Abstract
Septins are conserved, GTP-binding proteins that assemble into higher order structures, including filaments and rings with varied cellular functions. Using four-dimensional quantitative fluorescence microscopy of Ashbya gossypii fungal cells, we show that septins can assemble into morphologically distinct classes of rings that vary in dimensions, intensities, and positions within a single cell. Notably, these different classes coexist and persist for extended times, similar in appearance and behavior to septins in mammalian neurons and cultured cells. We demonstrate that new septin proteins can add through time to assembled rings, indicating that septins may continue to polymerize during ring maturation. Different classes of rings do not arise from the presence or absence of specific septin subunits and ring maintenance does not require the actin and microtubule cytoskeletons. Instead, morphological and behavioral differences in the rings require the Elm1p and Gin4p kinases. This work demonstrates that distinct higher order septin structures form within one cell because of the action of specific kinases.
INTRODUCTION
Septins are conserved guanosine triphosphate (GTP)-binding proteins that assemble into heteromeric, nonpolar polymers with varied cellular functions (Versele and Thorner, 2005; Weirich et al., 2008). Initially discovered in budding yeast based on their role in cytokinesis, septins have been shown to act as barriers to membrane diffusion, as scaffolds for proteins that regulate the cell cycle and cytoskeleton, and as players in cell migration (Hartwell, 1971; Barral et al., 2000; Takizawa et al., 2000; Gladfelter et al., 2001; Finger et al., 2003). Septin family members have been found in fungal, animal, and some algal genomes but not in plants (Pan et al., 2007). In all cases studied, septin function requires the assembly of septin subunits into higher order complexes in cells. Importantly, septin mutations or aberrant septin expression has been associated with a variety of neurodegenerative diseases as well as some cancers (Ihara et al., 2003; Hall and Russell, 2004; Hall et al., 2005; Russell and Hall, 2005; Ihara et al., 2007; Sitz et al., 2008).
Recently, oligomeric septin complexes were observed at the supramolecular level by electron microscopy for budding yeast, worm, and human in vitro-expressed complexes and at the atomic level by x-ray crystallography for three human septins (John et al., 2007; Sirajuddin et al., 2007; Bertin et al., 2008). All studies found that septins form symmetrical or apolar rods of repeating oligomers of septin subunits. The dimensions of the core rod varied depending upon the number of septins coexpressed. These rods can associate end-to-end to form nonpolar, striated filaments that are frequently paired in vitro (Frazier et al., 1998; Versele et al., 2004). However, very little is known about how the filaments seen in vitro relate to higher order septin structures observed by either light or electron microscopy in vivo. Septin complexes assemble into diverse higher order structures in cells, including filaments, rings, and “gauzes” of enmeshed filaments (Haarer and Pringle, 1987; Kinoshita et al., 2002; Rodal et al., 2005). There is still sizeable ambiguity regarding the basis for filament polymerization and how filaments may come together to build such varied higher order structures in vivo.
In budding yeast, five septins (Cdc3p, Cdc10p, Cdc11p, Cdc12p, and Shs1p/Sep7p) colocalize first as a patch and then form an hourglass-shaped collar between the mother and growing bud. At cytokinesis, the ring splits and then disassembles in both the daughter and mother cell (Caviston et al., 2003; Dobbelaere et al., 2003). Septin ring reorganizations are at least partially controlled by posttranslational modifications of the septin subunits that alter the subunit dynamics and appearance of higher order structures (Johnson and Blobel, 1999; Mortensen et al., 2002; Dobbelaere et al., 2003; Versele and Thorner, 2004; Egelhofer et al., 2008). How specific posttranslational modifications lead to changes in the polymerization of septin filaments or assembly of higher order structures in vivo is not well understood. At the end of each cell cycle, yeast septins are stripped of these modifications and the subunits are reused in subsequent cycles (McMurray and Thorner, 2008). In contrast, in differentiated mammalian cells such as neurons, higher order septin structures persist over extended times. Thus, the yeast model system is likely insufficient to identify mechanisms relating to long-term maintenance and maturation of the septin cortex because the structures are disassembled with the turning of each cell cycle.
We have established a model fungal system, Ashbya gossypii, for the study of higher order septin structures in vivo that complements work done both in other fungi and mammalian cells (Dietrich et al., 2004; Wendland and Walther, 2005; Helfer and Gladfelter, 2006). A. gossypii is a filamentous ascomycete that diverged at least 100 million years ago from a common ancestor with Saccharomyces cerevisiae (Dietrich et al., 2004). Notably, there is no uninucleate yeast form in the fungus and instead it grows exclusively as multinucleate hyphae that contain tens of nuclei sharing a common cytoplasm. This multinucleate state arises from asynchronous nuclear division without cell division (Gladfelter, 2006b; Gladfelter et al., 2006). In the genome, A. gossypii has the same set of septin genes as S. cerevisiae but with an additional, tandem duplication of CDC11. In A. gossypii, the septins are essential to establish septation sites (which form in response to stress and senescence), to promote mitosis, and to form asexual spores (Gladfelter, 2006a; Helfer and Gladfelter, 2006). Importantly, septins are not essential in A. gossypii. Therefore, we can perturb the structures and still monitor them in living cells. Finally, we are able to measure maturation of the septin cortex in the absence of cell cycle driven remodeling (Helfer and Gladfelter, 2006).
We use four-dimensional (4D) quantitative fluorescence microscopy to show that A. gossypii septins assemble into morphologically distinct classes of rings that vary in dimensions, intensities, lifetimes, and positions across a single cell. In these studies, we demonstrate that highly variable septin rings coexist in one cell and require different signaling pathways for their assembly and maintenance.
MATERIALS AND METHODS
Growth Conditions and Strain Construction
A. gossypii media, culturing, and transformation protocols are described in Wendland et al. (2000) and Ayad-Durieux et al. (2000) Strains generated and used in this study can be found in Table 1. All DNA manipulations were carried out according to Sambrook and Russell (2001). Plasmids used here are listed in Table 2. Polymerase chain reaction (PCR) was performed using standard methods with polymerases from Roche Diagnostics (Indianapolis, IN) and Invitrogen (Carlsbad, CA). Oligonucleotides (oligos) were synthesized by Integrated DNA Technologies (Coralville, IA), and all restriction enzymes came from New England Biolabs (Beverly, MA). Oligonucleotide primers are listed in Table 3. Plasmid isolation from yeast and sequencing were performed as described in Schmitz et al. (2006). All sequencing was carried out by Dartmouth College Core Facilities (Hanover, NH).
Table 1.
A. gossypii strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| wt | leu2Δ thr4Δ | Altmann-Johl and Philippsen (1996) |
| AG120 | Agelm1Δ::GEN3, AgSEP7-GFP-NAT1, leu2Δ thr4Δ | This study |
| AG121 | Aggin4Δ::GEN3, AgSEP7-GFP-NAT1, leu2Δ thr4Δ | This study |
| AG124 | AgSEP7-GFP-GEN3, leu2Δ thr4Δ | Helfer and Gladfelter (2006) |
| AG127 | AgSEP7-GFP-NAT1, leu2Δ thr4Δ | Helfer and Gladfelter (2006) |
| AG209 | Agnap1Δ::GEN3, AgSEP7-GFP-NAT1, leu2Δ thr4Δ | This study |
| AG231 | AgCDC11a-mCherry-NAT1, leu2Δ thr4Δ | This study |
| AG232 | AgCDC11a-mCherry-NAT1, AgSEP7-GFP-GEN3,leu2Δ thr4Δ | This study |
| AG245 | pAGB100 [AgCDC3-CFP-NAT1], leu2Δ thr4Δ | This study |
| AG306 | AgCDC11a-mCherry-NAT1, pAGB98 | This study |
| [AgCDC10-YFP-GEN3], leu2Δ thr4Δ | ||
| AG308 | AgCDC11a-mCherry-NAT1, pAGB136 | This study |
| [AgCDC12-Venus-GEN3], leu2Δ thr4Δ | ||
| AG311 | Agsep7Δ::GEN3, AgCDC11a-mCherry-NAT1, leu2Δ thr4Δ | This study |
| AG312 | Aggin4Δ::GEN3, AgCDC11a-mCherry-NAT1, leu2Δ thr4Δ | This study |
| AG313 | Agelm1Δ::GEN3, AgCDC11a-mCherry-NAT1, leu2Δ thr4Δ | This study |
| AG314 | Agnap1Δ::GEN3, AgCDC11a-mCherry-NAT1, leu2Δ thr4Δ | This study |
With the exception of plasmidic strains (brackets), all analyzed mycelia were homokaryotic (all nuclei have same genotype).
Table 2.
Plasmids used in this study
| Plasmid no. | Name | Vector | Relevant insert | Source |
|---|---|---|---|---|
| pUC19 | ||||
| pRS416 | Sikorski and Hieter (1989) | |||
| AGB011 | pAGT102 | pUC19 | CFP-NAT1 | Kaufmann (2009) |
| AGB021 | pAGT140 | pUC19 | GEN3 | Kaufmann (2009) |
| AGB023 | pAGT143 | pUC19 | YFP-GEN3 | Kaufmann (2009) |
| AGB024 | pAGT147 | pUC19 | Venus-GEN3 | Kaufmann (2009) |
| AGB048 | pAGT211 | pUC19 | mCherry-NAT1 | Kaufmann (2009) |
| AGB088 | pAGSep7-GFPGEN3 | pRS416 | SEP7-GFP-GEN3 | Helfer and Gladfelter (2006) |
| AGB098 | pAGCDC10-YFP | pRS416 | CDC10-YFP-GEN3 | This study |
| AGB100 | pAGCDC3-CFP | pRS416 | CDC3-CFP-NAT1 | This study |
| AGB112 | pAGCDC10 | pRS416 | CDC10 | This study |
| AGB122 | pTA CDC12 | pTA | CDC12 | This study |
| AGB123 | pAG CDC12 | pRS416 | CDC12 | This study |
| AGB125 | pAG CDC11A | pRS416 | CDC11a | P. Philippsen |
| AGB127 | pAG CDC3 | pRS416 | CDC3 | P. Philippsen |
| AGB136 | pCDC12-Venus:GEN3 | pRS416 | CDC12-Venus-GEN3 | This study |
| AGB139 | pTA CDC10 | pTA | CDC10 | This study |
| AGB141 | pAGCDC11a-Cherry:nat | pRS416 | CDC11a-mCherry-NAT1 | This study |
Table 3.
Oligonucleotide primers used in this study
| Primer no. | Name | Sequence 5′-3′ |
|---|---|---|
| AGO18 | Gin4D1 | CTGGCTGGCCTAAGCGTTACAAAAGGACGGTGCTGGCTGAACCT |
| Cgctagggataacagggtaat | ||
| AGO19 | Gin4D2 | CATTGCTACGGGTGGCAGCGGGAAGCGCAGGTATACAGAATAGT |
| Caggcatgcaagcttagatct | ||
| AGO20 | Gin4V1 | CTAAGCCAATCCAGCCCACCGAG |
| AGO21 | Gin4V2 | GGTCCGCGTGAGTCCGATTGTTG |
| AGO22 | Gin4IV1 | CTTGAAGGAGGTTCGCTCCATGC |
| AGO23 | Gin4IV2 | GACCCCACAAACGTCGATCCCTG |
| AGO24 | Elm1D1 | GACGACAATAGAAAACCAGTTGCATGGTGCTAGGGCATAGGGA |
| GCgctagggataacagggtaat | ||
| AGO25 | Elm1D2 | CAGACGATTACAAGCGCAGTAGTAACTGCGGTGTGTGTGGGGAT |
| Gaggcatgcaagcttagatct | ||
| AGO26 | Elm1V1 | CCCGTACGCCAACAGAGGAGATG |
| AGO27 | Elm1V2 | GTGCAGAACAAAGGGCTCTCGAC |
| AGO28 | Elm1IV1 | CAGATCTCGAGTGGCTCTACGTC |
| AGO29 | Elm1IV2 | GCGCAGCAAGAGAGAGTGTCTCC |
| AGO36 | VG5 | ggaggtagtttgctgattgg |
| AGO37 | VG3 | atgttggacgagtcggaatc |
| AGO107 | Cdc10 50R seq | TCGCTTACTGCAGGCCATC |
| AGO108 | Cdc10 50F seq | TCAAAGGAGCGCAAACAG |
| AGO109 | Cdc 10 F-2 | TTTCCCTGGCCCGTACAGCTTCTGTCTCC |
| AGO110 | Cdc 10 R-2 | GGAGTGAAGTGCTACCTCTGCCATTGGTTG |
| AGO111 | Cdc10 700F seq | TGATTGCCAAGGCAGACAC |
| AGO112 | Cdc10 700R seq | TTGCGGAGATACTGCGAGTG |
| AGO115 | Cdc12 F | ggcggaattcCCTCTAACCGCGTCATCAGC |
| AGO116 | Cdc12 R | ggcggatccGTGCAGGAGGAGGTGGATTC |
| AGO119 | Cdc12 50R seq | TGCGATCTGCTGTAAACC |
| AGO120 | Cdc12 700R seq | TGTCCTGCGCCTCAATCACC |
| AGO121 | Cdc12 700F seq | GAGTTAGTGGACGGACTATGC |
| AGO122 | Cdc12 50F seq | CCAACAGGAAACCTTCGGC |
| AGO174 | MHP1 F | GTAGACTGGGACTAGAGAACGGAGGCCTGAAGCAGCCAGGAGT |
| GTAgctagggataacagggtaat | ||
| AGO175 | MHP1 R | CTCCAGATCGTCTAGAACATCGTCATATGCGCGATCTATGAGAG |
| CCaggcatgcaagcttagatct | ||
| AGO176 | MHP1 V1 | AGTGAGTCCTTACGTAGGTTAG |
| AGO177 | MHP1 V2 | GTGCTCAAGAACATCGCTAGTTCC |
| AGO178 | MHP1 Int | CTGTCGGTGGAAAGCGACTG |
| AGO188 | 3′ Cdc3 25DS | GGGTAACAGGGGAGGGAAATAAAATGCATGCATAGAAGTAAAG |
| TGatgcctgcagccaaacagtgttc | ||
| AGO190 | 3′CDC11a | CTAGCGTATGATGCTCACATGTTTCAGCCATTCTTTTCTTGCTTCG |
| atgcctgcagccaaacagtgttc | ||
| AGO192 | 5′ CDC3 -stop | AGTTCTCAGTCTCCAGCACAGCCAAAGAAGAAAGGTTTCTTGCG |
| Caaaacgacggccagtgaattcg | ||
| AGO193 | 3′ CDC10 - stop | GCTACAGGCCACGGTGCCCAATCCTCCAGCACTAACATGCttcgtaaa |
| acgacggccagtgaattcg | ||
| AGO195 | 3′ CDC12 -stop | AAGAAGCTGGAGGAGCAGGTCAGAGCATTGCAACTAAGAAAGC |
| ACaaaacgacggccagtgaattcg | ||
| AGO197 | 5′ Cdc12 25ds F2a | GTAGTATCGCTGTATATCTTCAACATTGCGATCTGCTGTAAACCA |
| ctgcaggcatgcaagcttag | ||
| AGO198 | 5′ Cdc10 25ds F2a | GGGCTTTACAAAAGCAAGCCAAGAGGAGGAGAATAAAAGGCGC |
| ggcatgcaagcttag | ||
| AGO199 | 5′ 3CFP seq1 | AGCATCTCCACGAGGCCAAG |
| AGO200 | 5 ′ 3CFP seq2 | AGACACGTGCTGAAGTCAAG |
| AGO201 | 3′ 3CFP seq1 | CTGCCATTAGCAGACTAACG |
| AGO202 | 3′ 3CFP seq2 | GTCTTGTAGTTCCCGTCATC |
| AGO206 | 5′ 10 YFP seq1 | CGAGATTCCCGGGTAATAAC |
| AGO207 | 3 10 YFP seq1 | TTATCCCATTTGCCGTCGTG |
| AGO208 | 3 10 YFP seq2 | GCCCGAAGGTTATGTACAGG |
| AGO209 | 3 12 YFP seq1 | TAACTCGGCGGCTGCAGAAG |
| AGO234 | V3 NAT1 | ACATGAGCATGCCCTGCCCC |
| AGO235 | V2 NAT1 | GTGGTGAAGGACCCATCCAG |
| AGO270 | Nap1D1 | CAGCAAGCTTCTACAGCACACGCGGAGCAAGGCAAGGCAGAGAg |
| taaaacgacggccagtgaattcg | ||
| AGO271 | Nap1D2 | GAAACGGGACCTTAGCGCCGAGTGAACGCAACTACTGCTGCTTG |
| Ccatgattacgccaagcttgc | ||
| AGO278 | Nap1V1 | CAACGCTGGAGGAAAGATGCTGC |
| AGO279 | Nap1V2 | CGGTGCATTTGAAACCGCAACGC |
| AGO280 | Nap1IV1 | CTTCGACGACGAAGACTCTGACG |
| AGO281 | Nap1IV2 | GAAGCAATGTAGCCCGAGTCCTG |
| AGO299 | 5′Cherry 11a-stop | GAACGTCTGGAAAAAGAAGCCAAAACCAAGCAGGAAATTGAGG |
| ATatcggtgacggtgctggtttaattaac | ||
| AGO424 | Sep7D1 | GCACCACGTGTACCACAGACATGACCGCCGTCCAATATTCGAAC |
| Gaaaacgacggccagtgaattcg | ||
| AGO425 | Sep7D2 | GGGATATCTGCGTATAGACCTATTGTACCTGGCCTGTTGCGCGGG |
| catgattacgccaagcttgc | ||
| AGO426 | Sep7V1 | CTAGCTTGAAGGCTAAGCCCGAC |
| AGO427 | Sep7V2 | GCGCCCACAGTAGTGGTTTCAGA |
| AGO428 | Sep7IV1 | GCGGCTCTCAGGATAGGACGAA |
| AGO429 | Sep7IV2 | GATGAACTGTCCGTGCTCACCTC |
Lowercase letters denote areas of the oligonucleotide with homology to a different cassette or genomic sequence.
A. gossypii deletion mutants were generated using PCR-based, one-step gene targeting approach with dominant selection markers (McElver and Weber, 1992; Baudin et al., 1993; Wach, 1996; Wendland et al., 2000). For G418 resistance, the deletion cassettes were amplified off the pAGT140 template by using “gene name”-D1/D2 (or F/R for AGO174 and AGO175 only) primer pairs that had ∼45-bp homology to the termini of the gene of interest and ∼20-bp homology to the termini of a selection marker. For single deletion mutant strains, correct integration of selection markers into and removal of the gene of interest from the genome was verified by analytical PCR using combinations of oligonucleotides gene name-V1, V2, IV1, IV2, Int (gene/locus-specific sequences), VG3, VG5 (G418-specific sequences), and V2 NAT1 and V3 NAT1 (NAT1-specific sequences). Transformation of multinucleate mycelium leads to heterokaryotic cells containing a mixture of transformed and wild-type nuclei which may not display an apparent phenotype. For subsequent analysis, homokaryotic mycelia were obtained by isolating and growing single spores. To exclude phenotypes because of random mutation, at least two independent transformants were characterized for each strain.
To create strains AG231 (CDC11a-mCherry-NAT1) and AG245 (CDC3-CFP-NAT1), plasmids containing CDC11a and CDC3 (AGB125 and AGB127) were kindly provided by the Philippsen laboratory (University of Basel, Basel, Switzerland). Oligos AGO299, 190, 192, and 188 containing sequence homology to the 3′ of a septin open reading frame (ORF) were used to amplify either mCherry-NAT1 from AGB048 or CFP-NAT1 from AGB011. Yeast cotransformation of PCR products with plasmids AGB125 or 127 yielded AGB141 (CDC11a-mCherry-NAT1) and AGB100 (CDC3-CFP-NAT1). AGB141 was verified by restriction enzyme digest with SalI. AGB100 was verified by digestion with EcoRI and NdeI and sequenced with oligos AGO199, 200, 201, and 202. A. gossypii cells were transformed with AGB141 to make AG231. Strain AG232 (CDC11a-mCherry-NAT1, SEP7-GFP-GEN3) was created by digesting plasmid AGB088 (SEP7-GFP) with SacI and transforming this product into strain AG231. Verification of these strains was performed using fluorescence light microscopy.
To create strains AG306 (CDC11a-mCherry-NAT1, [pCDC10-YFP-GEN3]) and AG308 (CDC11a-mCherry-NAT1, [pCDC12-Venus-GEN3]), CDC10 and CDC12 were amplified along with 500 base pairs upstream and downstream of the ORFs out of the A. gossypii genome by using primer pairs AGO109/110 and AGO115/116. The products were ligated, using T4 ligase (Invitrogen), into pCR 2.1 vectors (Invitrogen) that had been cut with AflII by using TA Cloning kits (Invitrogen) to create plasmids AGB139 (CDC10) and AGB122 (CDC12). AGB139 was verified by restriction enzyme digest with BglII and HinDIII, and this product was sequenced using oligos AGO107, 108, 111, and 112. AGB122 was verified by digestion with EcoRI and NdeI and sequencing using oligos AG-O-119, 120, 121, and 122. AGB139 and AGB122 were digested with BamH1 and NotI and ligated into pRS416 linearized with BamHI and NotI to create plasmids AGB112 (CDC10) and AGB123 (CDC12). AGB112 was verified by digestion with NdeI and PvuII. AGB123 was verified by digestion with ScaI and XhoI. A PCR fragment that had homology to CDC10 and contained YFP-GEN3 was amplified from plasmid AGB023 (YFP-GEN3) by using oligos AGO193 and 198. Yeast cotransformation of this product and AGB112 created AGB098 (CDC10-YFP-GEN3). AGB098 was verified by digestion with XmaI and sequenced with oligos AGO202, 206, 207, and 208. This plasmid was then transformed into strain AG127 (SEP7-GFP-NAT1) to create strain AG-306. A PCR product with homology to CDC12 and containing Venus-GEN3 was amplified from plasmid AGB024 (Venus-GEN3) using oligos AGO195 and 197. This product and plasmid AGB123 were cotransformed into yeast to create plasmid AGB136 (CDC12-Venus-GEN3). AGB136 was verified by digestion with BamH1 and NdeI and sequencing using oligos AGO202, 206, 208, and 209. AGB136 was transformed into strain AG127 to create strain AG308. Verification of these strains was performed using fluorescence light microscopy.
Colony radial growth assays were performed by spotting equal volumes of spores from each mutant strain beside an isogenic strain carrying the Sep7-GFP-NAT1 construct on selective media. Measurements for four independent experiments for each genotype were collected each day for 10 d of growth at 30°C.
Microscope Setup and Image Processing
An AxioImager-M1 upright light microscope (Carl Zeiss, Jena, Germany) equipped with the following Zeiss oil immersion objectives was used: EC Plan-Neofluar 40×/1.3 numerical aperture (NA), Plan-Apochromat 63×/1.4 NA, Plan-Neofluar 100×/1.3 NA, and α Plan-Fluar 100×/1.45 NA. For visualization of green fluorescent protein (GFP), filter set 41025 (Chroma Technology, Brattleboro, VT) and filter set 38HE (Carl Zeiss) were used. For visualization of AlexaFluor 568, Chroma filter sets 41002B and 41043 were used. For visualization of Hoechst, Zeiss filter set 49 was used. An Exfo X-Cite 120 lamp was used as the fluorescent light source. Images were acquired with an Orca-AG (C4742-80-12AG; Hamamatsu, Bridgewater, NJ) charge-coupled device camera driven by either OpenLab 5 (Improvision, Lexington, MA) or Volocity 4 (Improvision). Z-stacks of still images were acquired at different slice sizes (0.2–0.5 μm) and processed by fast or iterative deconvolution using calculated point spread functions. OpenLab 5 was used to acquire time-lapse movies that were then processed in Volocity 4. All still images were linearly contrast enhanced in Volocity 4 and Photoshop CS2 (Adobe Systems, Mountain View, CA). All images and movies presented are maximum projections of three-dimensional volumes.
Time-lapse Analysis
All time-lapse movies presented here were acquired using the EC Plan-Neofluar 40×/1.3 NA objective, 10–20% fluorescence light transmission, exposed for 100 ms per z-step, through 14 1-μm steps, and then iteratively deconvolved identically using a calculated point spread function (45 iterations). Cells used in time-lapse experiments were germinated and grown on 12–25% Ashbya Full Medium (AFM) + 1–3% dextrose 1% agarose gel pads in a humid chamber for ∼13 h at 30°C and imaged in a humid chamber at room temperature (∼23°C). Volocity 4 was used to measure ring lengths and intensities. Ring length was defined as the distance between the visible (by Sep7-GFP) basal and apical boundaries of the ring, parallel with the growth axis. Measurements of individual ring lengths over a period of 4 h were initiated when separation from the growing tip was discernible. Initiation of measurements of Sep7-GFP intensity at the rings was done identically. To measure the intensity of a given ring, a region of interest was drawn around the boundaries of the ring, and the mean fluorescence value per pixel of the selected area was recorded. An adjacent region of the hypha (lacking a ring) was measured, and this average fluorescence value per pixel was subtracted from the ring area fluorescence value to yield the fluorescence per pixel that was contributed by the ring. The regions of interest were redrawn at every time point to compensate for any reorientation or movement of hyphae or gel pad drift. The variable presence of moving nuclei in the ring's region of interest contributes to noise in these measurements as nuclei have considerably less fluorescence than the cortex or the cytosol. Time-lapse movies were linearly contrast enhanced in Volocity 4.
Visualization of F-Actin, Microtubules, and Chitin
To view F-actin and maintain viability of the Sep7-GFP fluorophore, cells were fixed in a 2% paraformaldehyde (Fisher Scientific, Fair Lawn, NJ) and full media solution at 30°C (unless otherwise indicated). After fixing for 5–10 min, cell cultures were immediately washed with 1× phosphate-buffered saline (PBS) to prevent overfixation and concentrated by centrifugation. Cells were washed again and resuspended in 1× PBS. To stain for F-actin, cells were incubated with AlexaFluor-Phalloidin568 (Invitrogen) at a concentration of 6.6 μM and incubated at room temperature in the dark for 1 h. Cells were then washed twice with 1× PBS and resuspended in ProLong Gold antifade reagent (Invitrogen). To visualize chitin at septa, cells were grown in liquid culture for ∼14 h, collected via centrifugation, and washed with distilled H2O. Cells were then stained for 5 min with calcofluor white at a final concentration of 167 μg/ml. Cells were washed twice, resuspended in water, and mounted on a glass slide for observation under the microscope.
For cells treated with latrunculin-B (Lat-B; Alexis Biochemicals, San Diego, CA), liquid cultures were grown overnight for ∼13 h. These cells were incubated for 1 h at 30°C with latrunculin-B at a final concentration of 50 μM. For visualization of actin and Sep7-GFP, standard procedures were followed.
Treatment of cells with nocodazole (Sigma-Aldrich, St. Louis, MO) involved two separate fixation protocols. To view the Sep7-GFP fluorophore and nuclei response to nocodazole, cells were grown in liquid cultures for ∼14 h and treated with nocodazole for 2 h at a final concentration of 20 μg/ml at 30°C. Cells were fixed in a 2% paraformaldehyde and full media solution at 30°C for 15 min. The culture was then concentrated by centrifugation, washed and suspended with 1× PBS, and incubated at room temperature for 15 min with 1:500 Hoechst (1 mg/ml stock; Invitrogen). Stained cells were collected by centrifugation and resuspended in ProLong Gold antifade reagent. To confirm microtubules depolymerized in response to nocodazole, cells were grown and treated with nocodazole as described above. After incubation with nocodazole, cells were fixed for 1 h in formaldehyde (Fisher Scientific) at a final concentration of 3.7%. Cells were then processed for anti-tubulin immunofluorescence as described previously. Cells were incubated overnight at 4°C in a humid chamber with rat-anti-α-tubulin (Serotec, Oxford, United Kingdom) at a 1:50 dilution in 1× PBS + bovine serum albumin. After washing, cells were incubated 1 h in 2° antibody (1:200 dilution, Alexa Fluor 568-conjugated goat anti-rat antibody) and 1:500 dilution Hoescht (1 mg/ml stock solution). Cells were mounted on slides in ProLong Gold antifade reagent.
RESULTS
Diverse Higher Order Septin Structures Coexist in One Cell
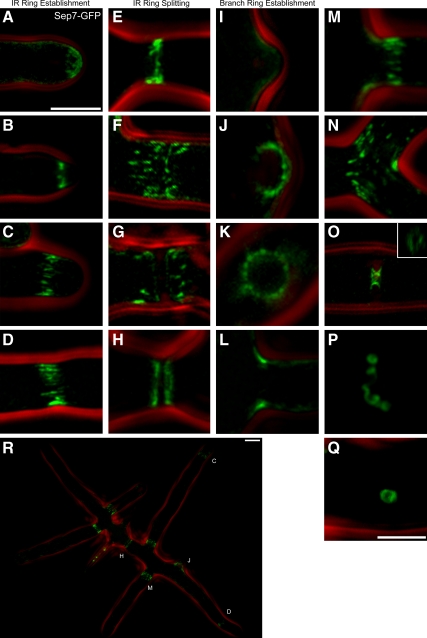
To visualize the septin cortex in living A. gossypii cells, Sep7p-GFP was expressed from the endogenous locus as the only copy of the gene and imaged by wide-field fluorescence microscopy followed by iterative deconvolution (see Materials and Methods). A. gossypii cells lacking Sep7p do not assemble septin rings as measured by Cdc11a-mCherry fluorescence (Supplemental Figure 1). Thus, Sep7p is an integral component of the septin cortex in these fungal cells and essential for higher order structures. Sep7-GFP strains have growth rates and morphology identical to the wild type, indicating that this fusion did not interfere with the function of Sep7p (Figure 1R; Helfer and Gladfelter, 2006; data not shown).
Figure 1.
Sep7-GFP localizes to diverse higher order septin structures. Sep7-GFP (AG127)–expressing cells were imaged live after growth overnight on media containing-agarose pad except where noted. All images are maximum projections of image stacks. (A and B) Sep7p at the hyphal tip. (C) A nascent IR ring. (D and E) IR rings. (F–H) Splitting of an IR ring and septation. (I) Sep7p at an incipient branch site. (J) Branch ring. (K) Cross section of branch ring. (L) Branch ring. (M) IR ring formed adjacent to branch ring. (N) IR ring traversing a Y-junction. (O) Hourglass-shaped septin structure at a septum. (P) Septin helix formed under stress. (Q) Cortically associated ∼1-μm ectopic ring. (R) Small mycelia in which some examples of the classes of rings shown in A–Q are noted highlighted by letters. (K and P) Cell was fixed with 2% paraformaldehyde. GFP fluorescence (green) and phase contrast outlines (red) are shown. Bars, 5 μm (A–J and L–O; in A); 2.5 μm (K, P, and Q; in Q); and 5 μm (R).
A diverse array of higher order septin structures is visible across developing single-celled mycelia (Figure 1, A–Q and R). Near tips of hyphae, Sep7p forms a diffuse cloud in which some faint, thin filaments can be distinguished (Figure 1A). Within ∼15 μm of growing tips, a cortical septin ring composed of discrete, thick filaments or “bars” is present in the majority of hyphae (Figure 1, B and C). We call these high order structures rings because they surround the cortex such that in cross section they look like circles (Figure 1K). Similar rings composed of Sep7p bars were observed at intervals ≥10 μm radiating away from the growing tip, toward the center of the cell (Figure 1D). Eighty-six percent of such rings were at this spacing frequency (N = 142). We call these rings interregion (IR) rings because they are in the middle region of the cell between the growing tip and the spore (Figure 1D). The dimensions of IR rings vary widely within and between cells. Specifically, the ring length, which was measured parallel to the growth axis, was on average 1.88 μm ± 0.622 (SD) long (N = 142). Additionally, double rings of short septin bars are found distant from the growing tips at the center of mycelia, which are septation sites (places where cell wall is deposited to generate two cell compartments, Figure 1, E–H).
Lateral branches emerge from the sides of hyphae in proximity to IR septin rings, indicating that one function of the rings may be in promoting branch emergence (Figure 1, I–L; 80% of branches within <10 μm of an IR ring; N = 109). Branches have a ring of septin filaments at their base, reminiscent of the septin structures found at the base of dendritic spines in mammalian neurons (Figure 1L; Tada et al., 2007; Xie et al., 2007). Branch rings have on average smaller dimensions than the IR rings but still are highly variable (1.44 ± 0.516 [SD] μm; N = 112). Additionally, the filaments of branch bases generally have much lower fluorescence intensity than IR rings. This makes maximum projections of three-dimensional (3D) images of branches seem to have concentrations of septins primarily at the edges and only faint signal across the hypha, but this is because of the projection. We could reliably count 14–18 bars/ring in branch base rings (Figure 1K) because branches can form perpendicular to the imaging plane, allowing full use of the high X-Y resolution of the microscope (N = 10). Often a new IR ring, deposited by the emerging branch tip, is found within 1.0 μm of these smaller branch rings (Figure 1M).
At lower frequency than the IR and branch base rings, additional distinct higher order structures can be found throughout cells (Figure 1, N–Q). In mature hyphae (20 h after germination) hyphal tips split into two forming a “Y-junction” and two new branches. Here, a variation on the IR ring is seen in which long filaments traverse the bent, “Y” shape extending from the original single axis into one of the two new axes (Figure 1N). Occasionally, an hourglass ring (Figure 1O) is observed that may be an alternative septation structure. In addition, helices seen in vitro with purified mammalian septins are also present here in vivo (Figure 1P) (Kinoshita et al., 2002). Finally, Sep7p can be found in ∼1-μm-diameter circles, of similar appearance and dimensions as those observed by Kinoshita and colleagues in mammalian cells when actin is disrupted (Kinoshita et al., 2002; Figure 1Q). These may represent a “reserve” pool of spontaneously forming septin structures that self-assemble into circles when they fail to be incorporated into organized higher order rings (see below).
These data demonstrate that morphologically diverse septin rings are found within a single cell (Figure 1R). Clearly, distinct classes of rings can coexist in a cell cortex that is in contact with a common cytoplasm. Many questions emerge about the organization of the septin cortex from these static images. Where are septin rings born? Can they assemble in the middle of hyphae? Do individual rings change dimensions or filament density with time after assembly? Do neighboring rings influence each other's organization? Do all rings persist for similar times? To address these questions, we performed time-lapse microscopy and followed septin rings in growing mycelia over many hours of development.
Septin Rings Assemble at Sites of Growth and Have Constant Dimensions
To measure the behavior of rings through time and space, Sep7-GFP cells were germinated on agar pads and followed for 12–16 h of growth by fluorescence microscopy. The time-lapse data demonstrated that all IR rings assemble at growing tips from clouds of diffuse filaments that initially travel with the growing tip (Supplemental Movies 1 and 2). Faint filaments detach from the tip and coalesce into discrete, resolvable septin filaments. These thick bars of septins are then anchored to the cortex and form a ring that is left in the “wake” of the growing tip (Supplemental Movies 1 and 2). Thus, IR rings never occur in the middle of hyphae but are exclusively assembled at sites of polarized growth. Similarly, side branch rings recruit Sep7 concurrently with the polarized growth generating branch emergence. However, as seen in the still images, branch rings seem to be fainter and shorter than IR rings (Supplemental Movies 1 and 2).
One possible mechanism by which diverse rings could be maintained on the cortex of a common cytoplasm would be if there were a maturation program or “timer” built into the nascent ring. Given the nucleotide binding capacity of septins, such a mechanism could be imagined based on GTP hydrolysis rates of individual subunits. In this case, we would expect that, upon assembly, there is a consistent time period from which a ring transitions from its initial appearance into different morphologies. However, splitting and disassembly of rings that were observed from their birth were rare events in the time-lapse movies of young, actively growing cells. We only observed splitting for six of 300 rings that were tracked from their assembly, so it was not possible to obtain significant measurements on the transition timing. Within this small population, no consistent timing pattern for transitions could be observed. We could see that when a cell showed signs of stress such as the appearance of vacuoles, there was a nearly instantaneous transition of many IR rings (>2 h old) to split double rings as well as the formation of ∼1-μm circles (Supplemental Movie 3). This indicates that rings of very different ages all have a similar potential to transition and change their higher order structures. These data suggest that an intrinsic timer that is based solely on the age of the septin complex is unlikely to be the primary basis for the diversity in septin ring organizations.
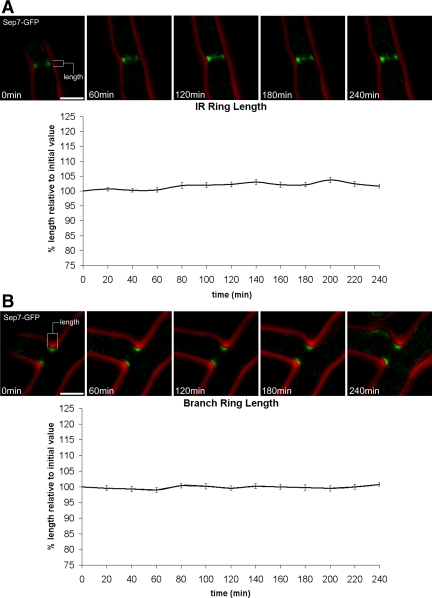
The time-lapse approach also enabled us to analyze the basis for variability in dimensions of the septin rings within different classes. The variability seen in the population could arise from heterogeneity in the initial dimensions of the ring at assembly or dynamics during maturation that led to the contraction or expansion (potentially polymerization or depolymerization) of filaments within the ring. Our data demonstrate that the initial dimensions of rings at their inception were maintained through the ring's lifetime until either disassembly or splitting (Figure 2, A and B). Individual rings did not change dimensions with time except on the rare occasion when a ring splits into two rings at septation sites (Figure 1, G and H, and Supplemental Movies 3 and 4). Thus, at birth, an individual ring's maximum size is established, however, the absolute size varies across rings. The fixed dimensions are indicative of a septin “cap,” potentially analogous to capping proteins for actin, that constrains the expansion of the cortex after assembly.
Figure 2.
Septin rings do not change dimensions with time. Single SEP7-GFP (AG127) cells were observed using 3D time-lapse fluorescence microscopy. Image stacks were acquired every 20 min. All images were acquired and processed identically. The sequences of images above the graphs are examples of an IR ring (A) and a branch ring (B) that were measured. (A) Individual IR septin rings were measured for 4 h after their establishment. N = 50. (B) Branch rings were measured for 4 h after their establishment. N = 30. The small fluctuations in these measurements reflect minor shifts in the orientation of the hypha containing the ring as it grows. GFP fluorescence (green) and phase contrast cell outlines (red) are shown. Bars, 5 μm.
Septin IR Rings Assimilate New Septin Subunits over Extended Time Periods
The constant dimensions of septin rings after assembly suggest that factors at the growing tip promote assembly and then rings are “static” until unknown cues trigger septation in a small subset of rings. Alternatively, rings may be constant in geometry but change in either the composition or dynamics of individual septin subunits and thus mature through time. These alternative models for maturation prompted us to measure the rate of incorporation of septin protein through the life of a ring.
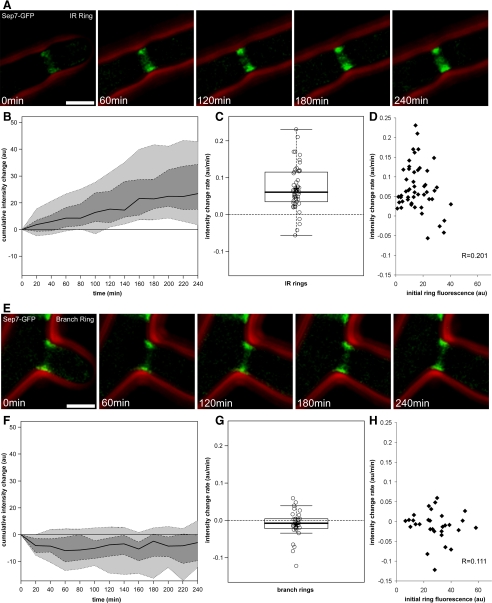
To assay Sep7p incorporation, the fluorescence intensity of Sep7-GFP at individual septin rings was determined for IR rings through time. We calculated an average fluorescence intensity from a three-dimensional reconstructed image at each time point (initial ring appearance to 240 min old). To control for machine noise and fluorescence in the cytosol underlying the septin cortex, a background fluorescence intensity from neighboring cytosol was measured at each time point and subtracted from the average fluorescence of the ring (see Materials and Methods). To measure the change in fluorescence of IR rings through time, the initial normalized fluorescence value for each ring was subtracted from each time point in the series. IR rings on average increase 2.5 times in their fluorescence intensity over 4 h; however, this increase varied from as little as 0.5 times to >7.0 times the starting intensity (Figure 3, A–C; N = 50 rings). The rate of increase in fluorescence we observed was significant based on a Student's t test (p < 0.001) in comparison with a null hypothesis of no increase in fluorescence. The rates of change in fluorescence vary significantly between rings (p < 0.01) but are nonetheless linear for all rings measured regardless of differences in their -fold change in fluorescence intensity (Figure 3, B and C). The linear behavior of the rate of fluorescence change in IR rings suggests that the incorporation of new subunits is likely due to filament polymerization or the stoichiometric association of additional free subunits with the existing polymers.
Figure 3.
Sep7-GFP intensity increases with time at IR septin rings but not at branch rings. Single SEP7-GFP (AG127) cells were observed using 3D time-lapse fluorescence microscopy. Image stacks were acquired every 20 min and were processed identically. (A and E) Images show examples of an IR ring (A) and a branch ring (E) that were measured. The GFP fluorescence (green) and phase contrast cell outlines (red) are shown. Bars, 5 μm. (B and F) Cumulative change in fluorescence intensity of Sep7-GFP at IR rings (B; N = 50) and branch rings (F; N = 30) was measured for 4 h after establishment. Change in fluorescence intensity was calculated by subtracting the intensity of the ring immediately after establishment (t = 0) from the intensity measured at a given time point. Intensity values were corrected for background fluorescence of the cytosol. Values are given in arbitrary fluorescence units (au). The black lines represent the median cumulative change for the population. Fifty percent of the measured rings are in the dark gray regions, and 80% of the rings fall in the combined light gray and dark gray regions. (C and G) The box and whisker plots show the distribution of slopes of the linear regression of fluorescence change at individual rings. The black line denotes the median value and the asterisk shows the mean value. The box is the middle 50% of the data, and the whiskers extend to the most extreme data point, no more than 1.5 times the interquartile range. Rates are given in arbitrary fluorescence units per minute (au/min). (D and H) The slopes of the linear regression of fluorescence changes for a ring are graphed against the initial fluorescence at that ring for each individual septin ring analyzed. Values are given in arbitrary fluorescence units and rates are given in arbitrary fluorescence units per minute. In both cases, R values are insignificant.
For an individual ring, the rates of intensity change are constant through time, indicating that addition of Sep7p to rings does not reach saturation in this interval. This suggests that subunit addition may be mechanistically similar both in very young rings and mature rings. Additionally, there is no correlation between the rate of fluorescence intensity change and the initial fluorescence intensity (Figure 3D; R = 0.20, N = 50). This indicates that the variation in rate of change between rings is not arising due to different starting amounts of Sep7p. Furthermore, the rate of fluorescence incorporation for a given ring does not correlate with its length (R = 0.11, N = 50) or due to its proximity to another septin ring (R = 0.19, N = 20). Thus, the rate of addition of new subunits is not determined by the amount of Sep7p in a preexisting ring or the length of that ring.
In striking contrast to IR rings, when similar intensity measurements were made on the rings at the bases of branches (such as the type in Figure 1, I–L) Sep7p intensity did not increase and thus had fluorescence change close to or at zero (Figure 3, E–G; N = 30 rings). The median rate of change in this class of rings is slightly negative, potentially because of photobleaching over time or loss of subunits. Branch rings fail to increase in intensity even when nearby (<10 μm away) IR rings do increase fluorescence intensity, indicating that this lack of incorporation is not likely because of local limitation in the concentration of free subunits.
Variable Septin Morphology Is Not Because of the Presence or Absence of Different Septin Subunits within a Ring
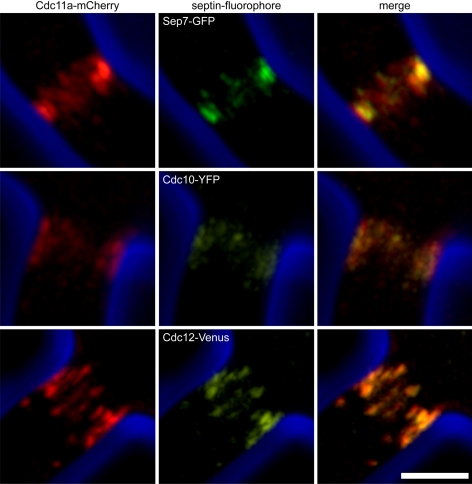
Given the variability in intensity and ring organization that we observed in one cell, we hypothesized that different septin structures within A. gossypii could arise because of the presence or absence of one or more of the six septin proteins in different classes of rings. This idea is attractive because of the ability of only two or three septin subunits to polymerize in vitro and assemble higher order structures in other species. To test this hypothesis, we tagged each individual septin subunit with a fluorescent protein and determined whether each septin subunit was found in all classes of septin rings. Each of the septins examined (Cdc3, Cdc10, Cdc11a, Cdc12, and Sep7) was found in all classes of rings, uniformly across the entire mycelia (Figure 4 and Supplemental Figure 2). To address whether two septin subunits precisely colocalized based on fluorescence signals, we coexpressed pairs of septin subunits that were tagged with different fluorescent proteins.
Figure 4.
All classes of septin rings contain all septin subunits. A. gossypii strains expressing Cdc11a-mCherry from the endogenous locus and either Sep7-GFP (AG232), Cdc10-YFP (AG306), or Cdc12-Venus(AG308) expressed using their endogenous promoter from a replicating plasmid were imaged in live cells grown overnight on agarose media pads. mCherry fluorescence (red), GFP fluorescence (green), Venus fluorescence (yellow), and phase contrast outlines (blue) are shown. Bar, 5 μm.
Representative colocalizations for Sep7-GFP and Cdc11-mCherry, Cdc10-YFP and Cdc11-mCherry, and Cdc12-YFP and Cdc11-mCherry are shown in Figure 4. In all pairwise combinations tested, the septins exactly colocalize in rings. Thus, as detectable by fluorescence, individual septin subunits are symmetrically and uniformly distributed in all classes of septin rings. These data indicate that all expressed septins are incorporated into all rings and that variable higher order organizations do not arise because of the incorporation or exclusion of specific septin subunits.
Septin Rings Are Maintained Independent of Actin or Microtubule Cytoskeletons
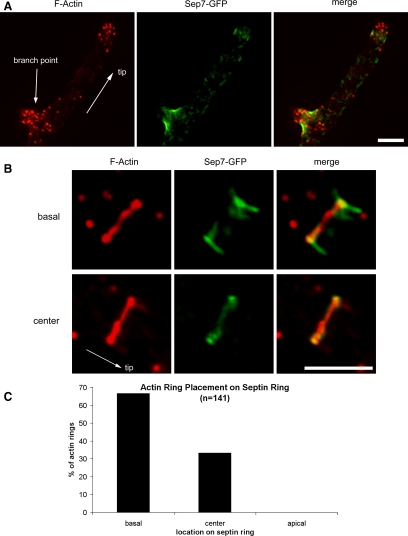
In mammalian cells, higher order septin organization has been linked to both the actin and microtubule cytoskeletons. We therefore tested the hypothesis that variability in ring structure arises because of associations with the actin or microtubule cytoskeletons. First, we observed the colocalization of F-actin and Sep7-GFP in normal growth conditions. The faint septin filaments form at growing tips where actin cables are nucleated; however, we could not resolve individual septin filaments juxtaposed to actin cables. New IR rings are located just behind the concentration of F-actin patches that congregate at growing tips (Figure 5A). Rings of F-actin were found at only a subset of IR and branch base rings. These F-actin rings are located at septin IR rings in the center of cells closest to the spores that are the most mature. Remarkably, the actin rings are not localized in the center of septin rings but instead were on the basal end (side closest to the center of the cell, away from the growing tip) in 67% of rings (N = 141). In the remaining cases, actin rings are in the center of the septin ring but never on the apical side of the septin ring. The asymmetric association related to the length of the ring such that rings with an actin ring at the basal side are on average 0.90 ± 0.19 (SD) μm long, whereas septin rings with actin rings in the middle are on average 0.51 ± 0.13 μm (N = 82; the smaller overall dimensions compared with the live imaging is because of fixation-induced cell shrinkage). These data indicate that there is some polarity within a ring as to the ability to recruit F-actin and that longer rings tend to be more asymmetric (Figure 5). Given that actin rings are observed only in mature septin rings that may already be many hours old, this septin ring asymmetry must be maintained for an extended time or established well after the initial assembly of the ring.
Figure 5.
F-Actin rings are asymmetrically localized at septin rings. (A) Colocalization of Sep7-GFP and F-actin. SEP7-GFP cells (AG127) were grown overnight in liquid media at 30°C. Cells were fixed with 2% paraformaldehyde and the F-actin was visualized using AlexaFluor-Phalloidin568. (B) Actin rings localized to the basal end (top) or center (bottom) of established septin rings. Arrows point to the hyphal tip and branch point. GFP fluorescence (green), Alexa568-Phalloidin (red), and phase contrast cell outlines (blue) are shown. Bar is 3 μm. (C) Actin ring position was scored relative to the associated septin ring (N = 141).
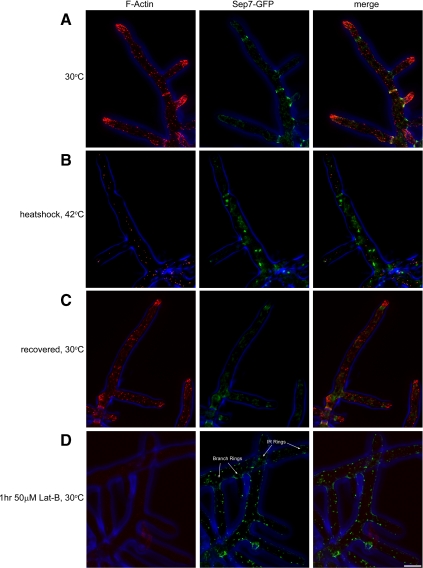
We perturbed the normal organization of F-actin with heat shock by transferring cells to 42°C for 20 min. This treatment leads to a rapid depolarization of the actin cytoskeleton in which cables and rings disassembled and F-actin patches redistributed uniformly along hyphae (Figure 6, A and B). On heat shock, Sep7-GFP fails to accumulate at growing tips (Figure 6B). All other higher order septin structures persist and are found with similar frequency to normal conditions (with the exception of an increase in the frequency of ∼1 μm circles; Figure 6B). When returned to 30°C, tip-associated septins accumulate when cell polarity is reestablished, usually within 20 min of recovery (Figure 6C). This indicates that only the initial tip-associated assembly of septin filaments requires polarized F-actin.
Figure 6.
F-actin is required for septin ring assembly but not maintenance. SEP7-GFP cells (AG127) were grown overnight in liquid AFM at 30°C. (A) Before heat shock, cells were fixed with 2% paraformaldehyde, and the F-actin was visualized using AlexaFluor-Phalloidin568. (B) Cells were transferred to 42°C for 20 min to depolarize the actin cytoskeleton and then processed for F-actin visualization. (C) Heat-shocked cells were returned to 30°C to recover for 20 min and then processed for F-actin visualization. (D) Cells were incubated for 1 h in growth media with latrunculin-B (final concentration, 50 μM) and then processed for F-actin visualization. GFP fluorescence (green), AlexaFluor-Phalloidin568 fluorescence (red), and phase contrast cell outlines (blue) are shown. Bar, 5 μm.
To determine whether the loss of septin filaments at tips is directly due to the loss of actin polarity or due to a more general response to temperature stress, we incubated cells in 50 μM Lat-B, which is sufficient to eliminate all F-actin in both cables and patches in cells (Figure 6D). Similar to heat shock, septin filaments are not assembled at tips in the presence of Lat-B, but all other classes of assembled rings persist throughout the cell (Figure 6D).
Additionally, during the incubations both at high temperature and in Lat-B, we observed a profusion of ∼1-μm septin circles. This suggests that these structures do not require actin polarity or cables for assembly. On return to 30°C after heat shock, the tip-associated septin filaments reestablish to all growing tips, concomitant with the disappearance of the small septin circles. This observation led us to speculate that these circles may form spontaneously at a sufficient concentration of free septins in the cytosol. In support of this idea, we have seen these circles in live cells when tip growth and IR ring assembly stop transiently. We have also observed them disappear just before a new branch ring forming (Supplemental Movies 4 and 5). We speculate that the ∼1-μm circles are likely self-assembling structures that form above a certain cytosolic concentration of septin oligomers or polymers. Thus, the majority of septin structures are maintained in the absence of polarized F-actin; however, assembly of nascent rings requires cell polarity.
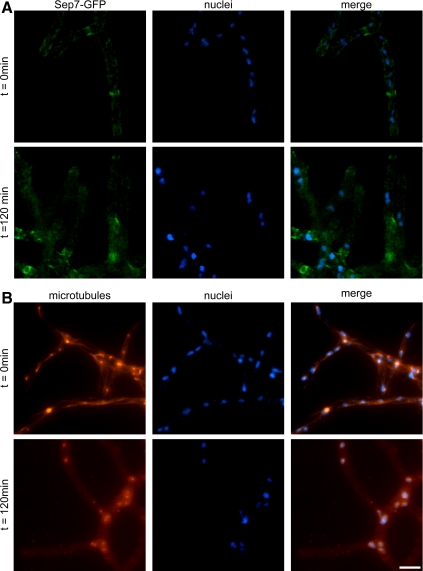
To determine whether microtubule dynamics or polymerization are required for septin organization, we incubated Sep7-GFP–expressing cells in the microtubule-destabilizing drug nocodazole. Under normal growth conditions, A. gossypii cells have intranuclear spindle microtubules, short cytosolic astral microtubules that emanate from the spindle pole body, and extensive cytosolic microtubules on which nuclei are transported for long-range migration toward the growing tip (Alberti-Segui et al., 2001; Gladfelter et al., 2006). Incubation in nocadazole leads to disassembly of all microtubules and induces an arrest of nuclei at G2/M in the division cycle, although polarized growth of hyphal tips continues at normal rates (Figure 7A; Gladfelter et al., 2006). On loss of microtubules, all higher order septin structures are retained, and new septin filaments are still visible at growing tips (Figure 7B). These data indicate that neither the progression of nuclei through the division cycle nor the microtubule cytoskeleton is required for any detectable feature of septin assembly or organization. This separation between septin organization and cell cycle progression is similar to what is observed in differentiated mammalian cells such as neurons where septins assemble and mature in G0.
Figure 7.
Microtubules are not required for septin ring assembly or maintenance. Cells expressing Sep7-GFP (AG127) were grown overnight in liquid AFM media at 30°C (t = 0) and then were treated with 20 μg/ml nocodazole for 120 min and fixed for visualization of Sep7-GFP (A) or anti-α-tubulin immunofluorescence (B) in combination with Hoechst staining of DNA. Sep7-GFP (green), anti-α-tubulin (red), and Hoechst (blue) stainings are shown. Bars, 5 μm.
Specific Ring Morphologies Require the Activity of Different Regulatory Proteins
The variable septin organization observed in A. gossypii is not because of an intrinsic timing program built into the septin ring, the presence or absence of specific septin subunits, or the actin or microtubule cytoskeletons. We hypothesized that distinct septin structures form at the cortex of a common cytoplasm because of the localized activity of septin regulators. Regulators such as kinases could deliver posttranslational modifications to septins that either lead to the assembly of different classes of rings or different maturation properties of individual rings. Ring-autonomous maturation steps that may be regulated include the initial anchoring to the cortex, the addition of new septin subunits to a pre-existing ring or the compression of filaments at a splitting ring. We predicted that mutation of such a regulator may disrupt only a subset of rings that require that activity for their genesis or maintenance, whereas the remaining classes are unaffected.
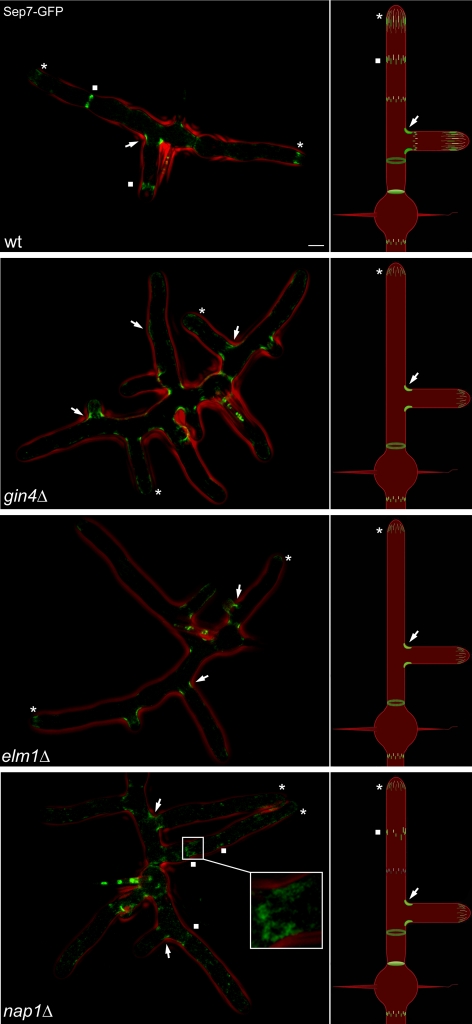
We tested this hypothesis by deleting the septin-associated kinases ELM1 and GIN4 and a septin-interacting factor encoded by NAP1 in cells expressing Sep7p-GFP. Notably, deletion of either GIN4 or ELM1 leads to complete loss of all IR rings found along main hyphae (N ≥500 hypha examined for each mutant). However, in both mutants there are still detectable fine filaments at tips, ∼1-μm circles, and branch base rings (Figure 8). Mutants lacking Elm1p have more intense tip associated filaments, whereas gin4Δ mutants have tip filaments comparable with wild type, indicating that potentially these kinases play different roles in the establishment of IR rings. In both mutants, the branch base rings had a similar spectrum of dimensions and fluorescence intensities as observed for wild-type cells, suggesting that these regulators are not required for any feature of this class of rings. We confirmed that these phenotypes were because of altered organization of the entire septin ring and did not simply delocalize Sep7p by observing the same phenotypes in mutants expressing a Cdc11a-mCherry (Supplemental Figure 3). Thus, these kinases are dispensable for the formation of branch base rings and filament assembly at tips but are required for IR ring morphogenesis.
Figure 8.
Deletion of septin regulators disrupts IR rings but not branch rings. Live cell fluorescence images of wild-type SEP7-GFP (AG127), gin4Δ SEP7-GFP (AG121), elm1Δ SEP7-GFP (AG120), and nap1Δ SEP7-GFP (AG209) cells grown overnight on agarose pads. Asterisks, squares, and arrows highlight the presence of tip-associated filaments, interregion rings, and branch rings, respectively. In the nap1Δ panel, an asymmetric interregion ring is highlighted and enlarged. GFP fluorescence (green) and phase contrast cell outlines (red) are shown. Bar, 5 μm.
Cells lacking these kinases showed several morphological defects in addition to loss of the septin IR rings. elm1Δ mutant hyphae (3.04 ± 0.03 [SEM] μm; N = 50) and gin4 Δ mutant hyphae (3.92 ± 0.04 μm; N = 55) are both significantly more narrow compared with wild-type cells (4.17 ± 0.05 μm; N = 66; p < 0.001 based Student's t test). Both elm1Δ and gin4Δ mutants lack any septation sites as monitored by calcofluor staining of the cell wall (Supplemental Figure 4). In response to localized cell stress or death, septa in wild-type cells act as “bulk-heads” and wall off the damaged cell from the rest of the mycelia (Supplemental Movie 3 and Supplemental Figure 5). In contrast, in the gin4Δ and elm1Δ mutants, the cell stress phenotype spreads across the entire mycelia and is not locally contained (Supplemental Figure 5). Finally, both mutants showed diminished colony radial growth rates. gin4Δ is 7.55 ± 0.22 (SEM) mm/d, elm1Δ is 7.39 ± 0.36 mm/d compared with wild type, which is 9.75 ± 0.31 mm/d as measured for four independent colonies over 9 d. These growth differences may be because of both the hyphal morphology defects and the inability to compartmentalize hypha with septa because of the lack of IR rings.
To determine whether Elm1p and Gin4p function in the assembly or maintenance of IR rings, we followed septin organization in elm1Δ and gin4Δ cells through time (Supplemental Movies 6 and 7). We observed that IR rings do not assemble in either mutant. Septins concentrate at the tips in diffuse filaments in both mutants. These filaments detach from the growing tip but only transiently coalesce into faint, thin bars that are associated with the cortex. In no case are normal IR rings observed at a concentrated zone behind the growing tip. In contrast, branch ring assembly and maintenance does not differ from wild-type cells. Thus, the assembly of rings fails in both of these mutants, with a clear defect in anchoring and construction of the higher order bars in IR rings.
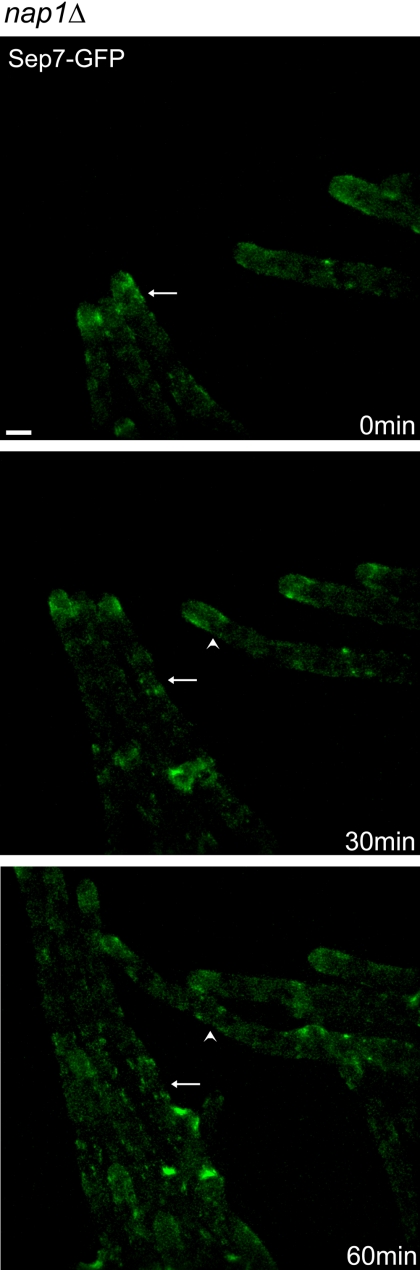
Deletion of NAP1 produces highly misorganized and asymmetric IR rings but, like the mutants described above, has no effect on the formation or organization of branch base rings. This suggests that Nap1p is important for the symmetric and constant dimensions that are imparted to a ring at assembly. Alternatively, symmetric rings may form transiently and then disassemble asymmetrically in the absence of Nap1p. Time-lapse analysis of Sep7-GFP in the nap1Δ mutant strains reveals that rings initially assemble with asymmetric dimensions, indicating a role for Nap1p in establishing IR ring dimensions and regular geometry (Figure 9).
Figure 9.
nap1Δ mutants assemble asymmetric IR rings. Images from a time-lapse series in which nap1Δ (AG314) cells were grown on an agar gel pad and imaged every 30 min. Sep7-GFP is asymmetrically deposited in IR rings as they form (arrow and arrowhead). Bar, 5 μm.
DISCUSSION
With the data presented here, we have established a new model system for the study of higher order septin structures. This fungal system complements current models for septin analysis in several important ways. Diverse, persistent, and distinct higher order structures form in the context of branching and polarized cell morphology (Tada et al., 2007; Xie et al., 2007). The coexistence and persistence of multiple, distinct septin structures in these branched fungal cells are remarkably similar to that of septins observed in the dendritic spines of mammalian neurons. We show that the long-lived rings are not static but rather change in their appearance and incorporate new subunits over many hours of existence. Importantly, these changes occur independently of the cell cycle. Thus, this is a powerful, genetically tractable model system in which to study mechanisms of in vivo assembly and maturation of the septin cortex without complication of cell cycle remodeling cues. Additionally, we show that IR and branch rings differ in their appearance, maturation and regulatory requirements. This indicates that spatially restricted signaling can produce functionally and morphologically distinct septin complexes within a single cell.
Septin complexes in A. gossypii initially assemble into fine filaments at growing hyphal tips and travel with the growth zone. After growing with the tip for 10–15 μm, an Elm1p/Gin4p kinase-dependent signal triggers the coalescence of filaments into thicker bundles and anchoring of complexes to the cortex to form stable IR rings. The role of these regulators may be to bundle filaments into higher order bars, to detach filaments from the site of polarized growth and/or to anchor septins to the plasma membrane. Interestingly, in Candida albicans hyphae, septins also initially congregate in a diffuse cloud at the growing hyphal tip (Sudbery, 2001; Gonzalez-Novo et al., 2004; Wightman et al., 2004; Gonzalez-Novo et al., 2008). Coordinately with the cell cycle, the septin clouds then coalesce into double rings to form septa that generate uninucleate compartments in the hyphae. Interestingly, Gin4p in C. albicans is dispensable for the tip-associated septin assembly and also the most basal septin structure formed at the germ bubble (Wightman et al., 2004). In C. albicans the dissociation of the cloud from the tip and formation of the septa is dependent upon Gin4p kinase, similar to what we have observed for A. gossypii, suggesting that the membrane anchoring event may be conserved among fungi and potentially animals. The abrupt kinase-dependent transition from faint, cytosolic filaments to bundles of thick bars fixed to the cortex, will enable future experiments to probe the molecular basis for septin/membrane connections. This is especially important because the mechanisms directing septin association to membranes are not known in any system, and we now have specific mutations that block this process.
The newly assembled IR rings vary in their initial size, intensity, and capacity to add subunits with time. However, each of these physical features function independently of one another because there is no correlation between any of these traits across populations of rings. Septin rings composed of bars similar to those in the IR rings are also seen in S. cerevisiae shmoos and gin4Δ mutants, and in C. albicans at the base of hyphal cells and in chlamydospores (Giot and Konopka, 1997; Sudbery, 2001; Wightman et al., 2004; Martin et al., 2005). However, none of these bar-structures require Gin4p kinase for assembly and/or are as persistent as the bars in IR rings that we observe. Therefore, we predict that there may be a different regulatory basis for the organization of septins into bars in the IR rings of A. gossypii. The septin bars we observe are spaced regularly around the cortex in a ring, and the number of bars per septin ring seems constant over an area, raising the question of how such uniform spacing is achieved. Potentially, there may be uncharacterized septin associated proteins that function similarly to microtubule-associated proteins and act as spacers to delineate the zones between septin bars.
After assembly and anchorage, the IR rings persist for indefinite periods (over multiple ≥16-h time series, only 6/300 rings disappeared). Notably, these IR rings continue to incorporate new septin subunits; in many cases rings more than double in their fluorescence intensity over 4 h of observation. This kind of maturation of the septin cortex has not been noted in other model fungi but may be a trait in higher eukaryotes where septin structures persist for extended periods. We predict that this additional protein incorporation could represent new filament polymerization or nonpolymerized Sep7p associating peripherally with septin filaments already organized in the ring. New filaments may polymerize in situ at a preassembled septin cortex or cytosolic filaments may intercalate into the preexisting higher order structure. It is also possible that the Sep7p subunits associate with the assembled structures without actually incorporating into filaments.
It is intriguing that new protein is added exclusively at bars and not in the spaces between the bars, suggesting that the preexisting structure is a template for the addition/polymerization of new protein. The rate of subunit addition varies but is always linear, indicating that the template/nucleator recruits a stoichiometric amount of either free subunit or polymer. Future experiments using photoactivatable GFP fused to septins and fluorescence recovery after photobleaching of the septin cortex will be useful in monitoring the position in the ring where addition takes place. Furthermore, there is no elongation of a septin bar with time suggesting that there maybe a capping protein prohibiting expansion from the ends of the existing polymer. The irregular alignment and variable lengths of filaments in nap1Δ mutants suggest that Nap1p may be required for capping the ends of the rings.
A notable trait of maturing IR rings is that they recruit F-actin rings asymmetrically and preferentially position actin at the basal (opposite from growth tip) end of the septin bars. This observation is striking given the known nonpolar nature of individual septin filaments formed in vitro (John et al., 2007; Sirajuddin et al., 2007; Bertin et al., 2008). Interestingly, in A. nidulans, the septin ring may disassemble in an asymmetric manner, with the basal side disappearing and the apical side persisting after septation (Westfall and Momany, 2002). In S. cerevisiae, dozens of proteins are asymmetrically localized to only one side of the septin collar (Gladfelter et al., 2001). In no case is the basis of the asymmetry known, although data in S. cerevisiae suggest that asymmetry is not due to the septins themselves but must be layered upon the septins by additional proteins (Vrabioiu and Mitchison, 2007). Our data further support that functional asymmetry can arise from a higher order septin structure in which septins are presumably symmetrically organized and suggest that this property is evolutionarily conserved.
Branch rings assemble near IR rings, indicating that the IR rings may serve as landmarks for the location of branch emergence. Cells lacking all septin structures or missing only IR rings still form branches but of aberrant morphology and positioning (Supplemental Figure 1 and Figures 8 and 9; Helfer and Gladfelter, 2006). Remarkably, the branch base septin rings do not add subunits with time and do not require the Elm1/Gin4 kinase for their assembly or maintenance. Therefore, the branch rings are morphologically distinct from IR rings, yet they generally form less than a few microns away from IR rings at the cortex of a common cytoplasm. New branch rings do not “poach” septin subunits from their neighboring IR rings, but in fact, neighboring IR rings will continue to increase in fluorescence intensity even while side branch rings form nearby. Presumably the different rings have access to common cytoplasmic pools of both septin subunits and regulatory proteins. This raises the question of how these different classes of septin rings are structurally and functionally distinguished despite their residence in the same space.
These data represent the debut of a new model system for the analysis of in vivo septin assembly and organization. We have demonstrated that the septin cortex shows great plasticity within a single cell. A. gossypii septin assembly, maturation, and organization do not require the nuclear cycle so this will be an important system for studying cell cycle-independent regulation of septins. The different septin structures and behaviors combined with the battery of mutants that we have developed make A. gossypii a great resource for biochemical and quantitative microscopy approaches to septin function in the future. We are well poised to understand how septin proteins may be modified to attach to membranes and to form diverse and persistent higher order structures in cells.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Harry Higgs, Elizabeth Smith, and Roger Sloboda for critically reading the manuscript; to Mark Borsuk and Cori D'Ausilio for statistical consultation; to the Gladfelter laboratory for helpful discussions; and to the Philippsen laboratory for advice and reagents. We thank Hanspeter Helfer for the original design of the scheme used in Figure 8. This work was supported by National Science Foundation grant MCB-0719126 (to A. G.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-12-1169) on February 18, 2009.
REFERENCES
- Alberti-Segui C., Dietrich F., Altmann-Johl R., Hoepfner D., Philippsen P. Cytoplasmic dynein is required to oppose the force that moves nuclei towards the hyphal tip in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 2001;114:975–986. doi: 10.1242/jcs.114.5.975. [DOI] [PubMed] [Google Scholar]
- Ayad-Durieux Y., Knechtle P., Goff S., Dietrich F., Philippsen P. A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 2000;113:4563–4575. doi: 10.1242/jcs.113.24.4563. [DOI] [PubMed] [Google Scholar]
- Barral Y., Mermall V., Mooseker M. S., Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell. 2000;5:841–851. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A., McMurray M. A., Grob P., Park S. S., Garcia G., 3rd, Patanwala I., Ng H. L., Alber T., Thorner J., Nogales E. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc. Natl. Acad. Sci. USA. 2008;105:8274–8279. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston J. P., Longtine M., Pringle J. R., Bi E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell. 2003;14:4051–4066. doi: 10.1091/mbc.E03-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich F. S., et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Gentry M. S., Hallberg R. L., Barral Y. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell. 2003;4:345–357. doi: 10.1016/s1534-5807(03)00061-3. [DOI] [PubMed] [Google Scholar]
- Egelhofer T. A., Villen J., McCusker D., Gygi S. P., Kellogg D. R. The septins function in G1 pathways that influence the pattern of cell growth in budding yeast. PLoS ONE. 2008;3:e2022. doi: 10.1371/journal.pone.0002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger F. P., Kopish K. R., White J. G. A role for septins in cellular and axonal migration in C. elegans. Dev. Biol. 2003;261:220–234. doi: 10.1016/s0012-1606(03)00296-3. [DOI] [PubMed] [Google Scholar]
- Frazier J. A., Wong M. L., Longtine M. S., Pringle J. R., Mann M., Mitchison T. J., Field C. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 1998;143:737–749. doi: 10.1083/jcb.143.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L., Konopka J. B. Functional analysis of the interaction between Afr1p and the Cdc12p septin, two proteins involved in pheromone-induced morphogenesis. Mol. Biol. Cell. 1997;8:987–998. doi: 10.1091/mbc.8.6.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A. S. Control of filamentous fungal cell shape by septins and formins. Nat. Rev. Microbiol. 2006a;4:223–229. doi: 10.1038/nrmicro1345. [DOI] [PubMed] [Google Scholar]
- Gladfelter A. S. Nuclear anarchy: mitosis in multinucleated cells. Curr. Opin. Microbiol. 2006b;9:547–552. doi: 10.1016/j.mib.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Gladfelter A. S., Hungerbuehler A. K., Philippsen P. Asynchronous nuclear division cycles in multinucleated cells. J. Cell Biol. 2006;172:347–362. doi: 10.1083/jcb.200507003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A. S., Pringle J. R., Lew D. J. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 2001;4:681–689. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Novo A., Correa-Bordes J., Labrador L., Sanchez M., Vazquez de Aldana C. R., Jimenez J. Sep7 is essential to modify septin ring dynamics and inhibit cell separation during Candida albicans hyphal growth. Mol. Biol. Cell. 2008;19:1509–1518. doi: 10.1091/mbc.E07-09-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Novo A., Jimenez J., Garcia M. J., Rios-Serrano I., Pla J., Jimenez A., Sanchez-Perez M. Dynamics of CaCdc10, a septin of Candida albicans, in living cells and during infection. Int. Microbiol. 2004;7:105–112. [PubMed] [Google Scholar]
- Haarer B. K., Pringle J. R. Immunofluorescence localization of the Saccharomyces cerevisiae CDC12 gene product to the vicinity of the 10-nm filaments in the mother-bud neck. Mol. Cell. Biol. 1987;7:3678–3687. doi: 10.1128/mcb.7.10.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall P. A., Russell S. E. The pathobiology of the septin gene family. J. Pathol. 2004;204:489–505. doi: 10.1002/path.1654. [DOI] [PubMed] [Google Scholar]
- Hall P. A., Todd C. B., Hyland P. L., McDade S. S., Grabsch H., Dattani M., Hillan K. J., Russell S. E. The septin-binding protein anillin is overexpressed in diverse human tumors. Clin. Cancer Res. 2005;11:6780–6786. doi: 10.1158/1078-0432.CCR-05-0997. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Helfer H., Gladfelter A. S. AgSwe1p regulates mitosis in response to morphogenesis and nutrients in multinucleated Ashbya gossypii cells. Mol. Biol. Cell. 2006;17:4494–4512. doi: 10.1091/mbc.E06-03-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M., Tomimoto H., Kitayama H., Morioka Y., Akiguchi I., Shibasaki H., Noda M., Kinoshita M. Association of the cytoskeletal GTP-binding protein Sept4/H5 with cytoplasmic inclusions found in Parkinson's disease and other synucleinopathies. J. Biol. Chem. 2003;278:24095–24102. doi: 10.1074/jbc.M301352200. [DOI] [PubMed] [Google Scholar]
- Ihara M., et al. Sept4, a component of presynaptic scaffold and Lewy bodies, is required for the suppression of alpha-synuclein neurotoxicity. Neuron. 2007;53:519–533. doi: 10.1016/j.neuron.2007.01.019. [DOI] [PubMed] [Google Scholar]
- John C. M., et al. The Caenorhabditis elegans septin complex is nonpolar. EMBO J. 2007;26:3296–3307. doi: 10.1038/sj.emboj.7601775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. S., Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann A. A plasmid collection for PCR-based gene targeting in the filamentous ascomycete Ashbya gossypii. Fung. Genet. Biol. doi: 10.1016/j.fgb.2009.05.002. In Press. [DOI] [PubMed] [Google Scholar]
- Kinoshita M., Field C. M., Coughlin M. L., Straight A. F., Mitchison T. J. Self- and actin-templated assembly of mammalian septins. Dev. Cell. 2002;3:791–802. doi: 10.1016/s1534-5807(02)00366-0. [DOI] [PubMed] [Google Scholar]
- Martin S. W., Douglas L. M., Konopka J. B. Cell cycle dynamics and quorum sensing in Candida albicans chlamydospores are distinct from budding and hyphal growth. Eukaryot. Cell. 2005;4:1191–1202. doi: 10.1128/EC.4.7.1191-1202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver J., Weber S. Flag N-terminal epitope overexpression of bacterial alkaline phosphatase and Flag C-terminal epitope tagging by PCR one-step targeted integration. Yeast. 1992;8(special issue):S627. [Google Scholar]
- McMurray M. A., Thorner J. Septin stability and recycling during dynamic structural transitions in cell division and development. Curr. Biol. 2008;18:1203–1208. doi: 10.1016/j.cub.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen E. M., McDonald H., Yates J., 3rd, Kellogg D. R. Cell cycle-dependent assembly of a Gin4-septin complex. Mol. Biol. Cell. 2002;13:2091–2105. doi: 10.1091/mbc.01-10-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Malmberg R. L., Momany M. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol. Biol. 2007;7:103. doi: 10.1186/1471-2148-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal A. A., Kozubowski L., Goode B. L., Drubin D. G., Hartwig J. H. Actin and septin ultrastructures at the budding yeast cell cortex. Mol. Biol. Cell. 2005;16:372–384. doi: 10.1091/mbc.E04-08-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S. E., Hall P. A. Do septins have a role in cancer? Br. J. Cancer. 2005;93:499–503. doi: 10.1038/sj.bjc.6602753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schmitz H. P., Kaufmann A., Köhli M., Laissue P. P., Philippsen P. From function to shape: a novel role of a formin in morphogenesis of the fungus Ashbaya gossypii. Mol. Biol. Cell. 2006;17:130–145. doi: 10.1091/mbc.E05-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M., Farkasovsky M., Hauer F., Kuhlmann D., Macara I. G., Weyand M., Stark H., Wittinghofer A. Structural insight into filament formation by mammalian septins. Nature. 2007;449:311–315. doi: 10.1038/nature06052. [DOI] [PubMed] [Google Scholar]
- Sitz J. H., Baumgartel K., Hammerle B., Papadopoulos C., Hekerman P., Tejedor F. J., Becker W., Lutz B. The Down syndrome candidate dual-specificity tyrosine phosphorylation-regulated kinase 1A phosphorylates the neurodegeneration-related septin 4. Neuroscience. 2008 doi: 10.1016/j.neuroscience.2008.09.034. [DOI] [PubMed] [Google Scholar]
- Sudbery P. E. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 2001;41:19–31. doi: 10.1046/j.1365-2958.2001.02459.x. [DOI] [PubMed] [Google Scholar]
- Tada T., Simonetta A., Batterton M., Kinoshita M., Edbauer D., Sheng M. Role of Septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr. Biol. 2007;17:1752–1758. doi: 10.1016/j.cub.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P. A., DeRisi J. L., Wilhelm J. E., Vale R. D. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- Versele M., Gullbrand B., Shulewitz M. J., Cid V. J., Bahmanyar S., Chen R. E., Barth P., Alber T., Thorner J. Protein-protein interactions governing septin heteropentamer assembly and septin filament organization in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:4568–4583. doi: 10.1091/mbc.E04-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele M., Thorner J. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J. Cell Biol. 2004;164:701–715. doi: 10.1083/jcb.200312070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele M., Thorner J. Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 2005 doi: 10.1016/j.tcb.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrabioiu A. M., Mitchison T. J. Symmetry of septin hourglass and ring structures. J. Mol. Biol. 2007;372:37–49. doi: 10.1016/j.jmb.2007.05.100. [DOI] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Weirich C. S., Erzberger J. P., Barral Y. The septin family of GTPases: architecture and dynamics. Nat. Rev. Mol. Cell Biol. 2008;9:478–489. doi: 10.1038/nrm2407. [DOI] [PubMed] [Google Scholar]
- Wendland J., Ayad-Durieux Y., Knechtle P., Rebischung C., Philippsen P. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene. 2000;242:381–391. doi: 10.1016/s0378-1119(99)00509-0. [DOI] [PubMed] [Google Scholar]
- Wendland J., Walther A. Ashbya gossypii: a model for fungal developmental biology. Nat. Rev. Microbiol. 2005;3:421–429. doi: 10.1038/nrmicro1148. [DOI] [PubMed] [Google Scholar]
- Westfall P. J., Momany M. Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol. Biol. Cell. 2002;13:110–118. doi: 10.1091/mbc.01-06-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman R., Bates S., Amornrrattanapan P., Sudbery P. In Candida albicans, the Nim1 kinases Gin4 and Hsl1 negatively regulate pseudohypha formation and Gin4 also controls septin organization. J. Cell Biol. 2004;164:581–591. doi: 10.1083/jcb.200307176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Vessey J. P., Konecna A., Dahm R., Macchi P., Kiebler M. A. The GTP-binding protein Septin 7 is critical for dendrite branching and dendritic-spine morphology. Curr. Biol. 2007;17:1746–1751. doi: 10.1016/j.cub.2007.08.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.