Abstract
Secretion and assembly of the extracellular matrix protein fibronectin regulates a number of normal cell and tissue functions and is dysregulated in disease states such as fibrosis, diabetes, and cancer. We found that mislocalized scaffolding by the plasma membrane Na-H exchanger NHE1 suppresses fibronectin expression, secretion, and assembly. In fibroblasts, wild-type NHE1 localizes to the distal margin of membrane protrusions or lamellipodia but a mutant NHE1-KRA2 lacking binding sites for PI(4,5)P2 and the ERM proteins ezrin, radixin, and moesin is mislocalized and found uniformly along the plasma membrane. Although NHE1 regulates intracellular pH homeostasis, fibronectin production is not regulated by changes in intracellular pH, nor is it attenuated in NHE1-deficient cells, indicating fibronectin expression is independent of NHE1 activity. However, fibronectin production is nearly absent in cells expressing NHE1-KRA2 because scaffolding by NHE1 is mislocalized. Additionally, secretion of active but not latent TGF-β is reduced and exogenous TGF-β restores fibronectin secretion and assembly. Our data indicate that scaffolding by NHE1-KRA2 dominantly suppresses fibronectin synthesis and TGF-β activation, and they suggest that NHE1-KRA2 can be used for obtaining a mechanistic understanding of how fibronectin production is regulated and speculatively for therapeutic control of dysregulated production in pathological conditions.
INTRODUCTION
The extracellular matrix protein fibronectin (FN) is a ligand for integrin receptors and has important functions in many physiological processes, including cell growth, differentiation, adhesion, and migration, and tissue remodeling and morphogenesis (reviewed in Larsen et al., 2006; Danen and Yamada, 2001; Miyamoto et al., 1998). Consistent with a role for FN in determining cell phenotype, increased FN expression is associated with differentiation of embryonic stem cells (Shirai et al., 2005). Increased FN deposition contributes to a number of pathological conditions, including hepatic (Kershenobich Stalnikowitz and Weissbrod, 2003) and pulmonary fibrosis (Crouch, 1990; Hetzel et al., 2005), and diabetic nephropathy, retinopathy, and macroangiopathy (Andresen et al., 1996; Hohenadel and van der Woude, 2004). In contrast, decreased FN deposition is seen in malignant cancers (reviewed in Ruoslahti, 1999; Labat-Robert, 2002), and FN expression is suggested as a prognostic indicator of lower risk for metastasis and longer survival (Christensen et al., 1988). Hence, a better understanding of how FN production is regulated will provide important information on its physiological and pathological functions.
The FN gene is a member of the “early-response” genes that rapidly increase expression to mediate cellular responses to growth factors. FN transcription is induced in response to the epidermal growth factor, platelet-derived growth factor and transforming growth factor-β (TGF-β; Blatti et al., 1988; Baron et al., 2006), most likely through direct effects on the FN promoter by the early growth response transcription factor Egr-1 (Liu et al., 2000; Baron et al., 2006). Cytokines also increase FN transcription by activation of the transcription factor NF-κB (Lee et al., 2002). However, regulated FN synthesis has primarily been studied in pathological conditions and little is known about the physiological regulation of FN production.
FN secreted by cells is soluble and is assembled into an insoluble fibrillar matrix that involves interactions with cell-surface receptors and extracellular matrix proteins (Wierzbicka-Patynowski and Schwarzbauer, 2003). FN binding by integrins is necessary to initiate and promote the assembly of FN into a fibrillar matrix (Wu et al., 1995; Wennerberg et al., 1996; Sechler et al., 2000). Integrin binding immobilizes dimeric FN, which induces conformational changes that expose FN-binding sites for dimerization to form fibrils (Schwarzbauer and Sechler, 1999; Pankov et al., 2000). Although the activation state of integrins regulates fibrillogenesis (Clark et al., 2004), interaction of the cytoplasmic domain of integrins with the actin cytoskeleton and signaling networks downstream of integrins involving Src and PI 3-kinase are also required (Wu et al., 1995; Hughes et al., 1996; Wierzbicka-Patynowski and Schwarzbauer, 2002). However, despite integrin activation and signaling being necessary for FN matrix assembly, they do not regulate FN production (Wu et al., 1995; Féral et al., 2007).
We now report an unexpected effect of mislocalized scaffolding by the Na-H exchanger NHE1 on inhibiting FN production that could provide insight on intracellular signals necessary for FN expression. NHE1, an integral plasma membrane protein, plays a central role in intracellular pH (pHi) and cell volume homeostasis by catalyzing an electroneutral exchange of extracellular Na+ for intracellular H+. NHE1 is activated by integrin engagement (Schwartz et al., 1991; Tominaga and Barber, 1998) and promotes the assembly (Tominaga and Barber, 1998; Denker et al., 2000; Denker and Barber, 2002) and turnover (Srivastava et al., 2008) of focal adhesions (FA). Independently of its ion transport activity, NHE1 also functions as a scaffolding platform for the assembly of signaling complexes by binding at least 12 distinct proteins (Baumgartner et al., 2006; Meima et al., 2007). Additionally, NHE1 binds phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2; Aharonovitz et al., 2000) and the ERM (ezrin/moesin/radixin) family of actin-binding proteins (Denker et al., 2000). In fibroblasts, ERM protein binding functions to cluster NHE1 at the distal margin of membrane protrusions or lamellipodia (Denker et al., 2000). We found that fibroblasts expressing a mutant NHE1 that lacks PI(4,5)P2 and ERM protein binding and is mislocalized along the plasma membrane do not produce or assemble FN. However, lack of FN production is due to mislocalized scaffolding by NHE1 and not mislocalized H+ efflux. Additionally, secretion of active but not latent TGF-β is suppressed by a mislocalized NHE1 scaffold, although exogenous TGF-β restores FN production and assembly. These data indicate that mislocalized NHE1 dominantly suppresses FN production and activation of latent TGF-β, perhaps by sequestering and mislocalizing necessary regulatory signals.
MATERIALS AND METHODS
Cell Culture
NHE1-deficient PS120 fibroblasts derived from hamster lung CCL39 fibroblasts (Pouyssegur et al., 1984) were obtained from J. Pouyssegur (INSERM, Nice, France). Wild-type and mutant NHE1 containing an hemagglutinin (HA)-epitope at the COOH-terminus and stably expressed in PS120 cells (WT, E266I, and KRA2 cells) were previously described (Denker et al., 2000). An NHE1-E266I-Myc construct was generated in pCDNA3.1 Hygro (+) and stably expressed in KRA2 cells (named K2E cells) by using Lipofectamine 2000 for transfection (Invitrogen, Carlsbad, CA). Heterologous expression of the System N1 transporter in PS120 cells (SN1 cells) was previously described (Chaudry et al., 1999). Fibroblasts were maintained in DME-H21 medium supplemented with 5% FBS and 1% Pen-Strep (growth medium) at 5% CO2. Mink lung epithelial cells (MLEC) expressing a truncated promoter of plasminogen activator inhibitor-1 (PAI-1; provided by R. Derynck, University of California, San Francisco) were maintained in DME-H21 medium supplemented with 10% FBS, 1% Pen-Strep, and 250 μg/ml geneticin (G-418; Invitrogen, Grand Island, NY) at 5% CO2. Where indicated, cells were treated with recombinant human (rh)TGF-β1 (PeproTech, Rocky Hill, NJ), and SB-431542 (Tocris Bioscience, Ellisville, MI) was used to inhibit activity of the TGF-β receptor type I (TGF-β-RI).
Immunolabeling
Unless otherwise indicated, cells grown for 48 h on glass coverslips were washed twice with PBS, fixed with 4% paraformaldehyde in PBS for 20 min, washed twice with PBS, permeabilized with 0.1% Triton X-100 in PBS for 10 min, and then blocked for 15 min with 10% FBS in PBS. Fixed cells were incubated with primary antibodies for 60 min, washed, and incubated with fluorescent-conjugated secondary antibodies for 45 min. For live-cell staining of extracellular FN, cells were incubated at 4°C with antibodies for FN for 1 h and fixed as described above without permeabilization. Primary antibodies were to paxillin (1:100; Zymed Laboratories, South San Francisco, CA), FN (1:100; Sigma, St. Louis, MO), β1 integrin (9EG7; 1:100; BD PharMingen, San Diego, CA), HA (12CA5; 1:100; Roche Molecular Biochemicals, Mannheim, Germany), and Myc (9B11; 1:200; Cell Signaling Technology, Danvers, MA). Secondary antibodies conjugated to FITC or Texas-red (Invitrogen) were used at 1:200 and nuclei were stained by incubating cells for 5 min with Hoechst 33342 (1:10,000; Invitrogen). Immunolabeling was visualized using a Zeiss Axiovert 35 (Thornwood, NY), and images were collected with a Spot RT cooled CCD (Diagnostic Instruments, Sterling Heights, MI).
For β1 integrin labeling, the Avidin/Biotin/Streptavidin Blocking Kit (Vector Laboratories, Burlingame, CA) was used. Cells were incubated with Avidin D for 15 min, rinsed with PBS, and then incubated with Biotin blocking solution for 15 min followed by incubating with anti-β1-integrin 9EG7 antibodies. The avidin conjugate was used for β1 integrin labeling, which was visualized as described above.
Immunoblotting
Unless otherwise indicated, immunoblotting used lysates from cells plated in 100-mm dishes and maintained in growth medium. Cells were washed three times in PBS and lysed in modified RIPA buffer (50 mM Tris-HCl, 135 mM NaCl, 3 mM KCl, 1% NP-40, protease inhibitors, 1 mM EGTA, 5 mM NaF, 10 mM sodium pyrophosphate, 1 mM glycerol phosphate, and 1 mM sodium vanadate, pH 7.2), and proteins in postnuclear supernatant fractions collected after centrifugation (850 × g for 5 min) were separated by SDS-PAGE and transferred to PVDF membranes. Membranes were probed with antibodies to FN (1:1000, Sigma), Egr-1 (1:500; Santa Cruz Biotechnology, Santa Cruz, CA), Smad3 (1:1000; Cell Signaling Technology), phosphorylated Smad3 (1:1000; Cell Signaling Technology), and β-actin (C4, 1:5000; Millipore, Temecula, CA). Bound antibody was detected by enhanced chemiluminescence (ECL; Amersham, Piscataway, NJ).
Immunoblotting for secreted FN was performed after chloroform-methanol precipitation of cell culture medium from cells 48 h after plating. Briefly, the medium was mixed methanol:chloroform (3:1 vol), and the mix was vortexed for 2 min and centrifuged at 10,000 × g for 5 min. The upper phase was discarded, leaving the interface intact, and the remaining mixture was washed with 300 μl methanol and centrifuged for 5 min, and pellets were resuspended in Laemmli buffer according to the concentration of the corresponding cell lysate. Proteins were separated by SDS-PAGE and processed for immunoblotting using antibodies to FN and β-actin.
Immunoblotting for NHE1-HA and TGF-β-RI used HA immune-precipitated complexes. Postnuclear supernatants from cell lysates were prepared as described for immunoblotting and incubated overnight with Sepharose-conjugated anti-HA antibodies (Roche Molecular Biochemicals). Immune complexes were recovered by centrifugation, separated by SDS-PAGE, transferred to PVDF membranes, and probed with antibodies to NHE1 (1:2000; Millipore), Myc (9B11, 1:1000), HA (12CA5; 1:1000), or TGF-β-RI (V-22, 1:500, Santa Cruz Biotechnology).
Immunoblotting for focal adhesion kinase (FAK) used lysates from cells maintained overnight in 0.2% FBS, trypsinized, resuspended in DMEM supplemented with 0.2% FBS, and plated in 100-mm dishes precoated with on FN (10 μg/ml). At the indicated times, postnuclear supernatants were collected as described above, and equal amounts of protein were precleared with protein G-Sepharose beads and incubated for 2 h with antibodies to FAK (2.5 μg; BD Biosciences, San Jose, CA), and for 1 h with protein G-Sepharose beads. Immune complexes were recovered by centrifugation, washed, separated by SDS-PAGE, and processed for immunoblotting with antibodies to phosphorylated FAK-pY397 (1:1000; Invitrogen). Membranes were stripped and probed with antibodies to FAK (1:1000; BD Biosciences).
Reverse Transcription-PCR
Collagen expression was determined by reverse transcription-PCR (RT-PCR) analysis for hamster collagen α1 (V) chain. Total RNA extracted using the RNeasy kit (Qiagen, Valencia, CA), and 2 μg was reverse transcribed using random decamer primers and amplified with PCR Supermix High Fidelity (Invitrogen) for 30 cycles at 94, 56, and 72°C. Oliogs included were F2 forward primer, 5′TACCCTGGAAGACAAGGGCC3′, and R2 reverse primer, 5′TCCTGGAGGGCCAGTCTTGC3′, which yielded a 322-base pair product, and F3 forward primer, 5′ATGGTGAACCTGGACAGACG3′, and R3 reverse primer, 5′TCCTTTGAGTCCAGGGAGTC3′, which yielded a 329-base pair product. Primers against 18S RNA were used as a control.
Northern Blotting
Cells were plated in 100-mm culture dishes to achieve 95, 50, and 25% confluency after 48 h. Total RNA was extracted using the RNeasy kit (Qiagen). RNA (20 μg) was separated on 1.0% agarose gels containing formaldehyde and transferred to a nylon membrane (Hybond-N+, Amersham). An FN DNA fragment (FN N29; provided by C. Damsky, University of California, San Francisco) was used as a template to prepare radiolabeled probe by using the Rad Prime DNA system (Invitrogen) according to the manufacturer's directions. Hybridization was performed under stringent conditions with 1× SSC and 0.1% SDS at 65°C for 2 h. GAPDH was used to confirm equal sample loading.
Luciferase Assays
The pGLFN105 promoter vector (Promega, Madison, WI) containing the FN promoter region between −105 and +14 was provided by D. Mercola (Sidney Kimmel Cancer Center, San Diego, CA) and previously described (Liu et al., 2000). pRK5 containing β-galactosidase under control of a human CMV promoter was provided by R. Derynck (University of California, San Francisco) and previously described (Feng et al., 1995). Cells were transfected using Transfast according to the manufacturer's recommendations (Promega). For each transfection the total quantity of transfected plasmid DNA pGLFN105 and pRK5 was kept constant. Forty-eight hours after transfection, cells were harvested and lysed in Reporter Lysis Buffer (Promega), and reporter activity was determined using a luciferase assay system (PharMingen) and β-galactosidase detection kit (Tropix, Bedford, MA) and measured using a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA). Data were normalized for transfection efficiency by β-galactosidase activity.
Secreted TGF-β was determined using MLEC cells stably expressing a truncated promoter of PAI-1 fused to the firefly luciferase reporter gene as described (Abe et al., 1994). MLEC cells were plated for 24 h in six-well tissue culture dishes, washed twice with PBS, and maintained for 4 h in DME-H21 medium without FBS. Conditioned medium collected from the indicated cells maintained for 18 h in DMEM supplemented with 0.2% FBS was directly applied to MLEC cells for determining active TGF-β or heated at 80°C for 10 min before applying for determining secreted latent TGF-β. In parallel cultures, MLEC cells were incubated in DMEM supplemented with (rh)TGF-β1 (0.1–5000 ng/ml; PeproTech). After 24 h MLEC cell extracts were prepared and assayed for luciferase activity using the Luciferase Assay System (Promega) according to the manufacturer's instructions, and luminescence was measured as described above. Luciferase units obtained from conditioned medium samples were normalized to cell number, and secreted TGF-β was quantified relative to luciferase units from samples supplemented with rhTGF-β1.
FITC-FN Labeling and Assembly Assays
Fibronectin assembly was determined by plating cells maintained in growth medium on glass coverslips coated with exogenous FITC-labeled bovine FN (15–50 μg/ml) or by adding exogenous FITC-labeled FN in the medium (McKeown-Longo and Mosher, 1995). Bovine plasma FN (Calbiochem-Novabiochem, La Jolla, CA) was labeled using the FluoReporter FITC protein labeling kit (Molecular Probes Invitrogen, Eugene, OR) according to the manufacturer's directions. Cells were maintained for 48 h, unless otherwise indicated, washed with PBS, fixed, and visualized as described for immunolabeling.
Integrin Expression and FITC-FN Binding
To determine β1 integrin expression, cells were biotinylated on ice for 90 min and lysed in modified RIPA buffer, and lysates were incubated with β1 integrin antibodies (9EG7, 1:100) for 60 min, followed by incubation for 60 min with protein A-Sepharose beads. Eluted proteins were probed for biotin with streptavidin-HRP. Integrin expression was confirmed by flow cytometry analysis. Cells were incubated in PBS-based dissociation buffer, washed twice with PBS, and kept in suspension in PBS containing 2% FBS. After incubating cells with anti-β1 integrin antibodies in PBS at 4°C for 1 h, cells were washed with PBS and incubated with anti-rabbit-Alexa Fluor 488 (A488) F(ab)2 (Invitrogen) at 4°C for 30 min. Cells were washed with PBS and analyzed using a FACS Vantage SE cell sorter (Becton Dickinson, San Jose, CA). Flow cytometry data were analyzed using CellQuest Pro4.0.1 software (Becton Dickinson). For β1 integrin binding to FN cells in suspension were mixed with the indicated concentrations of FITC-labeled FN for 60 min, and FITC-positive cells were analyzed as described above.
Intracellular pH Measurements
NHE1 activity and intracellular pH were determined in cells plated at 25 × 105 per well in 12-well plates and loaded with the fluorescent pH-sensitive dye 2,7-biscarboxyethyl-5(6)-carboxyfluorescein (BCECF; Invitrogen, Carlsbad, CA) by modifications of previously described methods (Denker et al., 2000). Ratios of BCECF fluorescence at Ex 490/Em 530 and Ex 440/Em 530 were acquired using a SpectraMax M5 plate reader (Molecular Dynamics). Fluorescence ratios were converted to pHi by calibrating each experiment with 10 μM nigericin (Invitrogen) in 105 mM KCl.
RESULTS
FN Expression and Transcription Are Decreased in Cells with Mislocalized NHE1
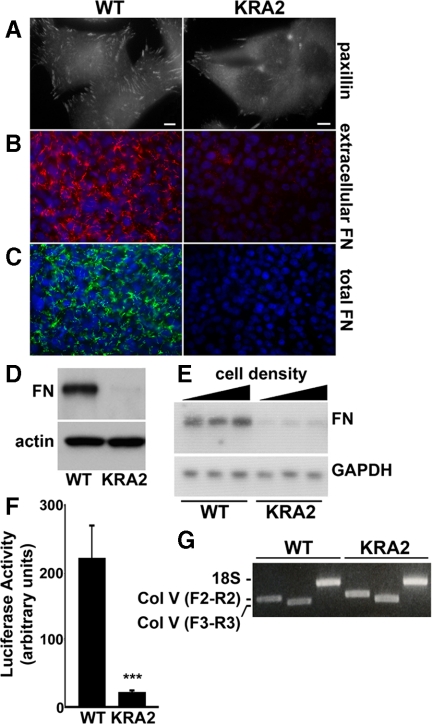
Wild-type NHE1, either endogenous in fibroblasts (Sardet et al., 1990; Grinstein et al., 1993) or recombinant expressed in NHE1-deficient PS120 fibroblasts (Denker et al., 2000; Denker and Barber, 2002), localizes to the distal margin of membrane protrusions or lamellipodia. The COOH-terminal cytoplasmic domain of NHE1 contains charged clusters of lysine and arginine residues in the juxtamembrane region that constitute binding sites for ERM proteins (Denker et al., 2000) and for PI(4,5)P2 (Aharonovitz et al., 2000). A mutant NHE1 with alanine substitutions in these charged clusters lacks ERM protein and PI(4,5)P2 binding and when stably expressed in NHE1-deficient PS120 fibroblasts is not clustered in lamellipodia but is uniformly distributed along the plasma membrane (Denker et al., 2000). PS120 fibroblasts stably expressing this mutant NHE1 (KRA2 cells; Table 1) have fewer and smaller FA complexes compared with PS120 fibroblasts stably expressing wild-type NHE1 (Figure 1A, WT) or CCL39 cells with endogenous NHE1 (data not shown), which are parental cells used for generating NHE1-deficient PS120 cells (Pouyssegur et al., 1984). In asking why FA complexes are attenuated in KRA2 cells, we found that FN is markedly attenuated compared with WT cells. Immunolabeling extracellular FN in live confluent fibroblasts showed a near absence of FN with KRA2 cells compared with abundant FN with WT cells (Figure 1B) or CCL39 fibroblasts (Supplemental Figure S1A). We also found that total FN (extracellular and intracellular) was nearly absent in KRA2 cells compared with WT cells (Figure 1C) and CCL39 cells (Supplemental Figure S1A), determined by immunolabeling permeabilized cells, and secreted FN was markedly attenuated (Figure 1D), determined by immunoblotting extracted culture medium (Figure 1D). Decreased total FN was not a clonal variation and was seen in all cell clones stably expressing NHE1-KRA2 (Supplemental Figure S1B).
Table 1.
Cell lines used in this study
| Cell name | Description | Reference |
|---|---|---|
| CCL39 | Clonal hamster lung fibroblasts with endogenous NHE1 | Pouyssegur et al. (1984) |
| PS120 | NHE1-deficient cells derived from CCL39 fibroblasts | Pouyssegur et al. (1984) |
| WT | PS120 cells stably expressing wild-type NHE1 | Denker et al. (2000) |
| KRA2 | PS120 cells stably expressing NHE1-KR/A, which contains alanine substitutions for COOH-terminal juxtamembrane lysine and arginine residues and does not bind PI(4,5)P2 or ERM proteins but does retain H+ efflux | Denker et al. (2000) |
| E266I | PS120 cells stably expressing NHE1-E266I, which contains an E266I substitution and does not have H+ efflux but does retain PI(4,5)P2 and ERM binding | Denker et al. 2000 |
| K2E | KRA2 cells stably co-expressing NHE1-E266I | This study |
| SN1 | PS120 cells stably expressing the System N1 glutamine-H+ transporter | Chaudry et al., (1999) |
Figure 1.
Fibronectin expression and transcription are decreased in fibroblasts expressing mislocalized NHE1-KRA2. (A) Paxillin immunolabeling reveals decreased abundance and size of FA complexes in KRA2 cells compared with WT cells. Bar, 5 μm. (B) Live-cell immunolabeling with FN antibodies reveals a marked decrease in secreted FN in KRA2 cells compared with WT cells (red, extracellular FN; blue, Hoechst staining for nuclei). (C) Total FN in permeabilized cells is decreased in KRA2 cells compared with WT cells (green, total FN; blue, Hoechst staining for nuclei). (D) Immunoblotting culture medium indicates secreted FN is attenuated in KRA2 cells compared with WT cells. Immunoblotting for β-actin in cell lysates was used to confirm equivalent cell numbers used to measure secreted FN. (E) Abundance of FN mRNA in KRA2 cells is attenuated compared with WT cells as determined by Northern blotting with GAPDH used as a loading control. FN transcription was not affected by cell density and was similar with 25, 50, and 95% confluency. Data are representative of blots from mRNA isolated from three separate cell preparations. (F) FN promoter activity, determined using a fragment of the human FN promoter between −105 and +14, is markedly decreased in the KRA2 cells compared with WT cells. Data represent means ± SEM of three separate cell preparations; *significant difference compared with WT; p < 0.001. (G) RT-PCR showing transcripts for hamster collagen α1 (V) chain in WT and KRA2 fibroblasts. Indicated are products using collagen primers F2 to R2 and F3 to R3, as described in Materials and Methods, and products of control primers for 18S RNA. Data are representative of two independent cell preparations.
We next asked whether decreased FN in KRA2 cells reflected changes in FN transcription. Because FN transcription can increase with greater cell density (Perkinson et al., 1996), we used cells at 25, 50, and 95% confluency. Northern blot analysis revealed a significant decrease in FN mRNA in KRA2 cells compared with WT cells at all three densities (Figure 1E; p < 0.001; n = 3). FN promoter activity also was significantly decreased in the KRA2 cells compared with WT cells, determined by expression of the pGLFN105 promoter vector containing a fragment (−105 and + 14) of the human FN promoter (Figure 1F; p < 0.001, n = 3). In contrast, expression of collagen V was similar in KRA2 and WT cells. Although antibodies to collagen I and V did not immunolabel hamster WT and KRA2 cells, RT-PCR confirmed both cell types express collagen V (Figure 1G). Because the hamster colI gene has not been sequenced, we used primers designed for rat and mouse collagen I; however, no signal was detected in WT or KRA2 cells (data not shown). These data indicate that in KRA cells FN production is decreased at the level of transcription, but collagen V expression is not impaired.
Fibronectin Assembly But Not Integrin Expression or FN Binding Is Impaired in KRA2 Fibroblasts
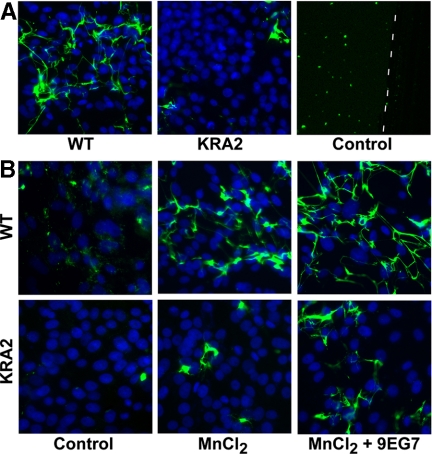
In addition to having decreased FN production and secretion, KRA2 cells were unable to assemble exogenous FN into a fibrillar matrix. When plated for 48 h on FITC-labeled FN (15 μg/ml) or with FITC-labeled FN in the medium (15 μg/ml), WT cells but not KRA2 cells assembled a fibrillar matrix (Figure 2A, Supplemental Figure S2). Control incubations without cells confirmed cell-dependent assembly of FN. KRA2 cells also did not assemble a fibrillar matrix with 30 or 50 μg/ml FITC-FN (data not shown) indicating impaired matrix assembly was independent of the exogenous FN concentration. Fibronectin assembly is dependent on integrin activation (Wu et al., 1995; Wennerberg et al., 1996; Sechler et al., 2000), and activating integrins with MnCl2 and integrin-activating 9EG7 antibodies increased FN assembly in KRA2 cells (Figure 2B). We plated cells on FITC-FN for 24 h and found that although limited matrix assembly was seen in KRA2 cells with the addition of MnCl2 (Figure 2B) or with 0.5 μg/ml 9EG7 antibody (data not shown) alone, there was a greater increase in assembly in both cells types with the addition of MnCl2 and 9EG7 antibodies. However, FN assembly by KRA2 cells was still less than that seen with WT cells.
Figure 2.
Fibronectin assembly is impaired in KRA2 fibroblasts. (A) WT cells but not KRA2 cells plated for 48 h assemble a matrix of exogenous FITC-labeled FN. FITC-FN–coated coverslips without cells (right panel) confirm cell-dependent FN assembly. White dashed line marks areas of the coverslip coated with FITC-FN (left of line) and scraped with a pipette tip (right of line). (B) Assembly of exogenous FITC-labeled FN in WT and KRA2 cells at 24 h after integrin activation with 0.05 mM MnCl2 alone or with 0.5 μg/ml integrin activating antibody 9EG7. MnCl2 plus 9EG7 accelerates FN assembly in WT cells and partially restores FN matrix assembly (green, exogenously added FITC-labeled FN; blue, Hoechst/nuclei).
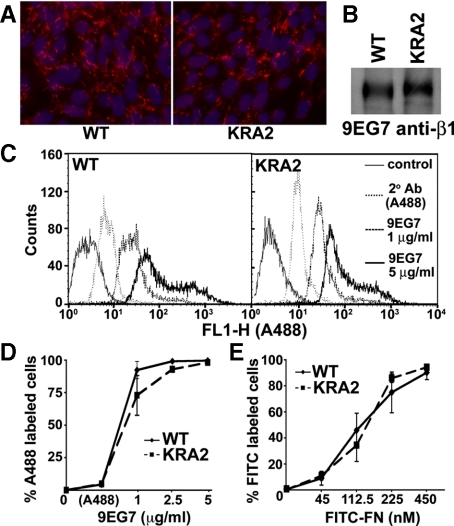
We used three approaches to show that expression of β1 integrin was similar in WT and KRA2 cells. We immunolabeled live, unpermeabilized cells with β1 antibodies (Figure 3A), we immunoprecipitated lysates of biotinylated cells with β1antibodies and immunoblotted immune complexes with streptavidin-HRP (Figure 3B), and we used flow cytometry of dissociated cells labeled with β1 antibodies (Figure 3C; Supplemental Figure S3). There also was no difference in total cell binding of FITC-labeled bovine FN (45–450 nM) by WT and KRA2 cells quantified by flow cytometry (Figure 3D, Supplemental Figure S4A; p > 0.1; n = 3). Specificity of FITC-FN binding was confirmed by blocking binding to WT cells with increasing concentrations of a FN RGD fragment (Supplemental Figure S4B). These data suggest that β1 expression and FN binding are not impaired in KRA2 cells, although attenuated integrin avidity, which we cannot directly measure in fibroblasts, might contribute to the inability of KRA2 cells to assemble a FN matrix. However, integrin activity is not necessary for FN biosynthesis (Wu et al., 1995; Féral et al., 2007), suggesting that decreased FN transcription in KRA2 cells is independent of possible reduced integrin avidity.
Figure 3.
β1 integrin expression and integrin affinity for FN are similar in WT and KRA2 fibroblasts. (A) Live-cell immunolabeling the surface of WT and KRA2 cells with β1 integrin antibodies (red, extracellular β1 integrin; blue, Hoechst staining for nuclei). (B) Cell surface expression of β1 integrin in WT and KRA2 cells determined after biotinylation, immunoprecipitation with 9EG7 anti-β1 antibodies, and immunoblotting the immune complex with streptavidin-HRP. (C and D) Cell surface expression of β1 integrin in WT and KRA2 cells determined by flow-cytometry analysis using the indicated concentrations of anti-β1 9EG7 antibodies. Secondary antibody (A488) alone was used to show nonspecific binding. (E) Binding of FITC-FN determined by flow-cytometry analysis indicates no difference between WT and KRA2 cells at each of the indicated concentrations of FN (p > 0.1; n = 3). Data are means ± SEM of three separate binding assays.
H+ Efflux by NHE1 Is Not Necessary for FN Production or Assembly
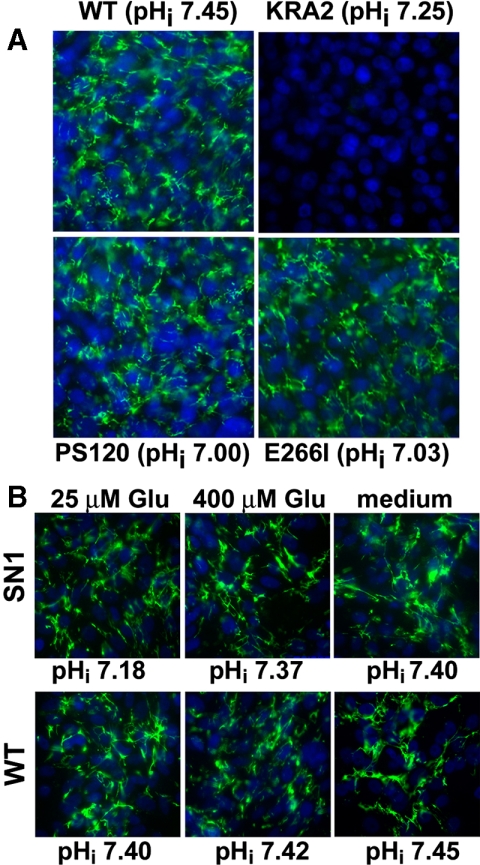
We next asked whether FN production and assembly are impaired in KRA2 cells because H+ efflux is decreased or mislocalized. Although KRA2 cells have regulated NHE1-dependent H+ efflux, NHE1 activity is attenuated compared with that in WT cells (Aharonovitz et al., 2000). However, live cell immunolabeling revealed that the abundance and organization of an extracellular FN matrix were not impaired in cells lacking H+ efflux by NHE1 (Figure 4A). This was confirmed in NHE1-deficient PS120 cells and in PS120 cells stably expressing a mutant inactive NHE1-E266I (E266I cells; Denker et al., 2000). These data also suggest that decreased steady-state pHi does not inhibit FN production because although the steady-state pHi of KRA2 cells was ∼7.25 and less than the ∼7.45 pHi of WT cells, the pHi of PS120 and E266I cells is ∼7.0 (Denker et al., 2000; Frantz et al., 2007).
Figure 4.
H+ efflux by NHE1 is not necessary for FN production or assembly. (A) NHE1-deficient PS120 cells and PS120 cells stably expressing mutant inactive NHE1-E266I lacking H+ efflux (E266I cells) have a FN matrix similar to that of WT cells. Steady-state pHi is given for indicated cell types. (B) Live-cell FN immunolabeling of WT cells and SN1 cells (PS120 fibroblasts stably expressing the SN1 amino acid transporter) maintained in glutamine-free DMEM supplemented with 25 and 400 μM extracellular glutamine or in growth medium containing 2 mM glutamine (green, extracellular FN; blue, Hoechst staining for nuclei). Steady-state pHi is indicated for each incubation condition. Data are representative of three separate cell preparations.
We previously showed that although NHE1-E266I is inactive, it retains binding to ERM proteins and PI(4,5)P2 and localizes like wild-type NHE1 at the distal margin of lamellipodia (Denker et al., 2000). Therefore, we speculated that FN production may be attenuated in KRA2 cells because H+ efflux is mislocalized. To test this prediction we used PS120 cells stably expressing the system N1 transporter (SN1 cells). SN1 is a plasma membrane amino acid transporter expressed in the CNS that couples uptake of extracellular glutamine with efflux of intracellular H+ efflux. When expressed in PS120 cells SN1 is uniformly localized along the plasma membrane, and increased H+ efflux can be driven acutely by increasing the concentration of extracellular glutamine (Chaudry et al., 1999). We found that extracellular FN secretion by SN1 cells was similar to that by WT cells at extracellular glutamine concentrations of 25 μM, which induces a pHi of 7.18 ± 0.03, 400 μM, which increases H+ efflux by SN1, resulting in a pHi of 7.37 ± 0.03, or when maintained in DMEM growth medium that contains 2 mM of glutamine and pHi is 7.40 ± 0.05 (Figure 4B). Incubating WT cells with extracellular glutamine concentrations between 25 μM to 2 mM had no effect on pHi or on the abundance of secreted FN secretion. These data confirm that FN production and matrix assembly is independent of pHi.
Localized Scaffolding by NHE1 Restores FN Production in KRA2 Cells
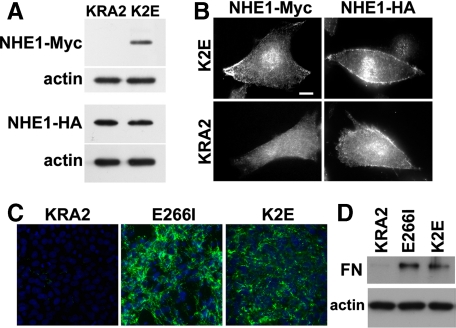
Our data suggested that attenuated FN production and assembly in KRA2 cells are not determined by decreased or mislocalized H+ efflux and that NHE1 expression is not necessary for FN production by PS120 cells. In addition to catalyzing Na+ and H+ exchange, NHE1 is recognized as a scaffolding platform by its COOH-terminal cytoplasmic domain directly binding more than 10 signaling proteins (Baumgartner et al., 2006; Meima et al., 2007). We predicted that NHE1-KRA2 might act in a dominant negative manner, perhaps by sequestering and mislocalizing a signaling molecule or complex necessary for FN production. To test this prediction we asked whether FN production in KRA2 cells could be restored by coexpressing NHE1 with normal localization in membrane protrusions. To confirm independence of H+ efflux we stably expressed NHE1-E266I-Myc in KRA2 cells (K2E cells). Expression of NHE1-E266I-Myc in K2E cells was confirmed by immunoblotting and had no effect on the abundance of NHE1-KRA2-HA expression (Figure 5A). Although we cannot rule out dimerization of NHE1-E266I-Myc and NHE1-KRA2-HA, the localization of NHE1-KRA2 was not visibly different in KRA2 and KE2 cells. Immunolabeling confirmed NHE1-E266I-Myc was clustered at membrane protrusions and NHE1-KRA2-HA retained a more uniform distribution along the membrane (Figure 5B). However, FN production, secretion, and assembly were restored in K2E cells. Immunolabeling unpermeabilized cells for extracellular FN (Figure 5C) and permeabilized cells for total FN (data not shown) and immunoblotting for secreted FN in the medium (Figure 5D) indicated FN production and assembly in K2E cells and was similar to E266I cells. There was no clonal variation in FN production, which was seen in all K2E clones (data not shown). We predict that FN production was restored because scaffolding by NHE1-E266I might suppress a dominant negative action of NHE1-KRA2.
Figure 5.
FN production in KRA2 cells is restored by coexpression of NHE1-E266I localized at membrane protrusions. Expression (A) and localization (B) of NHE1-E266I-Myc and NHE1-KRA2-HA in the indicated cells determined by immunoblotting and immunolabeling with antibodies to Myc and HA. Immunoblotting with antibodies to β-actin was used as a loading control. Bar, 5 μm. (C) Live-cell immunolabeling of FN in KRA cells, E266I cells, and K2E cells coexpressing NHE1-KRA2 and NHE1-E266I indicate coexpression of NHE1-E266I restores FN production and matrix assembly (green, extracellular FN; blue, Hoechst staining for nuclei). (D) Immunoblot of FN in media from KRA2, E266I, and K2E cells reveals K2E cells have restored FN secretion compared with KRA2 cells.
Exogenous TGF-β Restores FN Production and Increases Abundance of FA in KRA2 Cells
Several cell processes are impaired in KRA2 cells, including bundling of actin filaments into stress fibers and assembly of FA complexes (Denker et al., 2000; Figure 1A). However, because these processes are also impaired in NHE1-deficient PS120 cells (Denker et al., 2000; Denker and Barber, 2002) that express and assemble FN (Figure 4A), they likely do not determine attenuated FN biosynthesis in KRA2 cells. We found that increased autophosphorylation of FAK (pY397) in cells plated on FN was delayed in KRA2 cells compared with WT cells; however, this delay was also seen in PS120 cells and in E266I cells (Supplementary Figure S5). Additional signaling mechanisms associated with regulating FN production, including expression of the transcription factor CREB, phosphorylation of extracellular signaling kinase ERK, and increased glycolytic flux, are attenuated in E226I cells (Putney and Barber, 2004; Meima and Barber, unpublished observations) and are therefore unlikely causes of decreased FN synthesis in KRA2 cells.
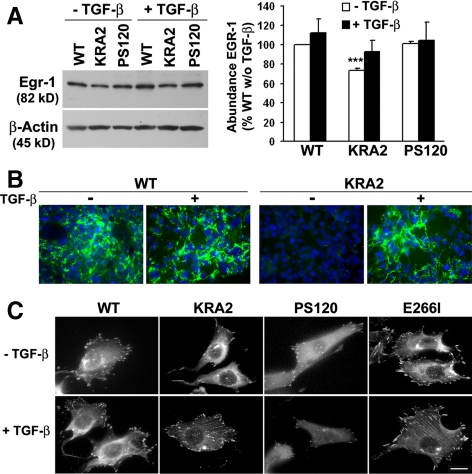
The transcription factor Egr-1 directly activates the FN gene (Liu et al., 2000), and we found that the abundance of Egr-1 was ∼30% less in total lysates of KRA2 cells compared with WT or NHE1-deficient PS120 cells (Figure 6A; p < 0.001, n = 3). Because the cytokine TGF-β induces expression of Egr-1 (Chen et al., 2006) and FN (Blobe et al., 2000), we tested the effect of treating KRA2 cells with exogenous TGF-β. Compared with untreated controls, WT, KRA2, and PS120 cells treated with TGF-β (5 ng/ml for 48 h) had increased expression of Egr-1, although expression in KRA2 cells remained less than that in WT or PS120 cells (Figure 6A). Additionally, TGF-β nearly restored FN production and assembly by KRA2 cells and appeared to increase fibrillogenesis in WT cells (Figure 6B). Increased FN production in KRA2 cells with TGF-β was completely blocked by SB-431542 (Supplemental Figure S6), an inhibitor of TFG-β-R1 kinase activity (Laping et al., 2002). In WT cells, SB-431542 inhibited FN production in the presence and absence of TGF-β (Supplemental Figure S6), indicating that FN production in WT cells is TGF-β-dependent. Moreover, the abundance and size of FA in KRA2 cells increased with TGF-β. As previously reported (Tominaga and Barber, 1998; Denker et al., 2000), KRA2 and PS120 cells have fewer and smaller FA compared with WT cells, and FA in E266I cells are more abundant compared with WT cells because lower pHi attenuates their turnover (Srivastava et al., 2008; Figure 6C). After 48 h with TGF-β, KRA2 cells were more spread and had more and larger FA, suggesting that the lack of FN likely limits FA assembly or stability. However, PS120 cells retained a fusiform morphology with no change in the abundance or size of FA, and FA in WT and E266I cells were similar in the absence and presence of TGF-β (Figure 6C). Hence, scaffolding by NHE1, even when mislocalized, is necessary for FA assembly.
Figure 6.
TGF-β1 increases Egr-1 expression and restores a secreted and assembled FN matrix and the abundance of FA in KRA2 cells. (A) Representative immunoblot (left) and means ± SEM of data from immunoblots of three separate cell preparations (right) indicate that expression of Egr-1 is decreased in KRA2 cells compared with WT and PS120 cells. Incubation with (rh)TGF-β1 (5 ng/ml for 48 h) increases Egr-1 expression in all three cell types. Immunoblotting for β-actin was used as a loading control. *Significant difference compared with WT and PS120 cells; p < 0.001. (B) FN production and assembly, although nearly absent in KRA2 cells, are similar to WT cells after treating with 5 ng/ml TGF-β for 48 h, as determined by immunolabeling for extracellular FN. Data are representative of three separate cell preparations. (C) FA, as determined by paxillin immunolabeling in the indicated cell types. Treating cells with rhTGF-β1 increases the abundance and size of FA in KRA2 cells but not in WT, PS120, or E266I cells. Bar, 10 μM.
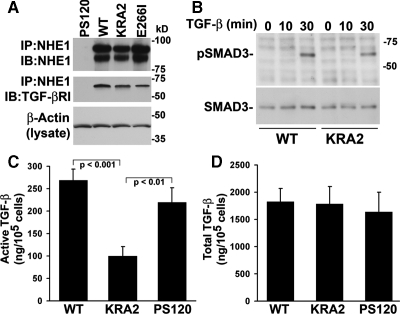
Because NHE1 (Bullis et al., 2002; Bourguignon et al., 2004) and TGF-β-RI (Razani et al., 2001) cosediment in caveolin-enriched plasma membrane fractions, we next asked whether mislocalized NHE1 might interfere with TGF-β signaling. We found that endogenous TGF-β-RI coprecipitates with wild-type and mutant NHE1-E266I and NHE1-KRA2 (Figure 7A), suggesting the proteins colocalize in a membrane microdomain. Moreover, the abundance of TGF-β-RI in cell lysates was similar in WT and KRA2 cells (data not shown). However, the ability of exogenous TGF-β to increase expression of Egr-1 and FN in KRA2 cells suggested that intracellular signaling by TGF-β was not impaired, which we confirmed by showing that with TGF-β, phosphorylation of SMAD3, a downstream effector of TGF-β-RI activation, was similar in KRA2 and WT cells (Figure 7B).
Figure 7.
Activation of TGF-β but not secretion of latent TGF-β or signaling by TGF-β-RI is inhibited in KRA2 cells. (A) TGF-β-RI coprecipitates with wild-type and mutant NHE1-HA. Immune complexes were immunoblotted with antibodies to NHE1, which recognize glycosylated (top band) and unglycosylated (bottom band) protein, and to TGF-β-RI. Aliquots of lysates were immunoblotted for β-actin to confirm equivalent amount of protein used for immunoprecipitation. Data are representative of two separate cell preparations. (B) Time-dependent increase in phosphorylated SMAD3 (pSMAD3) is similar in WT and KRA2 cells treated with 5 ng/ml rhTGF-β1. Immunoblotting for total SMAD3 indicates similar abundance in WT and KRA2 cells in the absence and presence of TGF-β. (C) Secreted active TGF-β determined by luciferase assays using MLEC cells expressing a truncated plasminogen activator inhibitor-1 (PAI-1) promoter and treated with conditioned medium from WT, KRA2, and PS120 cells. Luciferase units from MLEC cells treated with rhTGF-β1 were used to generate a standard curve and data, expressed as amount of TGF-β per 105 cells, represent means ± SEM of six cell preparations. (D) Secreted latent TGF-β determined as in C, but conditioned medium was incubated at 80°C for 10 min before adding to MLEC cells. Data are means ± SEM of four cell preparations.
We next asked whether secreted TGF-β was attenuated in KRA2 cells. Using a luciferase assay with MLEC cells expressing a truncated PAI-1 promoter that is activated by TGF-β (Abe et al., 1994), we found that active TGF-β in conditioned medium of KRA2 cells was significantly less compared with WT cells (p < 0.001, n = 6) or PS120 cells (p < 0.01, n = 6) but similar in WT and PS120 cells (p > 0.1, n = 6; Figure 7C). However, secreted total or latent TGF-β, determined in boiled samples of conditioned medium, was not significantly different in KRA2 cells compared with WT or PS120 cells (Figure 7B; p > 0.1; n = 4). Activation of latent TGF-β is regulated by a number of conditions including disruption of the ECM (Annes et al., 2003), and whether decreased activation by KRA2 cells is determined by lack of FN production or by other factors such as decreased abundance or activity of extracellular proteases remains to be determined. Moreover, because expression of Egr-1 and FN and also secreted active TGF-β were not attenuated in NHE1-deficient PS120 cells, our findings indicate that they are dominantly suppressed through mislocalized scaffolding by NHE1-KRA2.
DISCUSSION
The assembly of a FN matrix requires a number of recognized signaling mechanisms, including activation of integrins and integrin-regulated kinases (reviewed in Wierzbicka-Patynowski and Schwarzbauer, 2003), and the low-molecular-weight GTPase Rho, the Rho kinase ROCK, and Rho-regulated myosin contractility (Calì et al., 1999; Zhong et al., 1998; Yoneda et al., 2007). However, less is known about signals regulating FN synthesis. FN gene expression is increased by serum, growth factors, cytokines, and viruses, and is attenuated by many oncogenes (Kornblihtt et al., 1996), although these regulatory mechanisms primarily occur in pathogenic conditions, including fibrosis, inflammation, or malignant transformation. Our understanding of physiological regulation of FN production remains limited. Our data show that when NHE1 is mislocalized, FN production and assembly is nearly absent.
Previous reports indicate a bidirectional functional interaction between integrin signaling and NHE1. Activation and clustering of integrin receptors by FN binding stimulates NHE1 activity (Schwartz et al., 1991; Tominaga and Barber, 1998). Additionally, NHE1 is necessary for FN induction of the early response gene c-jun (Dike and Ingber, 1996) and for integrin-induced increases in FAK activity (Tominaga and Barber, 1998), which we confirmed (Supplemental Figure S5). However, our current findings indicate a previously unrecognized link between NHE1 and FN production and assembly, but neither NHE1 activity or NHE1 expression is necessary for FN synthesis, as indicated by normal FN abundance and organization in cells expressing an inactive NHE1-E266I that localizes in lamellipodia like wild-type NHE1 (Denker et al., 2000) or in NHE1-deficient PS120 cells. Rather, our current data suggest that mislocalized NHE1-KRA2 has a dominant-negative effect in suppressing FN synthesis. This possibility is supported by the ability of coexpressed NHE1-E266I to restore FN production in cells expressing NHE1-KRA2. We coexpressed NHE1-E266I instead of wild-type NHE1 to confirm pHi-independent effects on FN synthesis, which also was shown by pHi-independent effects on FN synthesis and assembly in cells expressing the SN1 transporter. Because the C-terminal cytoplasmic domain of NHE1 functions as a scaffolding platform that possibly assembles signaling complexes (Baumgartner et al., 2006; Meima et al., 2007), we predict that a mislocalized NHE1 scaffold in KRA2 cells might sequester and hence disrupt signaling mechanisms necessary for FN production.
In addition to decreased FN synthesis, KRA2 cells also have decreased expression of Egr-1 and activation of TGF-β, although secretion of latent TGF-β is not impaired. Egr-1, an early response transcription factor, is induced by growth factors and cytokines and activates TGF-β and FN gene expression (Liu et al., 2000). Although treating KRA2 cells with exogenous TGF-β restored FN synthesis and assembly and increased Egr-1 expression, the complex functional interplay between cytokines, Egr-1, and FN makes it difficult to understand the primary defect in KRA2 cells. One possibility is that decreased FN production, perhaps due in-part to decreased Egr-1 expression, is the primary defect that secondarily inhibits TGF-β activation. The latent TGF-β complex is covalently bound to as yet unidentified extracellular matrix proteins and senses perturbations of the extracellular matrix, which regulates cleavage to active TGF-β (Annes et al., 2003). Alternatively, decreased TGF-β activation could be the primary defect that secondarily suppresses Egr-1 and FN expression. Cleavage of latent to active TGF-β is regulated by proteases, low extracellular pH, integrins, and localization of the latent complex (Annes et al., 2003). Although TGF-β activation was not reduced in NHE1-deficient PS120 cells, mislocalized H+ efflux by NHE1-KRA2 might inhibit a pH-dependent cleavage of latent TGF-β. Additionally, TGF-β-RI coprecipitated in an immune complex with NHE1-wild type, -KRA2, and -E266I. We predict TGF-β-RI does not directly bind NHE1 but likely colocalizes with NHE1 in a plasma membrane “signalosome” microdomain, which when mislocalized in KRA2 cells might impair localization and hence cleavage of latent TGF-β complexes. However, KRA2 cells likely retain TGF-β-RI signaling, because the abundance of pSMAD3 in WT and KRA2 cells treated with exogenous TGF-β was similar. The hyaluronan receptor CD44 may be a link between NHE1 and TGF-β signaling. CD44 coprecipitates with NHE1 (Bourguignon et al., 2004) and TGF-β-RI, (Ito et al., 2004) and regulates the function of both proteins. Testing whether activation of TGF-β is restored in KRA2 cells plated on an assembled FN matrix might resolve whether decreased activation is a primary defect or is secondary to lack of FN production.
KRA2 cells were also unable to assemble an exogenous FN matrix, indicating an additional defect that is independent of decreased FN production. This defect is likely also due to a dominant-negative effect of mislocalized NHE1-KRA2 because matrix assembly was not impaired in E226I or PS120 cells, was partially restored in KRA2 cells treated with MnCl2 and integrin-activating antibodies, and was mostly restored in KRA2 cells coexpressing NHE1-E266I or treated with TGF-β. Assembly of FN into a fibrillar matrix requires tension generated by interaction of clustered FA proteins tethered to the actin cytoskeleton (Wierzbicka-Patynowski and Schwarzbauer, 2002). Although NHE1-KRA2 does not bind ERM proteins, which link the actin cytoskeleton to plasma membrane proteins, it remains enriched in P100 particulate fractions (unpublished findings), suggesting it retains cytoskeletal association. NHE1-KRA2 might dominantly mislocalize cytoskeletal proteins such as spectrin and cytoskeletal anchoring proteins such as ankyrin. Like ERM proteins, ankyrin binds directly to wild-type NHE1 (Denker and Barber, unpublished results), although whether it binds to NHE1-KRA2 is unknown. Hence, mislocalized scaffolding by NHE1-KRA2 could dominantly inhibit FN assembly through changes in cytoskeletal contractility or additionally by mislocalizing focal adhesion regulators such as ROCK, which directly phosphorylates NHE1 (Tominaga et al., 1998). As previously shown, assembly of FN into fibrils is blocked by inhibiting RhoA, an upstream regulator of ROCK, and similar to our findings, fibrillar FN is generated after treating cells with TGF-β (Calì et al., 1999). We show that attenuated FA size and abundance in KRA2 cells were restored with TGF-β and increased FN production, indicating that KRA2 cells retain cytoskeletal integrity for FA assembly.
The critical importance of a FN matrix and active TGF-β in morphogenesis and tissue remodeling underscores the importance of understanding their physiological regulation. Most remarkable of our findings is that FN production and assembly is nearly absent in KRA2 cells. Although further investigation is necessary to reveal how these processes are dominantly inhibited through mislocalized scaffolding by NHE1, our findings suggest that NHE1-KRA2 could be a valuable tool for obtaining a mechanistic understanding of how FN production and cleavage of active TGF-β are linked and regulated. More speculative is the possible use of NHE1-KRA2 for therapeutic control of increased FN production in pathological conditions such as fibrosis and inflammation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Caroline Damsky, Dean Sheppard, Torsten Wittmann, Dan Mercola, Dusko Ilic, and members of the Barber lab for technical advice and suggestions. We also thank Liyu Wu for help with luciferase assays to measure secreted TGF-β. This work was supported by National Institutes of Health Grant GM47413 to D.L.B. M.J.-V. was supported by a postdoctoral fellowship from the Spanish Ministry of Science and Education. Work was conducted in a facility constructed with support from Research Facilities Improvement Program Grant C06 RR16490 from the National Center for Research Resources, National Institutes of Health.
Abbreviations used:
- ERM
ezrin/moesin/radixin
- FA
focal adhesion
- FN
fibronectin
- NHE1
Na-H exchanger isoform 1
- PAI
plasminogen activator inhibitor-1
- PI(4,5)P2
phosphatidyl-4,5-biphosphate
- pHi
intracellular pH
- TGF-β
transforming growth factor-β
- TGF-β-RI
transforming growth factor-β receptor subtype I.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0842) on February 18, 2009.
REFERENCES
- Abe M., Harpel J. C., Metz C. N., Nunes I., Loskutoff D. J., Rifkin D. B. An assay for transforming growth factor-β using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal. Biochem. 1994;216:276–284. doi: 10.1006/abio.1994.1042. [DOI] [PubMed] [Google Scholar]
- Aharonovitz O., Zaun H. C., Balla T., York J. D., Orlowski J., Grinstein S. Intracellular pH regulation by Na(+)/H(+) exchange requires phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 2000;150:213–224. doi: 10.1083/jcb.150.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen J. L., Rasmussen L. M., Ledet T. Diabetic macroangiopathy and atherosclerosis. Diabetes. 1996;45(Suppl. 3):S91–S94. doi: 10.2337/diab.45.3.s91. [DOI] [PubMed] [Google Scholar]
- Annes J. P., Munger J. S., Rifkin D. B. Making sense of latent TGFbeta activation. J. Cell Sci. 2003;116:217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- Baumgartner M., et al. The Nck-interacting kinase phosphorylates ERM proteins for formation of lamellipodium by growth factors. Proc. Natl. Acad. Sci. USA. 2006;103:13391–13396. doi: 10.1073/pnas.0605950103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron V., Adamson E. D., Calogero A., Ragona G., Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006;13:115–124. doi: 10.1038/sj.cgt.7700896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatti S. P., Foster D. N., Ranganathan G., Moses H. L., Getz M. J. Induction of fibronectin gene transcription and mRNA is a primary response to growth factor stimulation. In AKR-2B cells. Proc. Natl. Acad. Sci. USA. 1988;85:1119–1123. doi: 10.1073/pnas.85.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobe G. C., Schiemann W. P., Lodish H. F. Role of transforming growth factor beta in human disease. N. Engl. J. Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- Bourguignon L. Y., Singleton P. A., Diedrich F., Stern R., Gilad E. CD44 interaction with Na+-H+ exchanger (NHE1) creates acidic microenvironments leading to hyaluronidase-2 and cathepsin B activation and breast tumor cell invasion. J. Biol. Chem. 2004;279:26991–27007. doi: 10.1074/jbc.M311838200. [DOI] [PubMed] [Google Scholar]
- Bullis B. L., Li X., Singh D. N., Berthiaume L. G., Fliegel L. Properties of the Na+/H+ exchanger protein. Detergent-resistant aggregation and membrane microdistribution. Eur. J. Biochem. 2002;269:4887–4895. doi: 10.1046/j.1432-1033.2002.03202.x. [DOI] [PubMed] [Google Scholar]
- Calì G, Mazzarella C., Chiacchio M., Negri R., Retta S. F., Zannini M., Gentile F., Tarone G., Nitsch L., Garbi C. RhoA activity is required for fibronectin assembly and counteracts beta1B integrin inhibitory effect in FRT epithelial cells. J. Cell Sci. 1999;112:957–965. doi: 10.1242/jcs.112.6.957. [DOI] [PubMed] [Google Scholar]
- Chaudry F. A., Reimer R. J., Krizaj D., Barber D., Storm-Mathisen J., Copenhagen D. R., Edwards R. H. Molecular analysis of system N suggests novel physiological roles in nitrogen metabolism and synaptic transmission. Cell. 1999;99:769–780. doi: 10.1016/s0092-8674(00)81674-8. [DOI] [PubMed] [Google Scholar]
- Chen S. J., Ning H., Ishida W., Sodin-Semrl S., Takagawa S., Mori Y., Varga J. The early-immediate gene EGR-1 is induced by transforming growth factor-beta and mediates stimulation of collagen gene expression. J. Biol. Chem. 2006;281:21183–21197. doi: 10.1074/jbc.M603270200. [DOI] [PubMed] [Google Scholar]
- Christensen L., Nielsen M., Andersen J., Clemmensen I. Stromal fibronectin staining pattern and metastasizing ability of human breast carcinoma. Cancer Res. 1988;48:6227–6233. [PubMed] [Google Scholar]
- Clark K., Pankov R., Travis M. A., Askari J. A., Mould P., Craig S. E, et al. A specific α5β1-integrin conformation promotes directional integrin translocation and fibronectin matrix formation. J. Cell Sci. 2004;118:291–300. doi: 10.1242/jcs.01623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch E. Pathobiology of pulmonary fibrosis. Am. J. Physiol. 1990;259:L159–L184. doi: 10.1152/ajplung.1990.259.4.L159. [DOI] [PubMed] [Google Scholar]
- Danen E. H., Yamada K. M. Fibronectin, integrins, and growth control. J. Cell Physiol. 2001;189:1–13. doi: 10.1002/jcp.1137. [DOI] [PubMed] [Google Scholar]
- Denker S. P., Huang D. C., Orlowski J., Furthmayr, Barber D. L. Direct binding of the Na-H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H+ translocation. Mol. Cell. 2000;6:1425–1436. doi: 10.1016/s1097-2765(00)00139-8. [DOI] [PubMed] [Google Scholar]
- Denker S. P., Barber D. L. Cell migration requires both ion translocation and cytoskeletal anchoring by the Na-H exchanger NHE1. J. Cell Biol. 2002;159:1087–1096. doi: 10.1083/jcb.200208050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dike L. E., Ingber D. E. Integrin-dependent inductions of early growth response genes in capillary endothelial cells. J. Cell Sci. 1996;109:2855–2863. doi: 10.1242/jcs.109.12.2855. [DOI] [PubMed] [Google Scholar]
- Feng X. H., Filvaroff E. H., Derynck R. Transforming growth factor-beta (TGF-beta)-induced down-regulation of cyclin A expression requires a functional TGF-beta receptor complex. Characterization of chimeric and truncated type I and type II receptors. J. Biol. Chem. 1995;270:24237–24245. doi: 10.1074/jbc.270.41.24237. [DOI] [PubMed] [Google Scholar]
- Frantz C., Karydis A., Nalbant P., Hahn K. M., Barber D. L. Positive feedback between Cdc42 activity and H+ efflux by the Na-H exchanger NHE1 for polarity of migrating cells. J. Cell Biol. 2007;179:403–410. doi: 10.1083/jcb.200704169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinstein S., Woodside M., Waddell T. K., Downey G. P., Orlowski J., Pouyssegur J., Wong D. C., Foskett J. K. Focal localization of the NHE-1 isoform of the Na+/H+ antiport: assessment of effects on intracellular pH. EMBO J. 1993;12(13):5209–5218. doi: 10.1002/j.1460-2075.1993.tb06216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzel M., Bachem M., Anders D., Trischler G., Faehling M. Different effects of growth factors on proliferation and matrix production of normal and fibrotic human lung fibroblasts. Lung. 2005;183:225–237. doi: 10.1007/s00408-004-2534-z. [DOI] [PubMed] [Google Scholar]
- Hohenadel D., van der Woude F. J. Gene expression in diabetic nephropathy. Curr. Diab. Rep. 2004;4:462–469. doi: 10.1007/s11892-004-0057-x. [DOI] [PubMed] [Google Scholar]
- Hughes P. E., Diaz-Gonzalez F., Leong L., Wu C., McDonald J. A., Shattil S. J., Ginsberg M. H. Breaking the integrin hinge. A defined structural constraint regulates integrin signaling. J. Biol. Chem. 1996;271:6571–6574. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- Ito T., Williams J. D., Fraser D., Phillips A. O. Hyaluronan attenuates transforming growth factor-beta1-mediated signaling in renal proximal tubular epithelial cells. Am. J. Pathol. 2004;164:1979–1988. doi: 10.1016/s0002-9440(10)63758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershenobich Stalnikowitz D., Weissbrod A. B. Liver fibrosis and inflammation. A review. Ann. Hepatol. 2003. 2003;2:159–163. [PubMed] [Google Scholar]
- Kornblihtt A. R., Pesce C. G., Alonso C. R., Cramer P., Srebrow A., Werbajh S., Muro A. F. The fibronectin gene as a model for splicing and transcription studies. FASEB J. 1996;10:248–257. doi: 10.1096/fasebj.10.2.8641558. [DOI] [PubMed] [Google Scholar]
- Labat-Robert J. Fibronectin in malignancy. Semin. Cancer Biol. 2002;12:187–195. doi: 10.1016/S1044-579X(02)00022-6. [DOI] [PubMed] [Google Scholar]
- Laping N. J., et al. Inhibition of transforming growth factor (TGF)-beta1-induced extracellular matrix with a novel inhibitor of the TGF-beta type I receptor kinase activity: SB-431542. Mol. Pharmacol. 2002;62:58–64. doi: 10.1124/mol.62.1.58. [DOI] [PubMed] [Google Scholar]
- Larsen M., Artym V. V., Green J. A., Yamada K. M. The matrix reorganized: extracellular matrix remodeling and integrin signaling. Curr. Opin. Cell Biol. 2006;18:463–471. doi: 10.1016/j.ceb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Lee B.-H., Park S.-Y., Kang K.-B., Park R.-W., Kim I.-S. NF-κB activates fibronectin expression in rat hepatocytes. Biochem. Biophys. Res. Commun. 2002;297:1218–1224. doi: 10.1016/s0006-291x(02)02356-2. [DOI] [PubMed] [Google Scholar]
- Liu C., Yao J., Mercola D., Adamson E. The transcription factor EGR-1 directly transactivates the fibronectin gene and enhances attachment of human glioblastoma cell line U251. J. Biol. Chem. 2000;275:20315–20323. doi: 10.1074/jbc.M909046199. [DOI] [PubMed] [Google Scholar]
- Meima M. E., Mackley J. R., Barber D. L. Beyond ion translocation: structural functions of the sodium-hydrogen exchanger isoform-1. Curr. Opin. Nephrol. Hypertens. 2007;16:365–372. doi: 10.1097/MNH.0b013e3281bd888d. [DOI] [PubMed] [Google Scholar]
- McKeown-Longo P. J., Mosher D. F. Interaction of the 70,000-mol-wt amino-terminal fragment of fibronectin with the matrix-assembly receptor of fibroblasts. J. Cell Biol. 1995;100:364–374. doi: 10.1083/jcb.100.2.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S., Katz B. Z., Lafrenie R. M., Yamada K. M. Fibronectin and integrins in cell adhesion, signaling, and morphogenesis. Ann. NY Acad. Sci. 1998;857:119–129. doi: 10.1111/j.1749-6632.1998.tb10112.x. [DOI] [PubMed] [Google Scholar]
- Perkinson R. A., Kuo B. A., Norton P. A. Modulation of transcription of the rat fibronectin gene by cell density. J. Cell Biochem. 1996;63:74–85. doi: 10.1002/(SICI)1097-4644(199610)63:1%3C74::AID-JCB6%3E3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Pouyssegur J., Sardet C., Franchi A., Allemain G., Paris A. A specific mutation abolishing Na+/H+ antiport activity in hamster fibroblasts precludes growth at neutral and acidic pH. Proc. Natl. Acad. Sci. USA. 1984;81:4833–4837. doi: 10.1073/pnas.81.15.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney L. K., Barber D. L. Expression profile of genes regulated by activity of the Na-H exchanger NHE1. BMC Genomics. 2004;16:46–59. doi: 10.1186/1471-2164-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B., Zhang X. L., Bitzer M., von Gersdorff G., Böttinger E. P., Lisanti M. P. Caveolin-1 regulates transforming growth factor (TGF)-beta/SMAD signaling through an interaction with the TGF-beta type I receptor. J. Biol. Chem. 2001;276:6727–6738. doi: 10.1074/jbc.M008340200. [DOI] [PubMed] [Google Scholar]
- Ruoslahti E. Fibronectin and its integrin receptors in cancer. Adv. Cancer Res. 1999;76:1–20. doi: 10.1016/s0065-230x(08)60772-1. [DOI] [PubMed] [Google Scholar]
- Sardet C., Counillon L., Franchi A., Pouysségur J. Growth factors induce phosphorylation of the Na+/H+ antiporter, glycoprotein of 110 kD. Science. 1990;247(4943):723–726. doi: 10.1126/science.2154036. [DOI] [PubMed] [Google Scholar]
- Sechler J. L., Cumiskey A. M., Gazzola D. M., Schwarzbauer J. E. A novel RGD-independent fibronectin assembly pathway initiated by α4β1 integrin binding to the alternatively spliced V region. J. Cell Sci. 2000;113:1491–1498. doi: 10.1242/jcs.113.8.1491. [DOI] [PubMed] [Google Scholar]
- Schwartz M. A., Lechene C., Ingber D. E. Insoluble fibronectin activates the Na/H antiporter by clustering and immobilizing integrin α5β1, independent of cell shape. Proc. Natl. Acad. Sci. 1991;88:7849–7853. doi: 10.1073/pnas.88.17.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer J. E., Sechler J. L. Fibronectin fibrillogenesis: a paradigm for extracellular matrix assembly. Curr. Opin. Cell Biol. 1999;11:622–627. doi: 10.1016/s0955-0674(99)00017-4. [DOI] [PubMed] [Google Scholar]
- Shirai T., Miyagi S., Horiuchi D., Okuda-Katayanagi T., Nishimoto M., Muramatsu M., Sakamoto Y., Nagata M., Hagiwara K., Okuda A. Identification of an enhancer that controls up-regulation of fibronectin during differentiation of embryonic stem cells into extraembryonic endoderm. J. Biol. Chem. 2005;280:7244–7252. doi: 10.1074/jbc.M410731200. [DOI] [PubMed] [Google Scholar]
- Srivastava J., Barreiro G., Groscurth S., Gringas A. R., Goult B. T., Critchley D. R., Kelly M.J.S., Jacobson M. P., Barber D. L. Structural model and functional significance of pH-dependent talin-actin binding for focal adhesion remodeling. Proc. Natl. Acad. Sci. USA. 2008;105:14436–14441. doi: 10.1073/pnas.0805163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T., Barber D. L. Na-H exchange acts downstream of RhoA to regulate integrin-induced cell adhesion and spreading. Mol. Biol. Cell. 1998;9:2287–2303. doi: 10.1091/mbc.9.8.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T., Ishizaki T., Narumiya S., Barber D. L. p160ROCK mediates RhoA activation of Na-H exchange. EMBO J. 1998;17:4712–4722. doi: 10.1093/emboj/17.16.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerberg K., Lohikangas L., Gullberg D., Pfaff M., Johansson S., Fassler R. β1 integrin dependent and independent polymerization of fibronectin. J. Cell Biol. 1996;132:227–238. doi: 10.1083/jcb.132.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicka-Patynowski I., Schwarzbauer J. E. Regulatory role for Src and phosphatidylinositol 3-kainase in initiation of fibronectin matrix assembly. J. Biol. Chem. 2002;277:19703–19708. doi: 10.1074/jbc.M200270200. [DOI] [PubMed] [Google Scholar]
- Wierzbicka-Patynowski I., Schwarzbauer J. E. The ins and outs of fibronectin matrix assembly. J. Cell Sci. 2003;116:3269–3276. doi: 10.1242/jcs.00670. [DOI] [PubMed] [Google Scholar]
- Wu C., Kevins V., O'Toole T. E., McDonald J. A., Ginsberg M. H. Integrin activation and cytoskeletal interaction are critical steps in the assembly of a fibronectin matrix. Cell. 1995;83:715–724. doi: 10.1016/0092-8674(95)90184-1. [DOI] [PubMed] [Google Scholar]
- Yoneda A., Ushakov D., Multhaupt H.A.B., Couchman J. R. Fibronectin matrix assembly requires distinct contributions from Rho kinases I and II. Mol. Biol. Cell. 2007;18:66–75. doi: 10.1091/mbc.E06-08-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C., Chrzanowska-Wodnicka M., Brown J., Shaub A., Belkin A. M., Burridge K. Rho-mediated contractility exposes a cryptic site in fibronectin and induces fibronectin matrix assembly. J. Cell Biol. 1998;141:539–551. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.