Abstract
L-DNA, the mirror image of natural D-DNA, can be readily self-assembled into designer discrete or periodic nanostructures. The assembly products are characterized by polyacrylamide gel electrophoresis (PAGE), circular dichroism (CD) spectrum, atomic force microscope (AFM) and fluorescence microscope. We found that the use of enantiomer DNA as building material leads to the formation of DNA supra-molecules with opposite chirality. Therefore, the L-DNA self-assembly is a substantial complement to the structural DNA nanotechnology. Moreover, the L-DNA architectures feature superior nuclease resistance thus are appealing for in vivo medical applications.
One of the central tasks of nanotechnology is to construct nano-scale structures with designed geometry, topology, and periodicity. Among various materials used to build designer nanoarchitectures, DNA distinguishes itself by its predictable Watson-Crick base-paring, structural stiffness and flexibility, ease to synthesize and biocompatibility. With a number of exciting breakthroughs in the past two decades, DNA based self-assembly has been recognized as one of the most efficient and reliable methods for bottom-up construction.1-19 The success of DNA as a generic building material makes it appealing to involve other members of the nucleic acid family in nano-scale construction. For example, tecto-RNA motifs have been used to form square-shaped tiles that further self-assemble into finite or infinite sized two-dimensional (2D) arrays.20 Unnatural nucleic acids, such as locked nucleic acid (LNA)21 and peptide nucleic acid (PNA),22 have also been incorporated into self-assembled DNA structures. Recently, a four-arm junction motif was synthesized using glycol nucleic acid (GNA).23 However, the structural (e.g. helical repeats and diameter), physical (e.g. thermal stability) and chemical properties (e.g. activity to intercalating fluorescent dyes) of these unnatural nucleic acid species are usually different from those of DNA, which adds extra design work and experimental uncertainty in the construction of more complicated nanostructures.
Here we report the facile preparation of well-defined discrete, one-dimensional (1D) and two-dimensional (2D) nanostructures using mirror-image DNA, L-DNA, the enantiomer of the natural DNA, D-DNA, by simply applying previously reported DNA self-assembly designs and protocols. As perfect mirror-images, L-DNA and D-DNA possess the same duplex conformation except for their opposite chirality.24,25 They are physically and chemically identical to each other under non-chiral circumstances. Given a rich toolbox of DNA-tile motifs,26-31 it is therefore a straight-forward and attractive idea to construct nanoarchitectures with L-DNA in substitution to the normally used D-DNA. Ideally, the geometry and periodicity of the resulting structures is expected to be identical to those composed of D-DNA. However, whether and how the difference in chirality of the DNA duplex is reflected on the supramolecular structure level remains mystery. Moreover, L-DNA strand is known to be resistant to various nucleases that only act on the D-DNA substrates. Therefore, for in vivo diagnostic and therapeutic applications using nucleic acids-based nanostructures, the nuclease-resistant property is highly desirable, which inspired us to explore L-DNA self-assembly.
As a pilot experiment, we tested the self-assembly of L-DNA into the well-known four-arm junction (J1) structure,32 which is composed of four duplex “arms” formed by four oligonucleotide strands (Figure 1a, sequence details see the supplemental information). First, stechiometric amounts of L-DNA strands were mixed in all possible combinations separately and annealed. As shown in Figure 1b, the correctly assembled partial or entire junction structure was resolved by the non-denaturing polyacrylamide gel electrophoresis (PAGE). The exonuclease resistance of the nano-junctions was also tested. As expected, L-DNA J1 is resistant to exonuclease, while D-DNA J1 is completely digested under the same conditions (Figure 1c). Circular dichroism (CD) measurements (Figure 1d) clearly reveal the opposite chirality of the junctions made from L- and D-DNA. In addition, both junctions exhibited similar thermal transition properties, with a melting temperature at approximately 45 °C (Figure 1e). These data strongly suggested that the L-DNA J1 junction can be efficiently assembled as the mirror-image of D-DNA J1.
Figure 1.
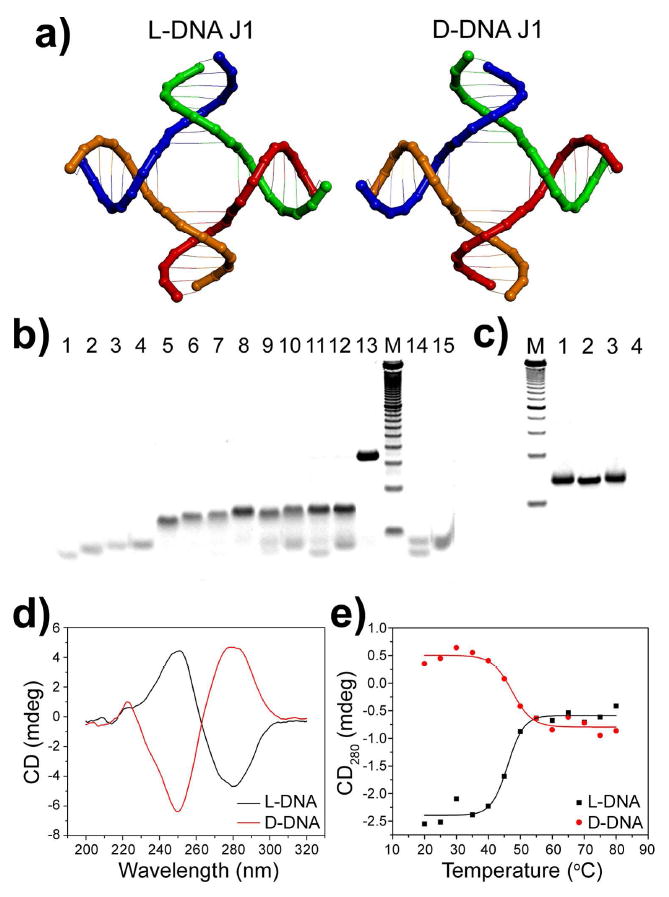
a) Structural models of L- and D-DNA J1 molecules. b) Non-denaturing PAGE of the assembly of the L-DNA J1. L-DNA oligonucleotide monomers, dimmers, trimers and tetramer are represented by the bands in lanes 1-4, 5-8, 9-12 and 13, respectively. Note that strands at the opposite corners of the junction cannot dimerize (lanes 14 and 15). Lane M contains 10-bp DNA ladder as a reference. c) Nuclease resistance of L-DNA J1. Lane M: 25-bp DNA ladder; lane 1 and 3: L-DNA J1 before and after exonuclease treatment; Lane 2 and 4: D-DNA J1 before and after exonuclease treatment. d) Circular dichroism spectra of L- and D-DNA J1. e) Thermal transition of L- and D-DNA J1 monitored by temperature dependent CD at 280 nm.
The success of constructing Holliday junction analogue using L-DNA made us wonder whether it is possible to apply this strategy to fabricate higher ordered nanostructures, such as nanotubes and 2D nanoarrays from L-DNA. Mao’s group recently reported the self-assembly of DNA nanotubes33 and 2D crystals34 each using a single oligonucleotide strand. We chose such structures as our model assembly targets because they are robust and cost efficient.
Figure 2a shows the assembly scheme of the one-strand L-DNA nanotube. At a first glance, one would think it is an exact duplicate of the D-DNA nanotube, except that the DNA duplexes within are all left-handed. However, when a piece of periodic 2D array wraps-up to form a tube, it has both possibilities to spiral either right-handedly or left-handedly. Although it has been reported that DNA tiles can be self-assembled into chiral nanotubes,35 their exact preference of chirality has never been revealed. Such chirality can be probed by atomic force microscope (AFM) imaging and examining the DNA nanotubes at the defect locations. Those defects, seen as the flat openings on the nanotubes under AFM, most likely arose from incomplete tube closing during tube formation. (One can imagine this as wrapping a narrow ribbon along a solid cylinder; nicks could arise where the ribbon is not tightly folded turn-by-turn.) Thus, the chirality in which a tube spirals can be easily determined by looking at the boundary between the single layer and double layer of the tube by AFM imaging. From these experiments, we discovered that L- and D-DNA nanotubes, assembled in the same experimental conditions, exhibit opposite chirality (see AFM images in Figure 2b and additional images in Figure S3 and S4 in the supplemental information). Specifically, the L-DNA tubes are exclusively right-handed while the D-DNA tubes are all left-handed, with no exceptions within our observation. Therefore, we can relate the overall chirality of the nanotubes to the intrinsic chirality of the DNA duplexes. This is a very interesting phenomenon, as it suggests that the chirality of the self-assembled DNA supramolecular structures can be controlled by choosing one of the enantiomer oligonucleotides as the fundamental building material. We speculate that the formation of left-handed chiral tubes by D-DNA may be attributed to the “relaxed” or underwound right-handed D-DNA duplexes under the in vitro conditions, which give arise to left-handed super-coils in the super-molecular structural level. Similarly, relaxed underwound left-handed L-DNA duplexes would lead to right-handed super-coils. Further study into this phenomenon regarding the mechanism of the chiral tube formation is guaranteed. Nevertheless, the formation of the right-handed L-DNA nanotubes spanning micrometers long was observed with no ambiguity.
Figure 2.
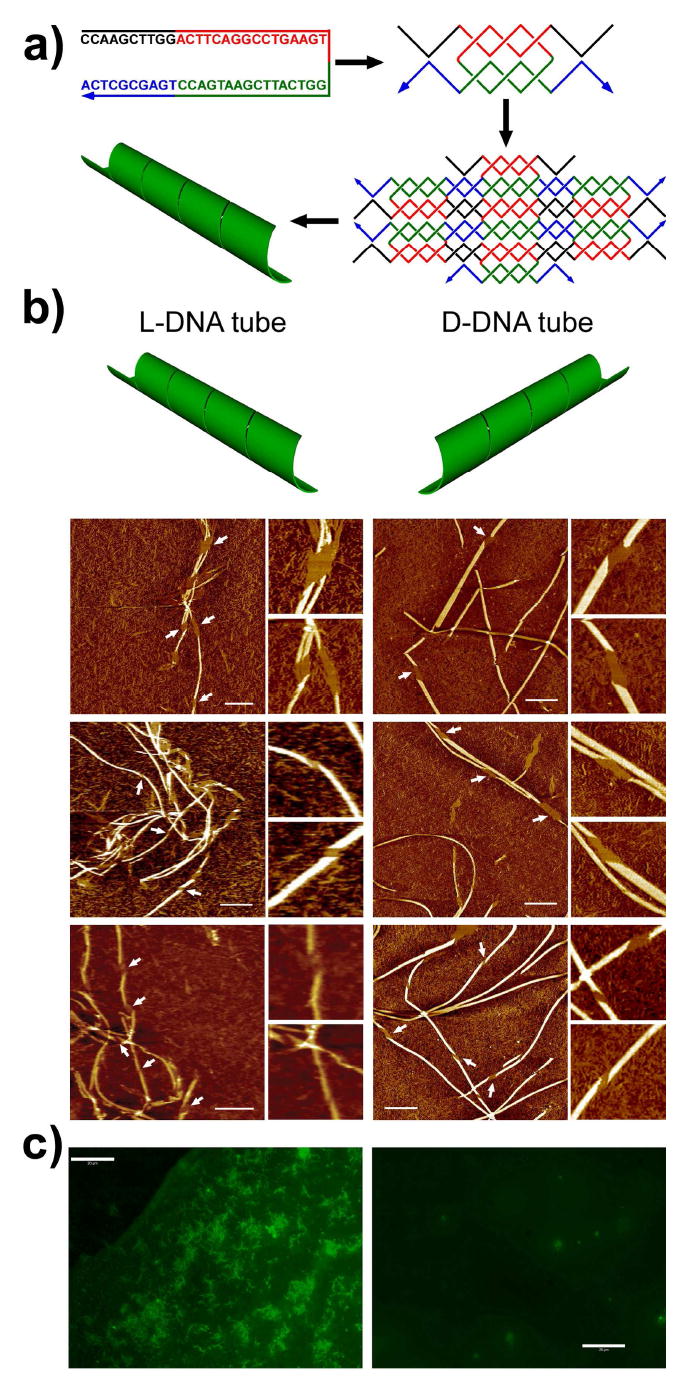
Self-assembly and characterization of L-DNA nanotubes and in comparison to D-DNA nanotubes. a) Assembly scheme. b) The opposite chirality of the L- (left-column) and D-(right column) DNA nanotubes revealed by the defects (pointed out by the arrow heads) on the tubes observed under AFM. Scale bar: 500 nm. Zoom-in images are 500×500 nm2. c) Fluorescence microscope images showing DNA nanotubes after exonuclease treatment. L-DNA tubes (left) persisted integrity while the D-DNA tubes (right) were digested. Scale bar: 20 μm.
Additionally, DNA exonuclease was incubated with the L- and D-DNA nanotubes. As shown in Figure 2c, the L-DNA nanotubes remained intact after enzyme digestion while the D-DNA ones were degraded. DNA nanotubes are known for its structural rigidity36. When properly labelled and modified, these exonuclease resistant L-DNA nanotubes could potentially serve as suitable bio-probing material for in vivo imaging applications.
As noted above, 2D arrays can also be assembled using a single DNA strand by aligning 1D D-DNA duplexes into 2D crystals.34 Briefly, palindrome segments on the strand allow it to self-hybridize and form a pseudo-continuous duplex which can further self-assemble into 2D arrays by sticky-end recognitions. By using an L-DNA strand with the same sequence, we expect the same assembly will result as using D-DNA, given the fact that L- and D-DNA duplexes have the same number of base-pairs in a helical repeat. Figure 3a depicts the schematic of the 2D array structure formed from L-DNA strand. AFM images shown in Figure 3b and Figure S5 (in supplemental information) clearly reveal that nice 2D arrays with high DNA density are formed, some of which curled-up to form tubes. Moreover, the rhombus-shape cavities were observed in zoom-in images, which are consistent with the crystal unit geometry and the lattice periodicity of the one strand D-DNA 2D array assembly reported by Mao’s group.34
Figure 3.
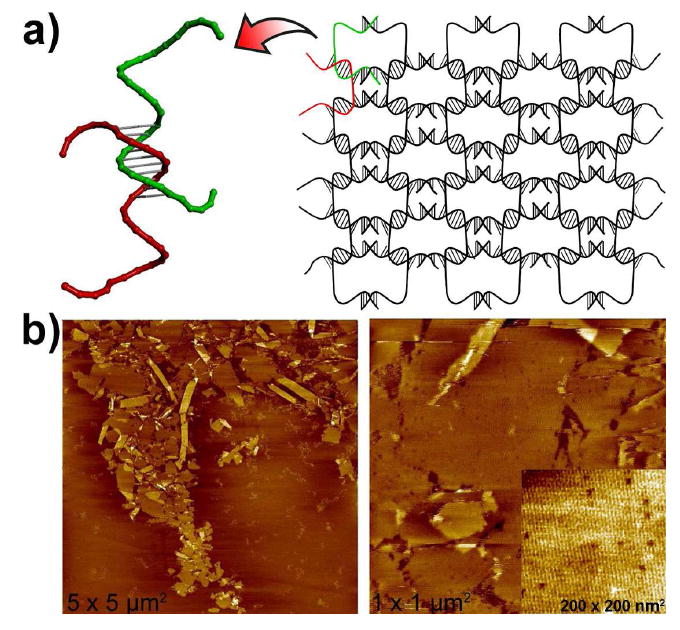
Two-dimensional nanoarrays self-assembled from L-DNA. a) Schematic drawings of the nanoarray structure. b) Representative AFM images of the self-assembled L-DNA nanoarrays.
In summary, we successfully constructed a series of designer nanoarchitectures using mirror-image DNA, L-DNA, an unnatural nucleic acid having the opposite helical chirality to the natural B-form DNA. By applying the well-established D-DNA self-assembly design principles and experimental conditions to L-DNA self-assembly, we have introduced a general method to build L-DNA nanostructures. One could easily relate this method to the “chiral pool synthesis”37 strategy that is widely applied in organic chemistry: a chiral starting material undergoes a series of successive reactions using achiral reagents which retain its chirality to obtain the desired target asymmetric molecule. This work on L-DNA self-assembly enriches the library of unnatural nucleic acids that are useful in nano-scale construction. More importantly, this strategy provides a convenient way to gain control over the chirality of the supramolecular DNA nanostructures such as helical nanotubes.
Finally, nuclease-resistance of L-DNA enables the L-DNA nanostructures to survive in a biological environment. It has been found that L-DNA can be designed or selected to bind with small molecules38 or peptides39. These properties open up opportunities for the in vivo medical applications of L-DNA nanostructures. For example, one can envision a self-assembled L-DNA nano-container delivering high density of small interfering RNA drugs to tumor cells. The supramolecular chiral nanostructure may also be used as a delivery vechile for functional chiral nucleic acid sequences such as spiegelmers.40 We expect L-DNA nanostructures to play an important role in addition to the other types of nucleic acid nanostructures for a wide range of bionanotechnology applications.
Supplementary Material
Experimental Section, DNA sequences, temperature dependent CD spectra and additional AFM images. This material is available free of charge via Internet at http://pubs.acs.org.
Acknowledgments
This research was supported by grants from the National Science Foundation (NSF), the Army Research Office (ARO) and the Technology and Research Initiative Fund from Arizona State University to Y.L.; and by grants from NSF, ARO, Air Force Office of Scientific Research, Office of Naval Research, and the National Institute of Health to H.Y.
References
- 1.Seeman NC. J Theor Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 2.Winfree E, Liu F, Wenzler LA, Seeman NC. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 3.Chen JH, Seeman NC. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 4.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 5.Shih WM, Quispe JD, Joyce GF. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 6.Goodman RP, Schaap IAT, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 7.Rothemund PW, Papadakis N, Winfree E. PLOS Biol. 2004;2:2041–2053. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 9.Aldaye FA, Sleiman HF. J Am Chem Soc. 2007;129:13376–13377. doi: 10.1021/ja075966q. [DOI] [PubMed] [Google Scholar]
- 10.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Nature. 2008;452:198–202. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 11.Le JD, Pinto Y, Seeman NC, Musier-Forsyth K, Taton TA, Kiehl RA. Nano Lett. 2004;4:2343–2347. doi: 10.1021/nl0515495. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Liu Y, Ke Y, Yan H. Nano Lett. 2006;6:248–251. doi: 10.1021/nl052210l. [DOI] [PubMed] [Google Scholar]
- 13.Zheng J, Constantinou PE, Micheel C, Alivisatos AP, Kiehl RA, Seeman NC. Nano Lett. 2006;6:1502–1504. doi: 10.1021/nl060994c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma J, Chhabra R, Liu Y, Ke Y, Yan H. Angew Chem Int Ed. 2006;45:730–735. doi: 10.1002/anie.200503208. [DOI] [PubMed] [Google Scholar]
- 15.Sharma J, Ke Y, Lin C, Chhabra R, Wang Q, Nangreave J, Liu Y, Yan H. Angew Chem Int Ed. 2008;47:5157–5159. doi: 10.1002/anie.200801485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams BAR, Lund K, Liu Y, Yan H, Chaput JC. Angew Chem Int Ed. 2007;46:3051–3054. doi: 10.1002/anie.200603919. [DOI] [PubMed] [Google Scholar]
- 17.He Y, Tian Y, Ribbe AE, Mao C. J Am Chem Soc. 2006;128:12664–12665. doi: 10.1021/ja065467+. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Lin C, Li H, Yan H. Angew Chem Int Ed. 2005;44:4333–4338. doi: 10.1002/anie.200501089. [DOI] [PubMed] [Google Scholar]
- 19.Malo J, Mitchell JC, Venien-Bryan C, Harris JR, Wille H, Sherratt DJ, Turberfield AJ. Angew Chem Int Ed. 2005;44:3057–3061. doi: 10.1002/anie.200463027. [DOI] [PubMed] [Google Scholar]
- 20.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 21.Rinker S, Liu Y, Yan H. Chem Commun. 2006:2675–2677. doi: 10.1039/b603872g. [DOI] [PubMed] [Google Scholar]
- 22.Lukeman PS, Mittal AC, Seeman NC. Chem Commun. 2004:1694–1695. doi: 10.1039/b401103a. [DOI] [PubMed] [Google Scholar]
- 23.Zhang RS, McCullum EO, Chaput JC. J Am Chem Soc. 2008;130:5846–5847. doi: 10.1021/ja800079j. [DOI] [PubMed] [Google Scholar]
- 24.Urata H, Shinohara K, Ogura E, Ueda Y, Akagi M. J Am Chem Soc. 1991;113:8174–8175. [Google Scholar]
- 25.Urata H, Ogura E, Shinohara K, Ueda Y, Akagi M. Nucleic Acids Res. 1992;20:3325–3332. doi: 10.1093/nar/20.13.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seeman NC. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 27.Deng ZX, Lee SH, Mao CD. J Nanosci Nanotechnol. 2005;5:1954–1963. doi: 10.1166/jnn.2005.504. [DOI] [PubMed] [Google Scholar]
- 28.Turberfield A. J Phys World. 2003;16:43–46. [Google Scholar]
- 29.Lin C, Liu Y, Rinker S, Yan H. ChemphysChem. 2006;7:1641–1647. doi: 10.1002/cphc.200600260. [DOI] [PubMed] [Google Scholar]
- 30.Feldkamp U, Niemeyer CM. Angew Chem Int Ed. 2006;45:1856–1876. doi: 10.1002/anie.200502358. [DOI] [PubMed] [Google Scholar]
- 31.Aldaye FA, Palmer AL, Sleiman HF. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 32.Kallenbach NR, Ma RI, Seeman NC. Nature. 1983;305:829–831. [Google Scholar]
- 33.Liu H, Chen Y, He Y, Mao C. Angew Chem Int Ed. 2006;45:1942–1945. doi: 10.1002/anie.200504022. [DOI] [PubMed] [Google Scholar]
- 34.Zhang C, He Y, Chen Y, Ribbe AE, Mao C. J Am Chem Soc. 2007;129:14134–14135. doi: 10.1021/ja075791k. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell JC, Harris JR, Malo J, Bath J, Turberfield AJ. J Am Chem Soc. 2004;126:16342–16343. doi: 10.1021/ja043890h. [DOI] [PubMed] [Google Scholar]
- 36.O’Neill P, Rothemund PWK, Kumar A, Fygenson DK. Nano Lett. 2006;6:1379–1383. doi: 10.1021/nl0603505. [DOI] [PubMed] [Google Scholar]
- 37.Hanessian S. Total synthesis of natural products, the “Chiron” approach. Pergamon Press; New York: 1983. [Google Scholar]
- 38.Dose C, Ho D, Gaub HE, Dervan PB, Albrecht CH. Angew Chem Int Ed. 2007;46:8384–8387. doi: 10.1002/anie.200703007. [DOI] [PubMed] [Google Scholar]
- 39.Williams KP, Liu XH, Schumacher TNM, Lin HY, Ausiello DA, Kim PS, Bartel DP. Proc Natl Acad Sci USA. 1997;94:11285–11290. doi: 10.1073/pnas.94.21.11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purschke WG, Radtke F, Kleinjung F, Klussmann S. Nucleic Acids Res. 2003;31:3027–3032. doi: 10.1093/nar/gkg413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental Section, DNA sequences, temperature dependent CD spectra and additional AFM images. This material is available free of charge via Internet at http://pubs.acs.org.


