CTR9 complemented the slow-growth phenotype of SV16 on rich media as well as the sensitivities to GdnHCl, hygromycin B, and neomycin sulfate (Figure 2B). Moreover, when we cloned the ctr9 allele out of the SV16 mutant into the same vector and tested it for complementation, we found that it failed to rescue any of the SV16 mutant phenotypes (Figure 2B).
Because CTR9 rescued the weakened nonsense suppression phenotype of SV16, we hypothesized that a mutation in CTR9 may be responsible for the weakened nonsense suppression in the SV16 mutant. We sequenced CTR9 from SV16 and the isogenic wild-type strain and found one difference that resulted in a change in amino acid (C227Y) in the SV16 mutant compared with wild-type.
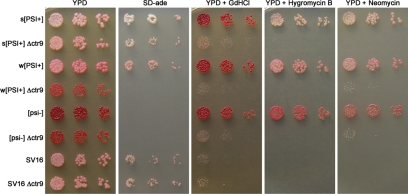
If the C227Y mutation in ctr9 was in fact causing the recessive phenotypes of the SV16 mutant, then we predicted that the complete deletion of the nonessential CTR9 gene would also weaken nonsense suppression. The entire CTR9 open reading frame was deleted in strong [PSI+], weak [PSI+], [psi−], and SV16 strains and the resulting strains were assessed phenotypically. Deletion of CTR9 did reduce the [PSI+]-mediated nonsense suppression of the ade1-14 allele (Figure 3). The null mutant produced darker pink colonies on YPD in the strong [PSI+] strain. In addition, Δctr9 in a strong [PSI+] strain grew significantly slower on SD-ade than the wild-type strong [PSI+] strain. When ctr9 was deleted in a weak [PSI+] strain, the colonies became red in color and did not grow on media lacking adenine. Because these colonies appeared phenotypically like [psi−], we wanted to determine if [PSI+] was still present. Deletion of CTR9 could cure the weak [PSI+] prion. Alternatively, the [PSI+] prion could still be present but phenotypically masked by more faithful translation termination at the premature ade1-14 stop codon in the CTR9 deletion. To genetically test for the presence of [PSI+], we crossed Δctr9 made in the weak [PSI+] strain to a [psi−] strain and examined the color of the resulting diploids on YPD. If [PSI+] was present but masked in the Δctr9 strains, then the diploid would appear pink because of the rescued nonsense suppression by one wild-type copy of CTR9 in the diploid. However, if [PSI+] had been cured by the deletion of CTR9, then the diploid would be red. The diploid was pink in color, indicating deletion of CTR9 did not cure weak [PSI+] (data not shown). We also tested for the presence of aggregated Sup35p in [PSI+] Δctr9. Sup35 protein aggregates were still detected by SDD-AGE (data not shown). Collectively, this data suggests that the [PSI+] prion was still present and that deletion of CTR9 reduced the nonsense suppression phenotype of [PSI+]. In addition, Δctr9 in strong [PSI+], weak [PSI+], and [psi−] strains were all temperature sensitive at 37°C and produced similar sensitivities to GdnHCl, hygromycin B, neomycin sulfate, and hydroxyurea as the SV16 point mutant (Figure 3 and data not shown).
Figure 3.
Δctr9 reduces nonsense suppression of strong and weak [PSI+] and exhibits phenotypes similar to SV16. CTR9 was deleted in strong (s) [PSI+], weak (w) [PSI+], [psi−], and SV16 strains. Liquid cultures were normalized to equal cell density, and fivefold serial dilutions were spotted onto the indicated media. Drug concentrations in the media are the same as those listed in the legend of Figure 2.
We also knocked out ctr9 from the SV16 mutant strain. Interestingly, we noted that the complete deletion of ctr9 in the SV16 strain was phenotypically similar to the SV16 point mutant (Figure 3), suggesting that the Ctr9-C227Y protein could be produced at a lower level, unstable, or severely impaired functionally. To test if the Ctr9-C227Y protein was present at a steady-state level similar to wild-type, lysates were prepared from wild-type and ctr9-C227Y [PSI+] strains and equal protein quantities were analyzed by SDS-PAGE and immunoblotting with antibodies to Ctr9p. The Ctr9-C227Y protein was barely detectable compared with wild-type Ctr9p (data not shown), suggesting less mutant protein is made or that it is very unstable.
Loss of Select Paf1C Members Also Reduces Nonsense Suppression
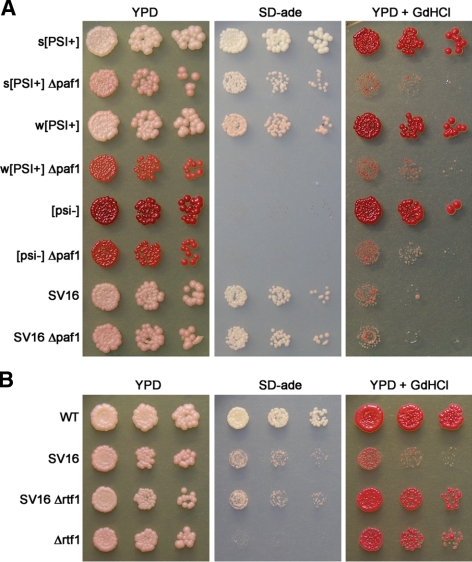
Because Ctr9p is part of a protein complex that contains Paf1p, Cdc73p, Leo1p, and Rtf1p (Koch et al., 1999; Krogan et al., 2002; Mueller and Jaehning, 2002; Squazzo et al., 2002), we tested whether other members of the complex also affected [PSI+]-mediated nonsense suppression. Deletions of CTR9 and PAF1 have been reported to have many phenotypes in common, whereas deletion of other members of Paf1C often produces fewer and weaker phenotypes (Betz et al., 2002; Mueller and Jaehning, 2002). Because CTR9 influences nonsense suppression, we predicted that the deletion of other Paf1C members would also affect the [PSI+] phenotype. We individually deleted the genes encoding members of Paf1C in strong [PSI+], weak [PSI+], [psi−], and SV16 strains. The deletion of PAF1 weakened nonsense suppression in both strong and weak [PSI+] strains and rendered the cells sensitive to GdnHCl (Figure 4A).
Figure 4.
Some components of Paf1C affect nonsense suppression of [PSI+]. (A) Deletion of PAF1 weakens [PSI+]-mediated nonsense suppression. Deletions of PAF1 were made in the following strains: strong (s) [PSI+], weak (w) [PSI+], [psi−], and SV16. Each deletion mutant and parental wild-type control strain was spotted as described in the legend of Figure 3. (B) Deletion of RTF1 suppresses the GdnHCl sensitivity of SV16 but not the reduction in nonsense suppression of ade1-14. A wild-type (wt) strong [PSI+] strain, SV16, SV16 Δrtf1, and strong [PSI+] Δrtf1 strain were spotted as described in the legend of Figure 3.
The individual deletion of RTF1 also weakened nonsense suppression in both strong and weak [PSI+] strains (Supplemental Figure S1). However, Δrtf1 strains were only mildly sensitive to GdnHCl (data not shown). Interestingly, the deletion of CDC73 resulted in an intermediate phenotype. Weakened nonsense suppression of ade1-14 by Δcdc73 was readily detected in a weak [PSI+] strain but not in a strong [PSI+] strain (Supplemental Figure S1). It is possible that Δcdc73 does weaken nonsense suppression in a strong [PSI+] strain but that a more sensitive assay is required to detect it. Deletion of LEO1 had no detectable effect on nonsense suppression of ade1-14 in either strong or weak [PSI+] strains by this assay (Supplemental Figure S1). Moreover, Δcdc73 and Δleo1 strains were not sensitive to GdnHCl (data not shown).
Deletion of RTF1 Suppresses Some of the SV16 Phenotypes
Loss of Leo1p or Rtf1p has been reported to suppress many of the phenotypes of Δctr9 and Δpaf1 mutants (Mueller and Jaehning, 2002). To test whether the loss of other components of Paf1C can suppress the ctr9-C227Y mutant, we assayed deletions of PAF1, CDC73, LEO1, and RTF1 in the SV16 mutant strain for rescue of the ctr9-C227Y phenotypes. The deletions of PAF1, CDC73, and LEO1 had no significant effect on the nonsense suppression of the ade1-14 allele in the [PSI+] SV16 strain (Figures 4A and Supplemental Figure S1). Interestingly, Δrtf1 suppressed some of the phenotypes associated with ctr9-C227Y in the SV16 mutant strain. Loss of Rtf1p suppressed the sensitivity of SV16 to 3 mM GdnHCl and 0.15 M hydroxyurea (Figure 4B and data not shown). However, it did not alter the level of nonsense suppression of ade1-14 (Figures 4B and Supplemental Figure S1).
To determine whether another member of Paf1C can functionally replace Ctr9p in translation termination, we assayed whether the overexpression of PAF1, CDC73, LEO1, or RTF1 could suppress the phenotypes of the SV16 mutant. None of the other Paf1C components expressed from a 2-μm plasmid restored the nonsense suppression of ade1-14 or the GdnHCl resistance in the [PSI+] SV16 mutant strain (data not shown).
Loss of CTR9 Alters Sup35p Aggregates
In [PSI+] cells, Sup35p is aggregated in a heritable, self-propagating conformation (Patino et al., 1996; Paushkin et al., 1996). One way mutants of CTR9 could affect nonsense suppression in [PSI+] cells is by altering the expression or aggregation of Sup35p. To determine if the level of Sup35 protein was altered by ctr9-C227Y or Δctr9, Sup35p levels in the SV16 and Δctr9 strains were compared with a wild-type [PSI+] strain by Western blot. No change in the steady-state Sup35p level was detected between the wild-type and mutant strains (Figure 5A), indicating that the weakened nonsense suppression in the CTR9 mutants cannot be explained simply by a difference in the amount of total Sup35p.
Figure 5.
Sup35p aggregates in SV16 and Δctr9 are similar to those of strong [PSI+]. (A) Mutations in CTR9 do not alter the steady-state expression level of Sup35p. Equal quantities of protein were loaded on a SDS-polyacrylamide gel, transferred to PVDF, and immunoblotted with antibodies recognizing Sup35p or Pgk1p. Pgk1p was used as a control for equal loading. (B) NM-GFP aggregates in the SV16 strain. NM-GFP was expressed in the indicated strains and visualized by direct fluorescence microscopy after a 6-h induction with 60 μM copper sulfate. (C) Sup35p aggregates in SV16 and Δctr9 strains migrate as a broader smear compared with those of strong [PSI+]. Equal amounts of protein were analyzed by SDD-AGE and immunoblotting with antibodies recognizing Sup35p. The positions of monomeric and aggregated Sup35p are indicated to the left of the blot. (D) Sup35p aggregates in SV16 and Δctr9 strains exhibit thermal stability similar to those from a wild-type strong [PSI+] strain. SDD-AGE was performed as in C except the lysates were heated at 65, 75, or 85°C before loading on the gel. (A–D) s[PSI+] and w[PSI+], strong and weak [PSI+] strains, respectively.
Another mechanism by which ctr9 could weaken nonsense suppression in [PSI+] cells is by altering the aggregation of the Sup35p. Mutants of CTR9 could alter the conformation of the aggregates, resulting in a change in the [PSI+] strain variant being propagated by the cells. In addition, the stability of the aggregates could be altered such that a smaller percentage of the Sup35p is aggregated, thereby generating a larger pool of soluble Sup35p for translation termination activity. We examined these possibilities by several complementary methods. First, we tested whether the Sup35p was still aggregated in the SV16 mutant by ectopically expressing the prion forming domain of Sup35 (NM) fused to GFP and visualizing the cells by direct fluorescence microscopy. NM-GFP gets incorporated into the preexisting Sup35p aggregates in [PSI+] cells, thereby acting as an indicator of the [PSI] state (Patino et al., 1996; Lindquist et al., 2001). One to three large, bright foci were typically observed in cells when NM-GFP was expressed in strong and weak [PSI+] strains (Figure 5B). In contrast, no foci were present in [psi−] cells, and NM-GFP was diffuse in the cytoplasm. When NM-GFP was expressed in the SV16 mutant, foci were observed, indicating that Sup35p was still aggregated (Figure 5B). In some cells, the amount of diffuse NM-GFP in the cytoplasm appeared to be increased in the SV16 mutant strain as compared with the wild-type strong and weak [PSI+] strains. However, this was not uniform among all cells examined (data not shown).
Second, we determined if there was a gross change in the size or conformation of the Sup35p aggregates by SDD-AGE. The difference in size of the aggregates between prion strain variants can be detected as a change in electrophoretic mobility by SDD-AGE (Kryndushkin et al., 2003). As seen previously (Kryndushkin et al., 2003), Sup35p from a strong [PSI+] strain migrated slightly farther into the gel than Sup35p from a weak [PSI+] strain (Figure 5C). In the SV16 and Δctr9 [PSI+] strains, the smear of Sup35p aggregates migrated even farther into the gel than strong [PSI+], and the smear covers a slightly broader range (Figure 5C). In addition, a small amount of monomeric Sup35p was sometimes detected in the SV16 and Δctr9 mutants but not in wild-type [PSI+] (data not shown). To further investigate a possible structural change in the Sup35p aggregates in the mutants, we examined their thermal stability. Protein lysates from strong [PSI+], weak [PSI+], SV16, and Δctr9 were heated at various temperatures before SDD-AGE (Figure 5D). At 65°C, the Sup35p aggregates appeared similar to those of the standard assay temperature of ∼25°C, but some change in size or stability was beginning to be apparent for strong [PSI+], SV16, and Δctr9 (compare 65°C in Figure 5D with standard assay in Figure 5C). At 75°C, aggregates from strong [PSI+], but not weak [PSI+], were broken down to monomer. By 85°C, aggregates from weak [PSI+] also began to break down to show some monomeric protein. In the SV16 and Δctr9 mutant strains, the Sup35p aggregates broke down to monomer at 75°C, similar to the strong [PSI+] strain. Taken together with our genetic data, this suggests that although there may be a small effect on the size of the Sup35p aggregates in the population within the SV16 and Δctr9 mutants, the heritable structure of the aggregates is not grossly changed from those of the original strong [PSI+] parental strain.
Δctr9 Alters Both the Aggregation of Sup35p and Nonsense Suppression
Finally, we used genetic analyses to distinguish the effects of CTR9 on the Sup35p aggregates in [PSI+] cells from those on translation termination and nonsense suppression. In [psi−] cells, mutations in the SUP35 C-terminal domain that reduce function create a nonsense suppression phenotype that mimics that of [PSI+] cells (Wickner et al., 1995). Furthermore, these Mendelian mutations cause inviability in combination with [PSI+] due to high levels of nonsense suppression (Cox, 1977; Liebman and All-Robyn, 1984; Zhou et al., 1999). We crossed one of these mutants, [psi−] 74-D694 sup35-Y351C, to SV16, sporulated the diploid, and analyzed the tetrads. Only two haploids were recovered from each tetrad (data not shown), further supporting the cell biological and biochemical results that the SV16 mutant still contains the [PSI+] prion. When we cured the diploid cells of [PSI+] by passaging them on media containing GdnHCl before sporulation, all four meiotic progeny were viable. Using these [psi−] progeny, we then asked whether the ctr9-C227Y mutant altered the nonsense suppression of a sup35 genetic mutant. As shown by a representative tetrad in Figure 6, the ctr9-C227Y sup35-Y351C double mutant exhibited less nonsense suppression of the ade1-14 allele than the sup35-Y351C mutant alone, suggesting that one way the ctr9 mutant is promoting more efficient termination is by genetically modifying nonsense suppression. However, the effect of ctr9-C227Y on [PSI+] appeared to be stronger than on sup35-Y351C, because the difference in color between [PSI+] ctr9-C227Y and the parental [PSI+] strain is slightly greater than it is between the ctr9-C227Y sup35-Y351C and sup35-Y351C strains (Figure 6, YPD). This is also consistent with our result that ctr9 has some effect on the size of the Sup35p aggregates in [PSI+] (Figure 5C). This suggests that the loss of Ctr9p may affect [PSI+]-mediated phenotypes in multiple ways (see below). We next tested whether the effects of Ctr9 on nonsense suppression were specific to Sup35. The nonsense suppression phenotype resulting from mutations in sup45 is also reduced in Δctr9 strains (Supplemental Figure S2). The extent to which the loss of Ctr9 alters nonsense suppression of sup45 mutants was not as striking as the effect on [PSI+]-mediated nonsense suppression.
Figure 6.
ctr9-C227Y reduces nonsense suppression of ade1-14 in the absence of Sup35p aggregates. SV16 [PSI+] was crossed to [psi−] sup35-Y351C, and the resulting diploid was cured of [PSI+] by growth on media containing 3 mM GdnHCl before sporulation. Fivefold serial dilutions of yeast cultures at equal cell density were spotted on YPD and SD-ade. The top four rows show control strong (s) [PSI+] and [psi−] strains and the [PSI+] SV16 ([PSI+] ctr9*) and [psi−] sup35-Y351C ([psi−] sup35*) parental strains used to generate the diploid. The bottom four rows show a representative tetrad with genotype indicated on the left. Presence of the ctr9-C227Y and sup35-Y351C alleles is designated with an asterisk adjacent to the gene name. Wild-type alleles are designated in capital letters. All progeny are [psi−].
Ctr9 Affects Transcript Processing and Translatability of mRNA
The [PSI+] prion causes suppression of all three stop codons in yeast (Tuite et al., 1983; Firoozan et al., 1991). We have shown that Δctr9 weakens nonsense suppression of the ade1-14 allele, which contains a premature UGA stop codon. To investigate whether the effect of ctr9 on nonsense suppression is specific to UGA and the ade1-14 allele, we expressed reporter constructs with each one of the stop codons fused in front of lacZ (Stansfield et al., 1995a). If the stop codon is read through in these reporters, then β-galactosidase is produced. Each reporter was expressed in wild-type [PSI+], [psi−], and Δctr9 [PSI+] strains. As expected, the strong [PSI+] strain exhibited a higher level of read-through of each stop codon than the isogenic [psi−] strain (Figure 7A). Furthermore, the weak [PSI+] strain produced a level of read-through that was intermediate to the strong [PSI+] and [psi−] strains. Deletion of CTR9 in strong [PSI+] caused a reduction in [PSI+]-mediated nonsense suppression of all three stop codons (Figure 7A), further supporting the idea that Δctr9 weakens nonsense suppression in general.
Figure 7.
Deletion of CTR9 reduces nonsense suppression of all three stop codons. Read-through of reporter constructs containing the indicated stop codon was assayed and expressed as the fold change of the [psi−] strain. Each bar in the graphs represents the mean and SD of three independent cultures assayed on the same day. Whereas the absolute fold change varied between days, the overall trend remained the same. (A) Δctr9 reduces nonsense suppression by [PSI+] at each of the stop codons. s[PSI+] and w[PSI+], strong and weak [PSI+] strains, respectively. (B) Δctr9 also reduces nonsense suppression by sup35-Y351C (designated sup35*). All strains are [psi−]. WT, wild-type.
To investigate whether this reduction in read-through by Δctr9 was specific to the prion, we introduced the same reporters into a [psi−] sup35-Y351C Δctr9 double mutant strain along with each single mutant and the wild-type control. The effect on read-through was not entirely specific to the [PSI+] prion for two reasons. First, the [psi−] Δctr9 mutant alone had significantly lower read-through than the [psi−] wild-type strain (Figure 7B; unpaired t test, p < 0.0003 for all stop codons), indicating that the loss of Ctr9p reduces basal read-through of UAA, UAG, and UGA stop codons. Second, the sup35-Y351C Δctr9 double mutant also had significantly lower read-through of all three stop codons than the sup35-Y351C mutant alone (Figure 7B; unpaired t test, p < 0.0024 for all stop codons). Interestingly, Δctr9 had a greater effect on [PSI+]-mediated read-through than on sup35-Y351C-mediated read-through for each of the stop codons (compare fold change in Figure 7, A and B). The effect is more striking on UAG and UGA than on UAA. This difference, along with the slight effect of Δctr9 on the size of the Sup35p aggregates (Figure 5C), supports the idea that Δctr9 affects [PSI+]-mediated nonsense suppression in multiple ways. Collectively, these results indicate that nonsense suppression of all three stop codons is reduced in the absence of Ctr9p.
Interestingly, we also noted a twofold reduction in the level of specific activity of β-galactosidase with the deletion of ctr9 using the nonstop control lacZ constructs. This suggests that Ctr9 and the Paf1 complex effects transcript stability or translatability of the mRNA. Indeed, qRT-PCR confirmed a reduction in the steady-state level of nonstop lacZ mRNA (eightfold), and the reduction was verified by Northern blot (data not shown).
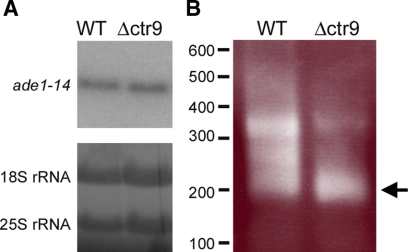
Next, we wanted to test if the level of ade1-14 transcript was reduced, hence accounting for the decreased amount of Ade1 product and change in phenotype in Δctr9. Curiously, we did not find a change in the steady-state levels of ade1-14 transcript by Northern blot analysis (Figure 8A, top). Loading of RNA was assessed by methylene blue staining of the 18S and 25S rRNA (Figure 8A, bottom). Because there was no change in the levels of ade1-14 transcript, another possibility that could account for the reduced amount of Ade1p in Δctr9 would be a change in the translatability of the ade1-14 mRNA. Supporting this hypothesis, a recent genome-wide analysis revealed that ADE1 transcript was predicted to have two or more poly(A) addition sites (Nagalakshmi et al., 2008). Because the Paf1C has been shown to play a role in posttranscriptional 3′end processing (Penheiter et al., 2005; Nordick et al., 2008), we wanted to test if the loss of CTR9 could affect the 3′ end processing of the ade1-14 transcript and consequently affect its translatability. Using 3′ rapid amplification of cDNA ends (RACE) analysis, we found that the shorter product of the ade1-14 3′ end was the predominant isoform in Δctr9 mutant as compared with a longer product in the wild-type strain (Figure 8B).
Figure 8.
Δctr9 alters the 3′ processing of the ade1-14 transcript. (A) Northern analysis of ade1-14 transcript levels are shown (top). Equal amounts of total RNA was loaded on the gel, and this was visualized by methylene blue staining of 18S and 25S rRNA (bottom). One representative blot is shown. (B) 3′ RACE analysis of total RNA isolated from wild-type (WT) and Δctr9 strains. The ade1-14 transcript has multiple isoforms in the WT and Δctr9 strain. Arrow indicates the predominant isoform that accumulates in the Δctr9 strain. The molecular-weight marker in base pairs is indicated on the left side of the gel.
Phenotypic Effects of ctr9 Are Not Limited to ade1-14
Isogenic [PSI+] and [psi−] yeast strains differ in numerous different growth phenotypes, many of which can be attributed to [PSI+]-mediated nonsense suppression (True and Lindquist, 2000; True et al., 2004). We wanted to determine if the loss of Ctr9p affected any of these [PSI+]-dependent traits. Few strong [PSI+]-dependent phenotypes were previously reported in the 74-D694 strain background (True and Lindquist, 2000). However, many [PSI+]-dependent phenotypes were identified in the 5V-H19 and 10B-H49 yeast strain backgrounds (True and Lindquist, 2000). Therefore, we made deletions of CTR9 in [PSI+] isolates of 5V-H19 and 10B-H49, cured the prion to obtain isogenic [psi−] Δctr9 strains, and examined the strain pairs for the effect of Δctr9 on some of the previously reported [PSI+]-dependent traits (True and Lindquist, 2000). Because Δctr9 alone is sensitive to many different growth conditions and compounds (Betz et al., 2002 and this study), we were limited in the number and range of phenotypes we could test.
Both 5V-H19 and 10B-H49 contain a premature UAA stop codon in ADE2 (the ade2-1 allele) that is suppressible by [PSI+]. Similar to our results with ade1-14 in 74-D694, Δctr9 weakened [PSI+]-mediated nonsense suppression of ade2-1 in both 5V-H19 and 10B-H49 (data not shown). 10B-H49 also contains a [PSI+]-suppressible allele of LYS1 (lys1-1), a component of the lysine biosynthetic pathway. 10B-H49 [psi−] cells grow poorly on media lacking lysine because they are unable to make enough full-length Lys1p. However, 10B-H49 [PSI+] cells make more Lys1p and can grow in the absence of lysine. When we deleted CTR9 in the 10B-H49 [PSI+] strain, the colonies grew slower than the wild-type 10B-H49 [PSI+] strain on media lacking lysine (Figure 9A), suggesting Δctr9 weakened the nonsense suppression of lys1-1. Another [PSI+]-dependent phenotype in 10B-H49 is resistance to cycloheximide. However, the gene target(s) of [PSI+] that contribute to this phenotype are not yet known. 10B-H49 [PSI+] cells grow better than isogenic [psi−] cells in the presence of 0.2 μg/ml cycloheximide (True and Lindquist, 2000 and Figure 8A). Surprisingly, the [psi−] Δctr9 strain was more resistant to cycloheximide than the wild-type [psi−] strain (Figure 9A).
Invasive growth and flocculation are two related [PSI+]-dependent phenotypes in 5V-H19 (True and Lindquist, 2000 and Figure 8B). [PSI+] cells invade the agar during growth on solid media and flocculate in liquid media, whereas [psi−] cells do not invade the agar and do not flocculate. When we deleted CTR9 from 5V-H19 [PSI+] cells, the cells invaded the agar much less than the wild-type [PSI+] strain on solid media and no longer flocculated in liquid media (Figure 9B and data not shown). Another [PSI+]-dependent trait in 5V-H19 that may be related to the flocculation and invasive growth phenotypes is a change in colony morphology. [PSI+] cells produce large colonies with a puckered and dimpled surface when grown on media containing 3% potassium acetate as the sole carbon source (True and Lindquist, 2000). In contrast, [psi−] cells produce smaller colonies with a smooth, rounded surface. The genes influenced by [PSI+] that are associated with this phenotype are unknown. However, the phenotype can be at least partially attributed to nonsense suppression (H. L. True, unpublished data). Loss of Ctr9p in the [PSI+] strain caused the cells to revert to a more [psi−]-like appearance (Figure 9C). Collectively, these results show that Δctr9 modifies multiple [PSI+]-dependent phenotypes.
DISCUSSION
We identified Ctr9p, a component of Paf1C, in a screen for factors that weaken [PSI+]-mediated nonsense suppression. Furthermore, we show that other members of the complex also affect [PSI+]-mediated nonsense suppression. In addition to Ctr9p and Paf1p, the complex is composed of Cdc73p, Leo1p, and Rtf1p (Koch et al., 1999; Krogan et al., 2002; Mueller and Jaehning, 2002; Squazzo et al., 2002). Paf1C is conserved from yeast to humans (Rozenblatt-Rosen et al., 2005; Zhu et al., 2005). However, the complex in humans does not appear to contain Rtf1p even though a homolog exists. We show that the loss of Paf1C weakens nonsense suppression and is a genetic modifier of [PSI+]-dependent phenotypes. How might Paf1C affect nonsense suppression? One possibility is that Paf1C is affecting transcript levels via initiation or elongation by RNA polymerase II. Paf1C is associated with RNA polymerase II from the promoters to the polyadenylation (polyA) sites of actively transcribed genes and is involved in the transcription and posttranscriptional processing of some mRNAs (Shi et al., 1996; Wade et al., 1996; Krogan et al., 2002; Pokholok et al., 2002; Squazzo et al., 2002; Mueller et al., 2004; Rondon et al., 2004; Penheiter et al., 2005; Nordick et al., 2008). Paf1C has been reported to influence the efficiency of transcription initiation and elongation (Pokholok et al., 2002; Squazzo et al., 2002; Rondon et al., 2004). However, it is unclear whether Paf1C directly affects these processes (Mueller et al., 2004; Penheiter et al., 2005). Paf1C is required for methylation of histone H3 (Krogan et al., 2003a,b; Ng et al., 2003), monoubiquitylation of histone H2B (Xiao et al., 2005), 3′ end formation of snoRNAs (Sheldon et al., 2005), and 3′ end formation of mRNAs in determining polyA site usage and polyA tail length (Penheiter et al., 2005). Recent work suggests that the influence of Paf1C on some histone modifications is likely to be an indirect consequence of reduced methyltransferase recruitment to RNA polymerase II in Paf1C mutants (Nordick et al., 2008).
The function of Paf1C in 3′ end formation of mRNAs is just beginning to be unraveled. Loss of Paf1C components leads to altered transcripts due to the change in usage of more distal polyA sites (Penheiter et al., 2005; Nordick et al., 2008). This effect on 3′ end formation has been characterized for specific transcripts (Penheiter et al., 2005; Nordick et al., 2008), but it is likely that more targets exist (Mozdy et al., 2008). Links between Paf1C and other factors that affect posttranscriptional 3′ end formation have recently been identified. Paf1C is required for the recruitment of cleavage and polyadenylation factors Cft1p and Pcf11p to RNA polymerase II (Mueller et al., 2004; Nordick et al., 2008). It was also shown that Paf1C directly binds Cft1p (Nordick et al., 2008).
One explanation for the influence of Paf1C on the phenotypic effects of [PSI+]-mediated nonsense suppression is via its effects on posttranscriptional 3′ end formation. Loss of Paf1C produces transcripts that have an altered 3′ end (Penheiter et al., 2005; Nordick et al., 2008). The 3′-extended MAK21 transcripts in Δpaf1 cells were targeted for degradation by the nonsense-mediated decay pathway, causing a reduction in the steady-state level of MAK21 transcript (Penheiter et al., 2005). This is likely to be true of additional targets of Paf1C; transcripts altered in 3′ end position and polyadenylation can be targeted for rapid degradation by the nonsense mediated decay pathway (Muhlrad and Parker, 1999). If the transcripts subjected to [PSI+]-mediated read-through are also targets of Paf1C-mediated processing, then we would expect that the loss of Paf1C components could lead to 3′-extended transcripts that are unstable. A smaller pool of transcript should result in less overall substrate for translation and translational read-through in [PSI+] cells that would thereby alter [PSI+]-mediated phenotypes. However, a change in the 3′ end processing may not alter transcript stability but may change translatability of the message. Indeed, the 3′ untranslated region has been shown to control polyadenylation of individual transcripts and translational efficiency (Beilharz and Preiss, 2007). A recent genome-wide study mapping transcribed regions in S. cerevisiae identified 540 genes that utilize more than one polyA site (Nagalakshmi et al., 2008). Interestingly, two sites of polyA tail addition were identified for ADE1. Our results demonstrate that individual deletions of CTR9, PAF1, RTF1, and CDC73 weaken [PSI+]-mediated nonsense suppression of the ade1-14 allele. This may be a consequence of a change in the utilization of the two polyA tail sites in ADE1. In support of this, we detected a significant and reproducible change in the ratio of the most abundant species of ade1-14 transcript in Δctr9 compared with the wild-type strain by 3′RACE. Interestingly, this change did not correspond to a reduced steady-state level of ade1-14 mRNA in cells deleted for ctr9. This suggests that there may be multiple ways by which the Paf1 complex influences mRNA translatability.
We identified four other [PSI+]-dependent phenotypes (cycloheximide resistance, flocculation, invasive growth, and colony morphology) that Δctr9 modifies. Because the loss of Ctr9p results in many drug sensitivities (Betz et al., 2002 and this study), we speculate that more slight alterations in the regulation of Paf1C might affect many other [PSI+]-dependent phenotypes. The genes involved in many [PSI+]-mediated phenotypes have not yet been identified. The cycloheximide-resistance phenotype is particularly interesting because [PSI+] grows better than [psi−], yet Δctr9 causes the [psi−] cells to become more resistant. This suggests that producing less of a specific transcript increases resistance to cycloheximide. If the ribosome reads through the normal stop codon in [PSI+] cells and reaches the 3′ end of the transcript without encountering another stop codon, this would trigger nonstop decay. In [psi−] cells, the transcript would be stable and translated, resulting in cycloheximide sensitivity. Loss of CTR9 in [psi−] cells could provide a different mechanism to destabilize the transcript and make cells more resistant to cycloheximide. Precedence exists for changes in nonstop decay modifying [PSI+]-dependent phenotypes (Wilson et al., 2005). Moreover, Paf1C itself may affect nonstop decay. The human PAF1C coimmunoprecipitates with the human SKI complex that mediates nonstop decay (Zhu et al., 2005). Furthermore, human PAF1C is required for the recruitment of the SKI complex to transcriptionally active genes (Zhu et al., 2005). Thus, different mechanisms to alter transcript stability could result in similar phenotypic consequences.
The strain 5V-H19 shows a [PSI+]-dependent increase in flocculation and invasive growth. Flocculation and invasive growth in yeast are regulated by the transcription factor FLO8 (Kobayashi et al., 1996, 1999; Liu et al., 1996). FLO8 is inactivated in many laboratory strains by a premature stop codon (Liu et al., 1996). However, FLO8 is not mutated in the 5V-H19 strain background (H. L. True, unpublished data) and is, therefore, not a likely target of read-through in 5V-H19 to produce the [PSI+]-dependent flocculation and invasive growth phenotypes. Loss of Paf1C reduces the effect of [PSI+] on flocculation and invasive growth. Interestingly, Paf1C affects the modification of histones at FLO8. Trimethylation of lysine 36 on histone 3 at FLO8 is reduced in Δpaf1 and Δctr9 mutants and acetylation of histones 3 and 4 on the 3′ end of FLO8 is increased in Δpaf1 and Δctr9 mutants (Chu et al., 2007). Although the genes involved in the [PSI+]-mediated changes in flocculation and invasive growth are unknown, it is plausible to speculate that [PSI+] and Δctr9 affect different genes in the flocculation and invasive growth pathways. However, for other phenotypes, both [PSI+] and Paf1C may affect the translation of the same transcript.
Transcriptional read-through of proximal polyA sites presumably occurs at a low rate. However, the use of alternative transcription termination sites may have biological functions, including the regulation of viral gene expression (Bousse et al., 2002; Poenisch et al., 2008). We describe another scenario, one in which changes in posttranscriptional processing modifies several [PSI+]-dependent phenotypes in yeast. The effect of Paf1C and 3′ end formation on translational nonsense suppression adds to the growing list of posttranscriptional processes that genetically modify [PSI+]-dependent traits. These links between transcriptional and translational control highlight the potential continuum for phenotypic diversity. If [PSI+]-mediated read-through provides an advantage to the organism whereby a phenotype can be altered in response to environmental changes, the ability to fine-tune the traits during transitions may enhance the organism's ability to adapt and survive. We have previously shown that it is possible to fix traits and become [PSI+]-independent (True et al., 2004). However, for an organism that must thrive in a fluctuating environment, fixation may not always be beneficial. Given that [PSI+]-mediated phenotypes are largely multifactorial, we postulate that there may be other global mechanisms for modulating the spectrum of phenotypes. One such mechanism may involve posttranscriptional processing through Paf1C.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yury Chernoff, Susan Liebman, Michael Ter-Avanesyan, Susan Lindquist (Whitehead Institute, Cambridge, MA), and Mick Tuite for providing yeast strains, plasmids, and antibodies. We thank John Cooper, Steve Johnson, and Naren Ramanan for use of equipment; Jason Weber and lab members for assistance with qRT-PCR and Northern blot analysis; and Michael Snyder for sharing the list of genes that utilize multiple polyadenylation sites. We thank members of the True lab and Melissa Brereton for helpful discussions and comments on the manuscript. This work was supported by the National Institutes of Health Grant GM072228 (H.L.T.), a USDA National Research Initiative Postdoctoral AREA Award 2005-35201-15383 (L.A.S.), and an Agency for Science, Technology and Research (A*STAR) fellowship (C.A.L.).
Abbreviations used:
- Δ
null
- GdnHCl
guanidine hydrochloride
- GFP
green fluorescent protein
- NM
prion-forming domain of Sup35p
- Paf1C
Paf1 complex
- SD
synthetic dropout
- s[PSI+]
strong [PSI+]
- SDD-AGE
semi-denaturing detergent agarose gel electrophoresis
- w[PSI+]
weak [PSI+]
- WT
wild-type
- YPD
yeast extract, peptone, and dextrose.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0813) on February 18, 2009.
REFERENCES
- Bagriantsev S., Liebman S. W. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J. Biol. Chem. 2004;279:51042–51048. doi: 10.1074/jbc.M410611200. [DOI] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier H., Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001;29:4767–4782. doi: 10.1093/nar/29.23.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz T. H., Preiss T. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA. 2007;13:982–997. doi: 10.1261/rna.569407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram G., Innes S., Minella O., Richardson J., Stansfield I. Endless possibilities: translation termination and stop codon recognition. Microbiology. 2001;147:255–269. doi: 10.1099/00221287-147-2-255. [DOI] [PubMed] [Google Scholar]
- Betz J. L., Chang M., Washburn T. M., Porter S. E., Mueller C. L., Jaehning J. A. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics. 2002;268:272–285. doi: 10.1007/s00438-002-0752-8. [DOI] [PubMed] [Google Scholar]
- Bousse T., Matrosovich T., Portner A., Kato A., Nagai Y., Takimoto T. The long noncoding region of the human parainfluenza virus type 1 f gene contributes to the read-through transcription at the m-f gene junction. J. Virol. 2002;76:8244–8251. doi: 10.1128/JVI.76.16.8244-8251.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley M. E., Bagriantsev S., Vishveshwara N., Liebman S. W. Guanidine reduces stop codon read-through caused by missense mutations in SUP35 or SUP45. Yeast. 2003;20:625–632. doi: 10.1002/yea.985. [DOI] [PubMed] [Google Scholar]
- Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., Liebman S. W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Chernoff Y. O., Uptain S. M., Lindquist S. L. Analysis of prion factors in yeast. Methods Enzymol. 2002;351:499–538. doi: 10.1016/s0076-6879(02)51867-x. [DOI] [PubMed] [Google Scholar]
- Chu Y., Simic R., Warner M. H., Arndt K. M., Prelich G. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 2007;26:4646–4656. doi: 10.1038/sj.emboj.7601887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson B., Couturier A., Chabelskaya S., Kiktev D., Inge-Vechtomov S., Philippe M., Zhouravleva G. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol. Cell. Biol. 2002;22:3301–3315. doi: 10.1128/MCB.22.10.3301-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B. [Psi], A cytoplasmic suppressor of super-suppressor in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- Cox B. S. Allosuppressors in yeast. Gen. Res. 1977;30:197–205. [Google Scholar]
- Czaplinski K., Majlesi N., Banerjee T., Peltz S. W. Mtt1 is a Upf1-like helicase that interacts with the translation termination factors and whose overexpression can modulate termination efficiency. Rna. 2000;6:730–743. doi: 10.1017/s1355838200992392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria M. J., Paushkin S. V., Han X., Weng Y., Perlick H. A., Dietz H. C., Ter-Avanesyan M. D., Peltz S. W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePace A. H., Santoso A., Hillner P., Weissman J. S. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- Derkatch I. L., Chernoff Y. O., Kushnirov V. V., Inge-Vechtomov S. G., Liebman S. W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doel S. M., McCready S. J., Nierras C. R., Cox B. S. The dominant PNM2- mutation which eliminates the psi factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics. 1994;137:659–670. doi: 10.1093/genetics/137.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone S. S., Cox B. S., Tuite M. F. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 1999;18:1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoozan M., Grant C. M., Duarte J. A., Tuite M. F. Quantitation of readthrough of termination codons in yeast using a novel gene fusion assay. Yeast. 1991;7:173–183. doi: 10.1002/yea.320070211. [DOI] [PubMed] [Google Scholar]
- Frolova L., et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- Giacomelli M. G., Hancock A. S., Masel J. The conversion of 3′ UTRs into coding regions. Mol. Biol. Evol. 2007;24:457–464. doi: 10.1093/molbev/msl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross T., Siepmann A., Sturm D., Windgassen M., Scarcelli J. J., Seedorf M., Cole C. N., Krebber H. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–649. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P., Kumar A., Lan N., Echols N., Snyder M., Gerstein M. A small reservoir of disabled ORFs in the yeast genome and its implications for the dynamics of proteome evolution. J. Mol. Biol. 2002;316:409–419. doi: 10.1006/jmbi.2001.5343. [DOI] [PubMed] [Google Scholar]
- Hatin I., Fabret C., Namy O., Decatur W. A., Rousset J. P. Fine-tuning of translation termination efficiency in Saccharomyces cerevisiae involves two factors in close proximity to the exit tunnel of the ribosome. Genetics. 2007;177:1527–1537. doi: 10.1534/genetics.107.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandl K. A., Munshi R., Ortiz P. A., Andersen G. R., Kinzy T. G., Adams A. E. Identification of a role for actin in translational fidelity in yeast. Mol. Genet. Genomics. 2002;268:10–18. doi: 10.1007/s00438-002-0726-x. [DOI] [PubMed] [Google Scholar]
- Keeling K. M., Salas-Marco J., Osherovich L. Z., Bedwell D. M. Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae. Mol. Cell Biol. 2006;26:5237–5248. doi: 10.1128/MCB.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi O., Suda H., Ohtani T., Sone H. Molecular cloning and analysis of the dominant flocculation gene FLO8 from Saccharomyces cerevisiae. Mol. Gen. Genet. 1996;251:707–715. doi: 10.1007/BF02174120. [DOI] [PubMed] [Google Scholar]
- Kobayashi O., Yoshimoto H., Sone H. Analysis of the genes activated by the FLO8 gene in Saccharomyces cerevisiae. Curr. Genet. 1999;36:256–261. doi: 10.1007/s002940050498. [DOI] [PubMed] [Google Scholar]
- Koch C., Wollmann P., Dahl M., Lottspeich F. A role for Ctr9p and Paf1p in the regulation G1 cyclin expression in yeast. Nucleic Acids Res. 1999;27:2126–2134. doi: 10.1093/nar/27.10.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochneva-Pervukhova N. V., Chechenova M. B., Valouev I. A., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. [Psi(+)] prion generation in yeast: characterization of the ‘strain’ difference. Yeast. 2001;18:489–497. doi: 10.1002/yea.700. [DOI] [PubMed] [Google Scholar]
- Kofuji S., Sakuno T., Takahashi S., Araki Y., Doi Y., Hoshino S., Katada T. The decapping enzyme Dcp1 participates in translation termination through its interaction with the release factor eRF3 in budding yeast. Biochem. Biophys. Res. Commun. 2006;344:547–553. doi: 10.1016/j.bbrc.2006.03.174. [DOI] [PubMed] [Google Scholar]
- Krishnan R., Lindquist S. L. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N. J., et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell. 2003a;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Krogan N. J., Kim M., Ahn S. H., Zhong G., Kobor M. S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J. F. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N. J., et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003b;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D. S., Alexandrov I. M., Ter-Avanesyan M. D., Kushnirov V. V. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Kwapisz M., Smagowicz W. J., Oficjalska D., Hatin I., Rousset J. P., Zoladek T., Boguta M. Up-regulation of tRNA biosynthesis affects translational readthrough in maf1-delta mutant of Saccharomyces cerevisiae. Curr. Genet. 2002;42:147–152. doi: 10.1007/s00294-002-0342-7. [DOI] [PubMed] [Google Scholar]
- Lawrence C. W. Classical mutagenesis techniques. Methods Enzymol. 1991;194:273–281. doi: 10.1016/0076-6879(91)94021-4. [DOI] [PubMed] [Google Scholar]
- Li L., Lindquist S. Creating a protein-based element of inheritance. Science. 2000;287:661–664. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- Liebman S. W., All-Robyn J. A. A non-Mendelian factor, [eta+], causes lethality of yeast omnipotent-suppressor strains. Curr. Genet. 1984;8:567–573. doi: 10.1007/BF00395701. [DOI] [PubMed] [Google Scholar]
- Lindquist S., Krobitsch S., Li L., Sondheimer N. Investigating protein conformation-based inheritance and disease in yeast. Philos. Trans. R. Soc. Lond B Biol. Sci. 2001;356:169–176. doi: 10.1098/rstb.2000.0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Styles C.A., Fink G.R. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J. Cryptic genetic variation is enriched for potential adaptations. Genetics. 2006;172:1985–1991. doi: 10.1534/genetics.105.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J., Bergman A. The evolution of the evolvability properties of the yeast prion [PSI+] Evolution. 2003;57:1498–1512. doi: 10.1111/j.0014-3820.2003.tb00358.x. [DOI] [PubMed] [Google Scholar]
- Mozdy A. D., Podell E. R., Cech T. R. Multiple yeast genes, including Paf1 complex genes, affect telomere length via telomerase RNA abundance. Mol. Cell. Biol. 2008;28:4152–4161. doi: 10.1128/MCB.00512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C. L., Jaehning J. A. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C. L., Porter S. E., Hoffman M. G., Jaehning J. A. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Parker R. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA. 1999;5:1299–1307. doi: 10.1017/s1355838299990829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O., Duchateau-Nguyen G., Hatin I., Hermann-Le Denmat S., Termier M., Rousset J. P. Identification of stop codon readthrough genes in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:2289–2296. doi: 10.1093/nar/gkg330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O., Duchateau-Nguyen G., Rousset J. P. Translational readthrough of the PDE2 stop codon modulates cAMP levels in Saccharomyces cerevisiae. Mol. Microbiol. 2002;43:641–652. doi: 10.1046/j.1365-2958.2002.02770.x. [DOI] [PubMed] [Google Scholar]
- Namy O., Galopier A., Martini C., Matsufuji S., Fabret C., Rousset J.P. Epigenetic control of polyamines by the prion [PSI(+)] Nat. Cell Biol. 2008;10:1069–1075. doi: 10.1038/ncb1766. [DOI] [PubMed] [Google Scholar]
- Ng H. H., Robert F., Young R. A., Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Nordick K., Hoffman M. G., Betz J. L., Jaehning J. A. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot. Cell. 2008;7:1158–1167. doi: 10.1128/EC.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino M. M., Liu J. J., Glover J. R., Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- Paushkin S. V., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Penheiter K. L., Washburn T. M., Porter S. E., Hoffman M. G., Jaehning J. A. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol. Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Poenisch M., Wille S., Staeheli P., Schneider U. Polymerase read-through at the first transcription termination site contributes to regulation of Borna disease virus gene expression. J. Virol. 2008;82:9537–9545. doi: 10.1128/JVI.00639-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok D. K., Hannett N. M., Young R. A. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- Roberts R. L., Fink G. R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Rondon A. G., Gallardo M., Garcia-Rubio M., Aguilera A. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 2004;5:47–53. doi: 10.1038/sj.embor.7400045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O., Hughes C. M., Nannepaga S. J., Shanmugam K. S., Copeland T. D., Guszczynski T., Resau J. H., Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol. Cell. Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M. E., Brown T. A., Trumpower B. L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon K. E., Mauger D. M., Arndt K. M. A Requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Finkelstein A., Wolf A. J., Wade P. A., Burton Z. F., Jaehning J. A. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol. Cell. Biol. 1996;16:669–676. doi: 10.1128/mcb.16.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. M., Liebman S. W. Allosuppressors that enhance the efficiency of omnipotent suppressors in Saccharomyces cerevisiae. Genetics. 1987;115:451–460. doi: 10.1093/genetics/115.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo S. L., Costa P. J., Lindstrom D. L., Kumer K. E., Simic R., Jennings J. L., Link A. J., Arndt K. M., Hartzog G. A. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I., Akhmaloka, Tuite M. F. A mutant allele of the SUP45 (SAL4) gene of Saccharomyces cerevisiae shows temperature-dependent allosuppressor and omnipotent suppressor phenotypes. Curr. Genet. 1995a;27:417–426. doi: 10.1007/BF00311210. [DOI] [PubMed] [Google Scholar]
- Stansfield I., Jones K. M., Kushnirov V. V., Dagkesamanskaya A. R., Poznyakovski A. I., Paushkin S. V., Nierras C. R., Cox B. S., Ter-Avanesyan M. D., Tuite M. F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995b;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Chien P., Yonekura K., Weissman J. S. Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell. 2005;121:49–62. doi: 10.1016/j.cell.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Tank E. M., Harris D. A., Desai A. A., True H. L. Prion protein repeat expansion results in increased aggregation and reveals phenotypic variability. Mol. Cell. Biol. 2007;27:5445–5455. doi: 10.1128/MCB.02127-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesyan M. D., Dagkesamanskaya A. R., Kushnirov V. V., Smirnov V. N. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesyan M. D., Kushnirov V. V., Dagkesamanskaya A. R., Didichenko S. A., Chernoff Y. O., Inge-Vechtomov S. G., Smirnov V. N. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol. Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- Toyama B. H., Kelly M. J., Gross J. D., Weissman J. S. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- True H. L., Berlin I., Lindquist S. L. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- True H. L., Lindquist S. L. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- Tuite M. F., Cox B. S., McLaughlin C. S. In vitro nonsense suppression in [psi+] and [psi−] cell-free lysates of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1983;80:2824–2828. doi: 10.1073/pnas.80.10.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite M. F., Mundy C. R., Cox B. S. Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uptain S. M., Sawicki G. J., Caughey B., Lindquist S. Strains of [PSI(+)] are distinguished by their efficiencies of prion-mediated conformational conversion. EMBO J. 2001;20:6236–6245. doi: 10.1093/emboj/20.22.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakov V. N., Valouev I. A., Lewitin E. I., Paushkin S. V., Kosorukov V. S., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. Itt1p, a novel protein inhibiting translation termination in Saccharomyces cerevisiae. BMC Mol. Biol. 2001;2:9. doi: 10.1186/1471-2199-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente L., Kinzy T. G. Yeast as a sensor of factors affecting the accuracy of protein synthesis. Cell Mol. Life Sci. 2003;60:2115–2130. doi: 10.1007/s00018-003-2334-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov K., Osipov K., Valouev I., Inge-Vechtomov S., Mironova L. N-terminal extension of Saccharomyces cerevisiae translation termination factor eRF3 influences the suppression efficiency of sup35 mutations. FEMS Yeast Res. 2007;7:357–365. doi: 10.1111/j.1567-1364.2006.00176.x. [DOI] [PubMed] [Google Scholar]
- Volkov K. V., Aksenova A. Y., Soom M. J., Osipov K. V., Svitin A. V., Kurischko C., Shkundina I. S., Ter-Avanesyan M. D., Inge-Vechtomov S. G., Mironova L. N. Novel non-Mendelian determinant involved in the control of translation accuracy in Saccharomyces cerevisiae. Genetics. 2002;160:25–36. doi: 10.1093/genetics/160.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T., Tuite M. F. Regulated translational bypass of stop codons in yeast. Trends Microbiol. 2007;15:78–86. doi: 10.1016/j.tim.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Alberti-Segui C., Rebischung C., Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wade P. A., Werel W., Fentzke R. C., Thompson N. E., Leykam J. F., Burgess R. R., Jaehning J. A., Burton Z. F. A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expr. Purif. 1996;8:85–90. doi: 10.1006/prep.1996.0077. [DOI] [PubMed] [Google Scholar]
- Weng Y., Czaplinski K., Peltz S. W. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol. Cell. Biol. 1996;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Masison D. C., Edskes H. K. [PSI] and [URE3] as yeast prions. Yeast. 1995;11:1671–1685. doi: 10.1002/yea.320111609. [DOI] [PubMed] [Google Scholar]
- Wilson M. A., Meaux S., Parker R., van Hoof A. Genetic interactions between [PSI+] and nonstop mRNA decay affect phenotypic variation. Proc. Natl. Acad. Sci. USA. 2005;102:10244–10249. doi: 10.1073/pnas.0504557102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T., Kao C. F., Krogan N. J., Sun Z. W., Greenblatt J. F., Osley M. A., Strahl B. D. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Derkatch I. L., Uptain S. M., Patino M. M., Lindquist S., Liebman S. W. The yeast non-Mendelian factor [ETA+] is a variant of [PSI+], a prion-like form of release factor eRF3. EMBO J. 1999;18:1182–1191. doi: 10.1093/emboj/18.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhouravleva G., Frolova L., Le Goff X., Le Guellec R., Inge-Vechtomov S., Kisselev L., Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Mandal S. S., Pham A. D., Zheng Y., Erdjument-Bromage H., Batra S. K., Tempst P., Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.