Abstract
The class C serine β-lactamase of Enterobacter cloacae P99 is irreversibly inhibited by O-aryloxycarbonyl hydroxamates. A series of these new inhibitors has been prepared to investigate the kinetics and mechanism of the inactivation reaction. A pH-rate profile for the reaction indicated that the reactive form of the inhibitor is neutral rather than anionic. The reaction rate is enhanced by electron-withdrawing aryloxy substituents and by hydrophobic substitution on both aryloxy and hydroxamate groups. Kinetics studies show that the rates of loss of the two possible leaving groups, aryloxide and hydroxamate are essentially the same as the rate of enzyme inactivation. Nucleophilic trapping experiments prove, however, that the aryloxide is the first to leave. It is likely, therefore, that the rate-determining step of inactivation is the initial acylation reaction, most likely of the active site serine, yielding a hydroxamoyl-enzyme intermediate. This then partitions between hydrolysis and aminolysis by Lys 315, the latter to form an inactive, cross-linked active site. A previously described crystal structure of the inactivated enzyme shows a carbamate cross-link of Ser 64 and Lys 315. Structure-activity studies of the reported compounds suggest that they do not react at the enzyme active site in the same way as normal substrates. In particular, it appears that the initial acylation by these compounds does not involve the oxyanion hole, an unprecedented departure from known and presumed reactivity. Molecular modeling suggests that an alternative oxyanion hole may have been recruited, consisting of the side chain functional groups of Tyr 150 and Lys 315. Such an alternative mode of reaction may lead to the design of novel inhibitors.
For decades now, β-lactams have been one of our most effective weapons against bacterial infections (1). These drugs, although still the first line of attack in many clinical situations, have been compromised to a considerable degree by bacterial resistance to them (2). Among various sources of resistance that have arisen in bacteria, the most generally troublesome is the production of β-lactamases. These enzymes very effectively catalyze the hydrolysis and thus destruction of β-lactams before they can reach their cellular targets (3).
The threat posed by β-lactamases to the efficacy of β-lactam antibiotics has been tackled by pharmaceutical companies in several ways. One approach that has been quite successful to date is that of including a β-lactamase inhibitor with a β-lactam antibiotic in combination therapies. For many years now, such combinations, using the now-classical β-lactamase inhibitors clavulanic acid, sulbactam and tazobactam, have been used to advantage (4). Since these inhibitors are themselves β-lactams, however, it is perhaps not surprising to find that certain β-lactamase mutants are capable of hydrolyzing them quite effectively. Such mutants have now been found in clinical settings and therefore the effectiveness of β-lactam antibiotics will continue to be threatened (5).
The circumstances described above explain the continuing interest in new β-lactamase inhibitors, and, in particular, in inhibitors not based on the β-lactam platform and/or that cannot be hydrolyzed by β-lactamases. To date, no generally effective small-molecule non-covalent inhibitors of β-lactamases have been found, although there are several types of non-β-lactam covalent inhibitors. The best known of the latter include the boronates (6–8) and phosphonates (9,10). Recently, we described an example of a new class of acylating agents, the O-aryloxycarbonyl hydroxamates or N,O-diacylhydroxylamines that appear to have affinity for the active site of class C β-lactamases. The lead compound, 1, interacted covalently with the active site producing a novel crosslinking of Ser 64 with Lys 315, 2 (11).

Several interesting questions arise with respect to the mechanism of action and the general structure-activity relationships of this class of compounds. In this paper we address these issues, making use of a new series of analogs 3 – 14. We find evidence that these compounds may in fact react differently with the active site of a class C β-lactamase than do normal substrates. This yields the promise of novel inhibitor design.
EXPERIMENTAL PROCEDURES
The class C P99 β-lactamase from Enterobacter cloacae was purchased from the Centre for Applied Microbiology and Research (Porton Down, Wiltshire, U.K.). Elemental analyses were carried out by Desert Analytics Laboratory. Electrospray mass spectra of enzyme complexes were obtained by the Mass Spectrometry Laboratory, School of Chemical Sciences, University of Illinois.
Synthesis
O-Aryloxycarbonyl Hydroxamates
These syntheses followed the general strategy of coupling N-hydroxycarbamates with chloroformates, as previously reported (11). Chloroformates, where not commercially available, were readily obtained from the reaction of a desired alcohol with phosgene in the presence of base (12). N-Hydroxycarbamates could be prepared from the corresponding chloroformates by the method of Defoin et al. (13). To then prepare the O-aryloxycarbonyl hydroxamates, the N-hydroxycarbamate was dissolved in methylene chloride and stirred at 0 °C under an atmosphere of dry nitrogen. Equimolar quantities of imidazole or pyridine in methylene chloride were added, followed by dropwise addition of the chloroformate in methylene chloride. The solution was maintained at 0 °C and stirred for 30 minutes. The resulting precipitate of imidazole or pyridine hydrochloride was then removed by filtration and the solvent removed from the filtrate by rotary evaporation, yielding the crude product. This was generally purified by silica chromatography and/or recrystallization.

N-(Benzyloxycarbonyl)-O-[(4-methoxyphenyl)oxycarbonyl]hydroxylamine (4)
Benzyl N-hydroxycarbamate (Aldrich; 0.79 g, 4.8 mmol) was acylated with 4-methoxyphenyl chloroformate (0.72 ml, 4.8 mmol) as described above, with dry pyridine (0.39 ml, 4.8 mmol) as the base. Ethyl acetate (7 ml) was added prior to filtration to aid the precipitation of pyridine hydrochoride. Following removal of solvent and the drying of the residue overnight on an oil pump, the resulting solid was recrystallized from benzene:cyclohexane to yield 1.04 g (68%) of colorless crystals (mp 88 °C). 1NMR: (d6-DMSO) δ11.70 (s, 1H), 7.39 (m, 5H), 7.16 (d, J = 9.4 Hz, 2H), 6.99 (d, J = 9.4 Hz, 2H), 5.20 (s, 2H), 3.77 (s, 3H). IR (KBr, cm−1) 1806, 1722. ES(-)MS 316.2. Anal. Calcd for C16H15NO6: C, 60.56; H, 4.76 ; N, 4.41. Found: C, 60.53 ; H, 4.53 ; N, 4.26.
Details of the synthesis and characterization of 3 –13 are provided as Supporting Information.
N-(Benzyloxycarbonyl)-O-(phenoxythionocarbonyl)hydroxylamine (14)
Benzyl N-hydroxycarbamate (99 mg, 0.593 mmol), dissolved in ethyl acetate (2 ml), was acylated with phenyl chlorothionocarbonate (0.0826 ml, 0.593 mmol) as described above, with dry pyridine (45 μl, 0.593 mmol) as base. Following removal of solvent, the resulting yellow solid was purified by chromatography on silica gel with 2:1 hexanes: ethyl acetate as eluent, and finally recrystallized from benzene:cyclohexane (1:1) to yield 14 mg (7.8 %) of colorless needle-like crystals (mp 84–86 °C). 1NMR: (CD3CN): δ 9.53 (s, 1H), 7.48 (t, J = 8 Hz, 2H), 7.40 (m, 6H), 7.14 (d, J = 7.2 Hz, 2H), 5.23 (s, 2H). IR (KBr, cm−1) 1721. ES(-)MS: m/z 302.07.
Analytical and kinetics methods
Enzyme concentrations were determined spectrophotometrically. Kinetics measurements were carried out at 25 °C, buffered in 20 mM 3-morpholinopropanesulfonic acid (MOPS) at a pH of 7.5 unless otherwise noted. The inhibitors were prepared in concentrated acetonitrile stock solutions and diluted to ≤ 5 % acetonitrile in reaction mixtures.
Determination of inhibitor pKa values
pKa values of 1, 3 – 5 and 9 were determined spectrophotometrically by observing the change in absorbance of 100 μM solutions of inhibitor at a suitable wavelength as a function of pH. The buffers were 20 mM formate for pH 3–4, 20 mM acetate for pH 4.5–5.5, and phosphate for pH 6.0–7.5, a constant ionic strength of 0.1 was maintained with NaCl. Analytical wavelengths used were 218 nm (1), 278 nm (3), 230 nm (4), 238 nm (5) and 230 nm (9).
1H NMR monitoring of the chemical shift of the benzyl methylene peak with respect to DSS in a solution of 1 (750 μM) in 50 mM KH2PO4 buffer in D2O over the pD range 5.7–7.7 was used to confirm the pKa of 1.
β-Lactamase inactivation – direct experiment
The P99 β-lactamase (0.25 μM) was incubated with inhibitor (0.5–400 μM) in buffer (500 μl) containing 1 mg/ml BSA. Aliquots of reaction mixture (15 μl) were diluted into cephalothin solution (1.0 ml, 200 μM) and hydrolysis of the latter was monitored spectrophotometrically at 278 nm. The initial rates, proportional to the amount of active enzyme, were plotted against time to obtain a progress curve that was fit to Scheme 1 (Results and Discussion) by the program Dynafit (14) to obtain the second order inactivation rate constant. No sign of saturation kinetics was observed at the inhibitor concentrations employed.
Scheme 1.

P99 β-lactamase inactivation – by competition with substrate
The enzyme (0.5–2.0 nM final concentration) was added to a buffered solution containing a mixture of the substrate cephalothin (200 μM) and the inhibitor (0 – 100 μM); hydrolysis of the substrate was monitored at 278 nm. The total progress curves were fit to Scheme 2 (Results and Discussion) by means of the Dynafit program (14).
Scheme 2.
Active site titration
Enzyme (final concentrations 0.25 μM for 1 and 3, 1.0 μM for 4, and 5 μM for 7 and 8) was incubated with inhibitor (0–1 μM for 1 and 3, 0–3 μM for 4, and 0–20 μM for 7 and 8) in buffer (500 μl), containing 1 mg/ml BSA. After 3 hours the reaction mixtures were assayed for active enzyme against 200 μM cephalothin, and the initial rates measured were plotted vs [I]/[E] to obtain the partition ratio k2/k3 from Scheme 1 (Results and Discussion) using eq 1.
| (1) |
pH dependence of inactivation
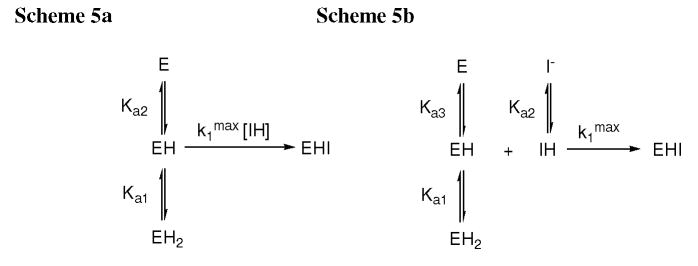
The pH profile of inactivation of the P99 β-lactamase (0.25 μM) by 1 (0.5 μM) was determined by direct loss of activity experiments as described above. The pH was controlled with acetate (pH 4–5), MES (pH 5.5–6.5), MOPS (pH 7–8), and AMPSO (pH 8.5) buffers, at 20 mM each. Equation 2, derived from Scheme 5a (Results and Discussion), was fit to the data by means of a nonlinear least squares program to obtain the pKa values.
Scheme 5.
| (2) |
Leaving group kinetics
Compound 3 (5 μM) was employed to monitor the release of the aryloxide leaving group upon reaction with the P99 β-lactamase (1 μM). Release of m-hydroxybenzoate was conveniently monitored by fluorescence emission at 398 nm following excitation at a wavelength of 294 nm. Compound 8 (1.5 μM) was used to investigate the release of the hydroxamate leaving group from the β-lactamase (0.5 μM) by monitoring the change in biphenyl fluorescence emission intensity at 315 nm after excitation at 240 nm. In each case, the initial fluorescence intensity was directly proportional to the compound concentration.
Aminolysis Experiments
The P99 β-lactamase (0.25 μM) was completely inactivated by 8 (2.0 μM) as indicated by its lack of activity against the substrate cephalothin. The inhibited enzyme solution was then treated, separately, with hydroxylamine and D-phenylalanine (1 mM each). After 2 hr, the reaction mixtures were again assayed for activity against cephalothin.
Aminolysis of the acyl-enzyme intermediate formed by 1, 8 and 12 was investigated using D-alanine, D-phenylalanine, L-phenylalanine and benzylamine. Enzyme (1 μM) was added to a solution of the inhibitor (50 μM) in 20 mM MOPS buffer containing D-alanine (0–170 mM), D-phenylalanine (0–40 mM), L-phenylalanine (0–40 mM) or benzylamine (0–20 mM). The reaction was monitored at 268 nm, 285 nm, and 228 nm for compounds 1, 8, and 12, respectively. The resulting progress curves were then fitted to Scheme 3 (Results and Discussion) by means of the Dynafit program (14). Note that D-phenyalanine had little or no effect on the rate of the spontaneous reaction of 1 in solution.
Scheme 3.
Isolation of the aminolysis product (18)
The product of the aminolysis reaction was obtained by reacting 8 (100 μM) with the P99 β-lactamase (1 μM) in MOPS buffer containing 40 mM D-phenylalanine (total volume 4.6 ml). The reaction was monitored spectrophotometrically at 285 nm until complete. The final solution was then acidified with 4 drops of concentrated HCl and then extracted with ethyl acetate (3x6 ml). The combined organic layers were washed with water (3 ml) and dried over NaSO4. After removal of the solvent, the resulting oil was dissolved in a minimum amount of methanol and purified by HPLC on a reverse phase 250/4 Nucleosil 100−5 C18 column, using a linear gradient of 50:50 aqueous MeOH containing 0.01% NH4HCO3 to 100% MeOH over 15 min. The product fraction was acidified to pH 4 and extracted twice with ethyl acetate. The extracts combined, dried with MgSO4, the solvent was removed and the residue subjected to ESMS analysis.
Mass spectroscopy of EI′
The enzyme (10 μM) was incubated with inhibitor (20 μM) in buffer (200 μl) for 15 min. The β-lactamase was then assayed against cephalothin to confirm inhibition and the reaction mixture quenched with cold 50 % aqueous trichloroacetic acid (20 μl). The quenched reaction mixture was held on ice for thirty min, centrifuged for 5 min. at 8000 rpm, and the supernatant decanted. The precipitated protein was then washed with 5% cold trichloroacetic acid (2 × 50 μl) followed by acetone (2 × 50 μl). The solid residue was dried on an oil pump and an electrospray mass spectrum obtained.
Computational Methods
Simulations were performed on a SGI workstation running the program Insight II, essentially as previously described (15–17). The crystal structure of the enzyme, inhibited by 1 [PDB entry 2p9v (11)] was modified to construct the anionic tetrahedral intermediate of cross-linking (24). The wild type Amp C β-lactamase crystal structure [PDB entry 2bls (18)] was the starting point for building the tetrahedral intermediate of acylation (28). The pH was set to 8.3 and the total charge on the complexes was 1.0. In both cases, the side chain of Tyr 150 was neutral and those of Lys 67 and 315 were cationic. The partial charges of the enzyme were assigned by Insight II. Partial charges (MNDO) of the inhibitor in the tetrahedral intermediate 24 were calculated from a model adduct with serine and lysine residues. The partial charges of 28 were determined in the same way on a model containing just the serine residue.
In both cases, 24 and 28, R and S isomers at the chiral reaction center were constructed. The active site was hydrated with a 15 Å sphere of water centered on Oγ of the nucleophilic serine 64. The models were then subjected to molecular dynamics runs of 100 ps. For each isomer, several typical snapshots from the molecular dynamics runs were selected. These structures were then each subjected to 1000 steepest descent energy minimization, followed by 2000 steps of conjugate gradients. Interaction energies between enzyme and ligand (Eint) were calculated for each of the minimized structures, as previously described (15).
The acyl enzyme model (25) was constructed by elimination of the lysine leaving group from 24. The active site Tyr150 side chain was taken as anionic in this case. The model was studied in the same way as 24 and 28.
RESULTS AND DISCUSSION
As described previously, the O-aryloxycarbonyl hydroxamate 1 irreversibly inactivates the class C Enterobacter cloacae P99 β-lactamase (11). A small amount of turnover, approximately two molecules of 1 per enzyme molecule, accompanied inhibition and thus the kinetics data were fitted to Scheme 1 (in Scheme 1, EI represents an intermediate that partitions between breakdown to free enzyme and inactivation; k0 represents the rate constant for the spontaneous hydrolysis of 1 in solution). It was assumed that conversion of E to EI was rate-determining; evidence for this will be presented below.
The compounds 3 – 14 also inhibited this enzyme irreversibly. Figure 1, for example, shows the results of experiments where, for compound 4, (a) the loss of enzyme activity with time at a particular enzyme and inhibitor concentration was determined, (b) a titration of residual activity vs the inhibitor/enzyme concentration ratio was conducted, and (c) total progress curves from the turnover of the good substrate cephalothin in the presence of various concentrations of the inhibitor were obtained. These data were simultaneously fitted to Scheme 1 (or 2) to obtain a fit yielding the lines shown in Figure 1, and thus k1 and k2/k3 values of (5.1 ± 0.8) x104 s−1M−1 and 2.0 ± 0.1, respectively. Similar experiments with the remaining compounds yielded the results given in Table 1. Also shown in Table 1 are the observed rate constants for the spontaneous hydrolysis of the inhibitors in MOPS-buffered solution at pH 7.5.
Figure 1.

A. Loss of activity of the P99 β-lactamase (0.25 μM) as a function of time in the presence of 4 (0.7 μM). B. Titration of the activity of the P99 β-lactamase (1.0 μM) on addition of 4; v/v0 is the fraction of enzyme activity remaining at a particular inhibitor/enzyme concentration ratio (I/E). C. Absorbance of the substrate cephalothin (0.2 mM) as a function of time on addition of the P99 β-lactamase (final concentration 1.9 nM) to a mixture of substrate and 4 (○, 0 μM;□, 9.5 μM;⋄, 28.7 μM;×, 47.9 μM). In all three graphs the points are experimental and the lines are derived from fitting the data to Schemes 1 and 2 (see text).
Table 1.
| k0 (s−1) | k1 (s−1M−1) | k2/k3 | |
|---|---|---|---|
| 1 | 2.5 × 10−4 | 6.1 ± 0.2 × 103 | 2.0 ± 0.1 |
| 3 | 2.78 × 10−4 | 5.4 ± 0.3 × 103 | 1.0 ± 0.1 |
| 4 | 1.19 × 10−4 | 5.1 ± 0.8 × 103 | 2.0 ± 0.1 |
| 5 | 9.84 × 10−4 | 2.2 ± 1.1 × 104 | 1.6 ± 0.1 |
| 7 | 4.45 × 10−4 | 1.8 ± 0.6 × 104 | 1.6 ± 0.1 |
| 8 | 3.59 × 10−4 | 3.1 ± 0.9 × 104 | 1.9 ± 0.2 |
| 9 | ND | 0.19 ± 0.01a | ND |
| 10 | 2.0 × 10−4 | 0.67 ± 0.03a | ND |
| 11 | 4.0 × 10−5 | 52 ± 30 | ND |
| 12 | 4.2 × 10−4 | 4.6 ± 1.5 × 103 | 2.2 ± 0.5 |
| 13 | 7.41 × 10−5 | 2.1 ± 0.8 × 103 | 15.0 ± 1.5 |
| 14 | 3.61x 10−4 | 2.7 ± 0.4 × 103 | 1.0 ± 0.1 |
Only rate of inactivation determined: ND = not determined.
It is evident from that data and from the derived Scheme 1 that a small amount of turnover, assumed to be hydrolysis (1–2 molecules hydrolyzed per inactivation event), accompanies inactivation. In this respect the aryloxycarbonyl hydroxamates resemble classical mechanism-based β-lactamase inhibitors such as clavulanic acid and sulbactam. The extent of such turnover is very variable, depending on enzyme and inhibitor, but it is certainly considerably larger for the clavulanic acid and sulbactam with the P99 β-lactamase than for the hydroxamates. The likely structural basis for turnover of the latter inhibitors is addressed below.
In view of the crystal structure of the inert complex and the observations described above, the inactivation involves a double acyl transfer reaction, which can be represented by Scheme 3 (11). In Scheme 3, A and B represent the Ser 64 side chain hydroxyl group and the Lys 315 side chain amine, and X and Y represent ArO and PhCH2OCONHO; the actual sequence of events, i.e. the identity of A, B, X and Y in a particular step will be discussed below.
The data of Table 1 indicate an electronic effect on k1, where the order of reactivity 4 (p-OMe) < 1 (p-H) < 5 (p-Cl) and the size of this effect (19), suggests departure of OAr as the leaving group in the rate-determining step. A similar order is observed in the spontaneous reaction, which represents, from 1H NMR observation, hydrolysis to form phenol and hydroxamic acid (and, presumably, carbon dioxide). The p-NO2 analog, 6, proved too unstable for proper kinetic analysis, but was shown to very rapidly inhibit the enzyme (the speed of this reaction also rather argues for X = OAr in Scheme 3, but stronger evidence is presented below). Loss of the hydroxamic acid, which must be the initial leaving group in the case of the symmetrical compound 12, can also, however, lead to an effective inhibitor—from the point of basicity, this leaving group is comparable to that of 1 but has a quite different distribution of polarity. The effective inhibition by 12 does show, however, that for any of these compounds, the hydroxamate could be, in principle, either X or Y.
The much poorer leaving groups OMe and NHPh present in 9 and 10, produce much poorer inhibitors. Addition of a hydrophobic substituent in either the aryloxy (7) or hydroxamate (8) moiety enhances reactivity, a not unexpected result at the class C β-lactamase active site (20,21).
Before further discussion of Scheme 3, it is useful to consider the structure and stability of the inhibitors in solution. 15N NMR spectroscopy of 15N-labelled 1 showed that this compound is an O-(rather than N-)acylated hydroxamate (11). This result is in accord with structural studies of similar O-acyl hydroxamates (22) and will be assumed to be true for all of the present compounds, as shown in the structural diagrams above. They are thus likely to be weak N-H acids (23). Spectrophotometric titrations (not shown) of 1, 3–5 and 9 yielded pKa values of 6.8 ± 0.1, 7.2 ± 0.1, 7.0 ± 0.1, 6.9 ± 0.3 and 7.1 ± 0.2, respectively. These results were supported by a 1H NMR titration of 1, where the upfield shift of the methylene protons with pH was monitored, yielding a pKa of 6.73 ± 0.02. The structural differences between 1, 3 – 5 and 9 have little effect on the pKa values, as would be expected on the basis of their remoteness from the site of dissociation. At pH 7.5, where the data of Table 1 was obtained, the inhibitors would be largely anionic in solution.
pH rate profiles for the hydrolysis of 1 and 13 are shown in Figure 2 (note that buffer concentrations had little or no effect on these rate constants). These were fitted to Schemes 4a and 4b, respectively, yielding rate constants for 1 of kH2O = 6.4 × 10−5s−1, k−H2O = 2.0 × 10−4s−1, k−OH = 9.3 s−1M−1, and for 13 of kH2O = 3.9 × 10−5 s−1M−1 and kOH = 19.6 s−1M−1. The kH2O values of 1 and 13 are quite similar. As would be expected, the rate of hydroxide attack on I (13) is greater than that on I− (from 1). The apparent rate of water attack on I−, however, is greater than that on IH. This probably reflects some degree of intramolecular catalysis, nucleophilic or general base, in the former case (23). The slow hydrolysis of 11 at pH 7.5 probably stems from its likely much higher (sulfonamide) acidity than 1 and the lower reactivity of its anion at this pH. These results show that both the neutral forms of the inhibitors and their anions (excluding 13) are potentially reactive as acylating agents. This is significant since the active sites of β-lactamases are known to have affinity for anions.
Figure 2.
pH-rate profile for the hydrolysis of 1 (●) and 13 (○) in aqueous solution. The points are experimental and the lines are derived from fitting the data to Schemes 4a and 4b (see text).
Scheme 4.
The pH-rate profile for the inactivation of the P99 β-lactamase by 1 was determined (Figure 3). This data was fitted to Scheme 5a, yielding a k1max value of (3.9 ± 0.8) × 10 4 s−1M−1, and pKa1 and pKa2 values of 5.62 ± 0.17 and 6.78 ± 0.18, respectively. The value of pKa2 strongly suggests that the second dissociation in the pH-rate profile of 1 (Ka2) represents dissociation of the inhibitor rather than that of the enzyme, as represented in Scheme 5a. Although normal substrates also display a second dissociation associated with loss of enzyme activity at high pH, pKa values for this dissociation are typically around 9 (25,26). It seems likely, therefore, that the active inhibitor in the present case is the neutral compound rather than the anion. Scheme 5b, therefore, kinetically equivalent to Scheme 5a, is probably correct. The value of pKa3 is probably larger that that of pKa2, as indicated above. The lower pH dissociation, pKa1, presumably reflects activation of the active site towards 1, perhaps by dissociation of a residue required in the basic form for activity. Similar pKa values have been reported for reactions of the P99 β-lactamase (kcat/Km) with substrates, e.g. 5.85 ± 0.09, 5.92 ± 0.16, 6.21 ± 0.11, 5.89 ± 0.07 and 6.29 ± 0.04 (24,25). The important consequence of the pH profile of Figure 3, which is presumably very similar to those for 3–8 because of their similar pKa2 values (see above), is that the maximal activity of these inhibitors is expressed at around pH 6.2 and thus the data of Table1, taken at pH 7.5, does not reflect their absolute potency [e.g. compare 1 at pH 7.5 (k1 = 6.1 × 103 s−1M−1) with pH 6.2 (k1 = 2.5 × 104 s−1M−1) and k1max = 3.9 × 104 s−1M−1]. The low inhibitory activity of 11 at pH 7.5 is probably largely due to its lower pKa2 than that of 1 and thus to the absence of significant quantities of IH at the above-mentioned pH.
Figure 3.
pH-rate profile for the inactivation of the P99 μM β-lactamase by 1. The points are experimental and the line is derived from fitting the data to Schemes 5a or 5b (see text).
The time-dependence of the loss of leaving groups (X and Y of Scheme 3) on reaction of these inhibitors with the enzyme was investigated by fluorescence measurements. The departure of the m-hydroxybenzoate from 3 and biphenylmethyl N-hydroxycarbamate from 8 were thus investigated (Figure 4). The data of Figure 4a could be fitted to Scheme 1 from which the second order rate constant for reaction of enzyme with 3 was (6.8 ± 1.7) × 103 s−1M−1. Similarly, the data for 8 from Figure 4b could be fitted to give a rate constant of (4.0 ± 0.2) × 104 s−1M−1. Comparison of these values with those from Table 1 suggests that the two leaving groups depart at a rate essentially the same as loss of enzyme activity. In terms of Scheme 3, this is strong evidence that (k2 + k3) ≫ k1 [X-CO-Y] under the concentration conditions employed in these experiments. Thus, it is likely that the rate-determining step for the inactivation is the first acylation step with rate constant k1. The enzyme isolated after inactivation contained only an additional CO moiety, as revealed by mass spectrometry and the crystal structure (11).
Figure 4.

A. Fluorescence intensity change at 398 nm (excitation at 294 nm) on reaction of 3 (5.0 μM) with the P99 β-lactamase (1.0 μM). The fluorescence change reflects release of m-hydroxybenzoate. B. Fluorescence intensity change at 315 nm (excitation at 240 nm) on reaction of 8 (1.5 μM) with the P99 β-lactamase (0.5 μM). The fluorescence change reflects release of p-phenylbenzyl N-hydroxycarbamate. In both graphs the points are experimental and the line is derived from fitting the data to Scheme 1 (see text).
A major remaining unknown is the order of departure of the leaving groups - what are the identities of × and Y in Scheme 3? This was determined by means of a trapping experiment. It is known that the acyl-enzyme derived from the depsipeptide 17 can be intercepted by specific amines and amino acids as acyl-acceptors (26) to yield amide (peptide) products. Small aliphatic amines were ineffective as acceptors, as were L-amino acids, but D-amino acids were specifically active. The effect of added amine acceptors on the reaction of O-aryloxycarbonyl hydroxamates with the P99 β-lactamase was assessed by the effect of such acceptors on the initial rates of turnover of these inhibitory substrates (Scheme 3, where Y′ is the product of aminolysis of the initially formed acyl-enzyme by the amine acceptor). In the presence of an effective acceptor, X-CO-Y would be a substrate rather than an inhibitor.

The inhibited enzyme, 16 (Scheme 3), was inert to common nucleophiles such as hydroxylamine, benzylamine and D-phenylalanine, i.e. no restoration of enzyme activity was observed after their addition. On the other hand, addition of D-phenylalanine to the initial reaction mixture containing the inhibitor 1 led to enhanced turnover prior to inactivation (Figure 5). The fitting of these data to Scheme 3 yielded a k4/k2 value of (4.0 ± 2.0) × 104. Similar experiments with both 1 and 12 and with D-alanine as the added nucleophile, yielded a k4/k2 value of 3300 ± 470 from a combined fit of the data to Scheme 3 (not shown). This result supports a common acyl-enzyme derived from both 1 and 12 which thus must contain Y= PhCH2OCONHO. The very similar values of the partition ration k2/k3 for 1, 3–7, and 12 also support a common intermediate. In contrast, L-phenylalanine, to 40 mM, produced no enhanced turnover of 1 and thus D-amino acids may be specific acyl acceptors on reaction with 15. Interestingly, this result correlates with those for depsipeptide substrates of the P99 β-lactamase, such as 17, where D-amino acids were found to be specific acceptors (27). This point will be revisited below.
Figure 5.
Absorbance changes with time on addition of the P99 β-lactamase (final concentration 1.0 μM) to mixtures of 1 (50 μM) and D-phenylalanine (▵, 0 mM;⋄, 5.0 mM;□, 10.0 mM;×, 20.0 mM;○, 40.0 mM). The points are experimental and the lines derived from fitting the data to Scheme 3 (see text).
Unambiguous and definitive identification of 15 as ROCONHOCO-E was achieved by isolation of the product Y′ from reaction mixtures containing 8 and D-phenylalanine. After hplc purification of this product, negative ion electrospray mass spectroscopy yielded a mass of 433.0, which corresponds to the structure 18.
Thus, reaction of these inhibitors, except perhaps for 9 and 10, with the P99 β-lactamase probably initially produces a hydroxamoyl-enzyme intermediate, 15, which then partitions between hydrolysis and the cross-linking reaction. We cannot be sure at present whether the first nucleophile (A in Scheme 3) is the active site serine (64) or lysine (315) but will henceforward assume it is the former, based on all precedent.
Of the new compounds tested, perhaps the most interesting results arose from 13 and 14. First, we will consider 13. Crystal structures of complexes of β-lactamases with substrates (18, 28,29) and transition state analogues (30,31) show that the amide group of the typical side chain of β-lactams is hydrogen-bonded to active site functional groups. In particular, the side chain amide nitrogen atom is normally within hydrogen-bonding distance of a specific backbone carbonyl group of the β-2 strand that makes up one wall of the active site. Methylation of this side chain nitrogen atom of a substrate appears to lead to loss of affinity for the active site of β-lactamases and the structurally related bacterial DD-peptidases. For example, N-methyl-benzylpenicillin 19 has no antibiotic activity (32), and 20 (unlike 17) is not a β-lactamase substrate (R.F. Pratt, unpublished). Compound 13, however, inhibits the P99 β-lactamase only slightly less effectively than the unsubstituted analog 1 (Table 1). [Note, however, that this comparison reflects the situation at pH 7.5; when the pH dependence of the activity of 1 is taken into account (see above), 13 is some 20-fold less active than 1, but still unexpectedly reactive]. This result suggests that 13, and by implication, the other inhibitors of Table 1, may interact with the enzyme active site in an orientation different from that of normal substrates. It should be noted, however, that turnover (hydrolysis) of 13 at the active site (as reflected in the k2/k3 values of Table 1 and Scheme 1) occurs to a significantly greater extent than with the other compounds. Thus, the acyl-enzyme 15 derived from 13 is more accessible to water than that from 1, or, perhaps more likely, is less reactive in the cross-linking reaction.

Compound 14, a specific thiono analogue of 1, also inhibited the P99 β-lactamase only slightly less effectively than 1 (Table 1). Serine β-lactamases, like serine proteases, are believed to employ an “oxyanion hole”, a hydrogen bond donor site to stabilize anionic tetrahedral intermediates and transition states during catalysis. Evidence for this derives from crystal structures, and, functionally, from the effect of thiono substitution on substrate turnover rates (33,34). For example, the thiono-β-lactam 21 (X = S) is some 106 times poorer (kcat and kcat/Km) as a substrate of the P99 β-lactamase than is the oxo analog (21, X = O) (33). Similarly the thionoester 22 (X = S) is much poorer a substrate than 22 (X=O) (34). These results are believed to arise from the imperfect fit of sulfur in the oxyanion hole during the tetrahedral transition states of catalysis (35). The observation that 14 is only a slightly poorer inhibitor of the P99 β-lactamase than 1 strongly suggests that the oxyanion hole is not involved in the acylation steps leading to inhibition by these compounds.

An interesting aspect of the reaction of the P99 β-lactamase with 14 was that a mass spectrum of the inhibited enzyme showed a mass increase above that of the native enzyme of 31 amu. This is consistent with incorporation of CO, but not of CS, to produce the bridged structure 16. Sulfur to oxygen exchange must therefore have occurred. We speculate that the exchange occurs during reaction of the initial acyl-enzyme with the Lys 315 amine by a mechanism such as shown in Scheme 6. There is literature precedent for loss of sulfur in thionoester hydrolysis and aminolysis (36,37).
Scheme 6.
Finally, it has already been noted (11) that the similar reactivity of 1 and 3 indicates that the aryloxy leaving group of these compounds does not interact with the P99 β-lactamase active site in the same way as do classical substrates, including 17, where a correctly placed carboxylate group affords significant rate enhancement (16). These observations, taken together with those above, strongly indicate that 1 and its analogues react with the β-lactamase active site in an orientation quite different from that of even a very close structural analog, the substrate 17.
Although the conclusion reached above seems sound, a related issue that should be addressed is that of why 1 does not react in the classical mode of 17 and β-lactams. This may well be because of the greater rigidity of 3, particularly with respect to the N-X-C-OAr dihedral rotation (23). Molecular dynamics simulations suggest that this rotation is much more restricted when X = O (and the oxygen is predominantly sp2 hybridized as a carbonate) than when X = CH2. The corresponding N-C6-C7-N4 dihedral angle of ca. 120° found in penicillins (38), for example, would therefore be only accessible to 1 after surmounting an energy barrier of several kilocalories (39). We have previously reported that the X = NH analogues of 1, where the barrier would be even higher, are not substrates or inhibitors of the P99 β-lactamase or of the structurally similar Streptomyces R61 DD-peptidase (24). The limited conformational accessibility of the β-lactamase active site has been previously discussed (40). The structural issues relating to the alternative binding of 1 are explored below.

First, a model of the tetrahedral intermediate 24 for the cross-linking reaction was constructed directly from the crystal structure (11). Both S and R constructs were considered. Molecular dynamics simulations suggested that neither structure had significant flexibility while cross-linked, and both achieved minimum energy structure (by the criterion of the lowest interaction energy Eint (15)) with the oxyanion interacting with the polar side chain functional groups of Tyr 150 and Lys 67. It was not possible to position the oxyanion in the classical oxyanion hole (30, 33). The R and S structures differed in the positioning of the hydroxamate leaving group – in the R form, it is directed out of the active site in much the same orientation as leaving groups in models of tetrahedral intermediates from classical substrates (16,17), while in the S form it is directed into the protein. The lower energy, and more structurally intuitively appealing, R form is shown in Figure 6B. As mentioned above, the oxyanion appears to be stabilized by a hydrogen bond to the Tyr 150 hydroxyl group. It is noteworthy in this regard, that in the crystal structure of the inhibited enzyme, the side chain of Tyr 150 has moved to a position where it can donate a hydrogen bond to the oxygen of the cross-linking carbonyl group (11). In the tetrahedral intermediate model, the hydroxamate leaving group does not appear to interact with any particular active site residue except perhaps with Thr 316, which may donate a hydrogen bond through its side chain to the hydroxamate carbonyl oxygen. The hydroxamate would be a good leaving group, however, and may not require general acid catalysis. Notably, no hydrogen bond acceptor/potential general base lies in contact with the Lys 315 side chain nitrogen, which is cationic in this model. This, however, is in accord with the likely first tetrahedral intermediate/transition state in the aminolysis of an acyl derivative with a good leaving group, where nucleophilic attack is thought to be rate-determining and no general base catalysis is observed (41).
Figure 6.
Stereoviews of energy-minimized tetrahedral intermediate structures formed on reaction of the P99 β-lactamase with 1. A. During formation of the acyl-enzyme. B. During aminolysis of the acyl-enzyme by Lys 315, the crosslinking reaction. Only heavy atoms are shown.

A model of the acyl-enzyme 25 was then constructed by elimination of Lys 315 from 24. A number of starting hydroxamate conformations were examined by molecular dynamics. It was clear that the acyl enzyme is very mobile, with the reactive carbonyl moving between two extremes, one where it could be hydrogen-bonded to Tyr 150 and Lys 67, where it would presumably be susceptible to attack by Lys 315 (see above), and the other where it was positioned in the oxyanion hole, where it would not be susceptible to attack by Lys 315, for steric reasons, but presumably would be susceptible to specific attack by water and D-phenylalanine (see above) in the same way as classical substrates. The latter orientation would then lead to free enzyme by the k2 and k4 branches of Scheme 3. Noteworthy, however, is the absence of interactions between the hydroxamate side chain and the enzyme in 25 (not shown), an observation that possibly relates to the limited flexibility of 1, discussed above.
It is interesting that nucleophilic attack by D-phenylalanine on the acyl-enzyme generates the aminolysis product 18 rather than the presumably more inert acyl enzyme 26. Departure of serine from 27 must be sufficiently facilitated by the Tyr 150/Lys 67 couple to overcome the innate leaving group ability of the hydroxamate. The hydroxamate can act as a leaving group in the initial acylation, however, presumably if appropriately placed in the tetrahedral intermediate, as shown by the activity of 12 as an inhibitor.
Finally, a model of the initial acylation tetrahedral intermediate 28 was constructed. A large range of conformations was possible. On the basis of the experimental evidence presented above, conformations where the oxyanion occupied the oxyanion hole were excluded as starting points in favor of conformations where, as in the models of 24, the oxyanion was placed in the vicinity of the Tyr 150 and Lys 67 side chains. As with 24, both R and S configurations were possible at the tetrahedral atom. The R species contained the phenoxide leaving group directed out of the active site and the hydroxamate in the classical acyl side chain site, adjacent and parallel to the β-2 strand; the S species contained the converse disposition of phenoxide and hydroxamate. The lowest energy (Eint) structure found from several molecular dynamics runs on both the R and S species was one of the intuitively more appealing R structures and is shown in Figure 6A. In this structure, the oxyanion is stabilized by direct interaction with Tyr 150, Lys 67 and Lys 315. The only close interaction with the phenoxide and hydroxamate leaving groups is a possible hydrogen bond between the hydroxamate carbonyl and the amido nitrogen of the side chain of Asn 152. Again there is the suggestion that with two good leaving groups, only stabilization of the tetrahedral oxyanion intermediate is necessary.
Preliminary experiments with other relevant enzymes were also conducted. These showed that 1,3 – 14 also inactivated the class A TEM-2 and class D OXA-1 β-lactamases although less effectively (k1 ≤ 100 s−1M−1) than class C enzymes. The Actinomadura R39 DD-peptidase was inhibited at similar low rates although, notably, the Streptomyces R61 DD-peptidase was not inactivated; the latter enzyme has a histidine residue in place of Lys 315 of the class C β-lactamases (42). Compound 1 was a poor substrate of α-chymotrypsin (Km ≥ 1 mM, kcat/Km = 18 s−1M−1).
The results of experiments presented in this paper demonstrate that O-aryloxycarbonyl hydroxamates are quite effective covalent inhibitors of a class C β-lactamase. Structural evidence shows that this is achieved by covalent cross-linking of the active site, between the nucleophilic serine and the lysine of the KTG motif (11). Structure-reactivity studies of the new series of compounds strongly suggest that they react with the active site in a different orientation than normal substrates and substrate/transition state analogue inhibitors, at least as currently believed. A new way for the active site to stabilize transition states opens the door to a new class of inhibitors, a goal that we will pursue.
Supplementary Material
Details of the syntheses of O-aryloxycarbonyl hydroxamates 3 – 13. This material is available free of charge via the Internet at http://pubs.acs.org.
Footnotes
This research was supported by National Institutes of Health, Grant R01 AI-17986 Received Date
Abbreviations: AMPSO, 3-[(1,1-dimethyl-2-hydroxyethyl)amino]-2-hydroxypropanesulfonic acid; BSA, bovine serum albumin; DMSO, dimethyl sulfoxide; ESMS, electrospray mass spectrometry; IR, infra-red; MES, 2-(N-morpholino)-ethanesulfonic acid; MOPS, 3-(N-morpholino)propanesulfonic acid; NMR, nuclear magnetic resonance.
References
- 1.Abraham EP. The beta-lactam antibiotics. Sci Am. 1981;244:76–86. doi: 10.1038/scientificamerican0681-76. [DOI] [PubMed] [Google Scholar]
- 2.Jacoby G, Bush K. β-Lactam resistance in the 21st century. In: White DG, Alekshun MN, McDermott PF, editors. Frontiers in Antimicrobial Resistance. Am. Soc. Microbiol; Washington, D.C: 2005. pp. 53–65. [Google Scholar]
- 3.Fisher JF, Meroueh SA, Mobashery S. Bacterial resistance to β-lactam antibiotics: compelling opportunism, compelling opportunity. Chem Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 4.Buynak JD. Understanding the longevity of the β-lactam antibiotics and of antibiotic/β-lactamase inhibitor combinations. Biochem Pharmacol. 2006;71:930–940. doi: 10.1016/j.bcp.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Babic M, Hujer AM, Bonomo RA. What’s new in antibiotic resistance? Focus on beta-lactamases. Drug Resist Updates. 2006;9:142–156. doi: 10.1016/j.drup.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Crompton IE, Cuthbert BK, Lowe G, Waley SG. β-Lactamase inhibitors. The inhibition of serine β-lactamases by specific boronic acids. Biochem J. 1988;251:453–459. doi: 10.1042/bj2510453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin R, Jones JB. Rational design and synthesis of a highly effective transition state analog inhibitor of the RTEM-1 β-lactamase. Tetrahedron Lett. 1995;46:8399–8402. [Google Scholar]
- 8.Weston GS, Blazquez J, Baquero F, Shoichet BK. Structure-based enhancement of boronic acid-based inhibitors of AmpC β-lactamase. J Med Chem. 1998;41:4577–4586. doi: 10.1021/jm980343w. [DOI] [PubMed] [Google Scholar]
- 9.Pratt RF. Inhibition of a class C β-lactamase by a specific phosphonate monoester. Science. 1989;246:917–919. doi: 10.1126/science.2814513. [DOI] [PubMed] [Google Scholar]
- 10.Rahil J, Pratt RF. Mechanism of inhibition of the class C β-lactamase of Enterobacter cloacae P99 by phosphonate monoesters. Biochemistry. 1992;31:5869–5878. doi: 10.1021/bi00140a024. [DOI] [PubMed] [Google Scholar]
- 11.Wyrembak PN, Babaoglu K, Pelto RB, Shoichet BK, Pratt RF. O-Aryloxycarbonyl hydroxamates: new β-lactamase inhibitors that cross-link the active site. J Am Chem Soc. 2007;129:9548–9549. doi: 10.1021/ja072370u. [DOI] [PubMed] [Google Scholar]
- 12.Nagel M, Hansen HJ. Synthesis of polyalkylphenyl prop-2-ynoates and their flash vacuum pyrolysis to polyalkylcyclohepta[b]furan-2(2H)-ones. Helv Chim Acta. 2000;83:1022–1048. [Google Scholar]
- 13.Defoin A, Pires J, Streith J. From 1-(silyloxy)butadiene to 4-amino-4-deoxy-DL-erythrose and to 1-amino-1-deoxy-DL-erythritol derivatives. Hetero-Diels-Alder reactions with acylnitroso dienophiles. Helv Chim Acta. 1991;74:1653–1670. [Google Scholar]
- 14.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 15.Curley K, Pratt RF. Effectiveness of tetrahedral adducts as transition-state analogs and inhibitors of the class C β-lactamase of Enterobacter cloacae P99. J Am Chem Soc. 1997;119:1529–1538. [Google Scholar]
- 16.Ahn YM, Pratt RF. Kinetic and structural consequences of the leaving group in substrates of a class C β-lactamase. Bioorg Med Chem. 2004;12:1537–1542. doi: 10.1016/j.bmc.2003.12.042. [DOI] [PubMed] [Google Scholar]
- 17.Adediran SA, Zhang Z, Nukaga M, Palzkill T, Pratt RF. The D-methyl group in β-lactamase evolution: Evidence from the Y221G and GC1 mutants of the class C β-lactamase of Enterobacter cloacae P99. Biochemistry. 2005;44:7543–7552. doi: 10.1021/bi050136f. [DOI] [PubMed] [Google Scholar]
- 18.Beadle BM, Trehan I, Focia PJ, Shoichet BK. Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC β-lactamase. Structure. 2002;10:413–424. doi: 10.1016/s0969-2126(02)00725-6. [DOI] [PubMed] [Google Scholar]
- 19.Jencks WP, Gilchrist M. Non-linear structure-activity correlations. The reactivity of nucleophilic reagents towards esters. J Am Chem Soc. 1968;90:2622–2637. [Google Scholar]
- 20.Powers RA, Shoichet BK. Structure-based approach for binding site identification on AmpC β-lactamase. J Med Chem. 2002;45:3222–3234. doi: 10.1021/jm020002p. [DOI] [PubMed] [Google Scholar]
- 21.Kaur K, Pratt RF. Mechanism of reaction of acyl phosph(on)ates with the β-lactamase of Enterobacter cloacae P99. Biochemistry. 2001;40:4610–4621. doi: 10.1021/bi002243+. [DOI] [PubMed] [Google Scholar]
- 22.Schraml J, Sýkora J, Fiedler P, Roithová J, Mindl I, Blechta V, Císa3ová I, Exner O. N,O-diacylhydroxylamines – structures in crystals and solutions. Org Biomol Chem. 2004;2:2311–2314. doi: 10.1039/B403728F. [DOI] [PubMed] [Google Scholar]
- 23.Exner O, Simon W. Acyl derivatives of hydroxylamine. XII Dissociation constants of hydroxamic acids and their functional derivatives. Coll Czech Chem Commun. 1965;30:4078–4093. [Google Scholar]
- 24.Cabaret D, Garcia Gonzalez M, Wakselman M, Adediran SA, Pratt RF. Synthesis, hydrolysis, and evaluation of 3-acylamino-3,4-dihydro-2-oxo-2H-1,3-benzoxazinecarboxylic acids and linear azadepsipeptides as potential substrates/inhibitors of β-lactam-recognizing enzymes. Eur J Org Chem. 2001:141–149. [Google Scholar]
- 25.Page MI, Vilanova B, Layland NJ. pH dependence of and kinetic solvent isotope effects on the methanolysis and hydrolysis of β-lactams catalyzed by class C β-lactamase. J Am Chem Soc. 1995;117:12092–12095. [Google Scholar]
- 26.Adediran SA, Pratt RF. β-Secondary and solvent deuterium kinetic isotope effects on catalysis by the Streptomyces R61 DD-peptidase: comparisons with a structurally similar class C β-lactamase. Biochemistry. 1999;38:1469–1477. doi: 10.1021/bi982308x. [DOI] [PubMed] [Google Scholar]
- 27.Pazhanisamy S, Govardhan CP, Pratt RF. β-Lactamase-catalyzed aminolysis of depsipeptides: amine specificity and steady state kinetics. Biochemistry. 1989;28:6863–6870. doi: 10.1021/bi00443a013. [DOI] [PubMed] [Google Scholar]
- 28.Oefner C, D’Arcy A, Daly JJ, Gubernator K, Charnas RL, Heinze I, Hubschwerlen C, Winkler FK. Refined crystal structure of β-lactamase from Citrobacter freundii indicates a mechanism for β-lactam hydrolysis. Nature. 1990;343:284–288. doi: 10.1038/343284a0. [DOI] [PubMed] [Google Scholar]
- 29.Strynadka NCJ, Adachi H, Jensen SE, Johns K, Sielecki A, Betzel C, Sutoh K, James MNG. Molecular structure of the acyl-enzyme intermediate in β-lactam hydrolysis at 1.7 Å resolution. Nature. 1992;359:700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 30.Lobkovsky E, Billings EM, Moews PC, Rahil J, Pratt RF, Knox JR. Crystallographic structure of a phosphonate derivative of the Enterobacter cloacae P99 cephalosporinase: mechanistic interpretation of a β-lactamase transition state analog. Biochemistry. 1994;33:6762–6772. doi: 10.1021/bi00188a004. [DOI] [PubMed] [Google Scholar]
- 31.Hess S, Martin R, Kindler AM, Paetzel M, Gold M, Jensen SE, Jones JB, Strynadka NCJ. Structure-based design guides the improved efficacy of deacylation transition state analogue inhibitors of TEM-1 β-lactamase. Biochemistry. 2000;39:5312–5321. doi: 10.1021/bi992505b. [DOI] [PubMed] [Google Scholar]
- 32.Leigh T. N-Alkyl derivatives of penicillin V. J Chem Soc. 1965:3616–3619. doi: 10.1039/jr9650003616. [DOI] [PubMed] [Google Scholar]
- 33.Herzberg P, Moult J. Bacterial resistance to β-lactam antibiotics: Crystal structure of β-lactamase from Staphylococcus aureus PC1 at 2.5 Å resolution. Science. 1987;236:694–701. doi: 10.1126/science.3107125. [DOI] [PubMed] [Google Scholar]
- 34.Murphy BP, Pratt RF. Evidence for an oxyanion hole in serine β-lactamases and DD-peptidases. Biochem J. 1988;256:669–672. doi: 10.1042/bj2560669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curley K, Pratt RF. The oxyanion hole in serine β-lactamase catalysis: interactions of thiono substrates with the active site. Bioorg Chem. 2000;28:338–356. doi: 10.1006/bioo.2000.1184. [DOI] [PubMed] [Google Scholar]
- 36.Smith SG, O’Leary M. The kinetics of the acidic and alkaline hydrolysis of ethyl thionbenzoate. J Org Chem. 1963;28:2825–2828. [Google Scholar]
- 37.Reynaud P, Moreau RC, Samana J-P. Thioamides IV. Réaction des thioesters de O-ethyle avec les amines aliphatiques et aromatiques. Préparation de thioamides mono et di-substitués. Bull Soc Chim Fr. 1965:3623–3628. [Google Scholar]
- 38.Dexter DD, van der Veen JM. Conformations of penicillin G: crystal structure of procaine penicillin G monohydrate and a refinement of the structure of potassium penicillin G. J Chem Soc Perkin Trans I. 1978:185–190. doi: 10.1039/p19780000185. [DOI] [PubMed] [Google Scholar]
- 39.Montanari B, Ballone P, Jones RO. Density functional study of crystalline analogs of polycarbonates. Macromolecules. 1998;31:7784–7790. [Google Scholar]
- 40.Murphy BP, Pratt RF. N-(Phenylacetyl)glycyl-D-aziridine-2-carboxylate, an acyclic amide substrate of β-lactamases: The importance of shape of the active site in β-lactamase evolution. Biochemistry. 1991;30:3640–3649. doi: 10.1021/bi00229a008. [DOI] [PubMed] [Google Scholar]
- 41.Adediran SA, Kumar I, Pratt RF. Deacylation transition states of a bacterial DD-peptidase. Biochemistry. 2006;45:13074–13082. doi: 10.1021/bi061341d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knox JR, Moews PC, Frère JM. Molecular evolution of bacterial β-lactam resistance. Chem Biol. 1996;3:937–947. doi: 10.1016/s1074-5521(96)90182-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of the syntheses of O-aryloxycarbonyl hydroxamates 3 – 13. This material is available free of charge via the Internet at http://pubs.acs.org.