SUMMARY
The breast cancer susceptibility protein, BRCA2, is essential for recombinational DNA repair. BRCA2 delivers RAD51 to double-stranded DNA (dsDNA) breaks through interaction with eight conserved, ~35 amino acid motifs, the BRC repeats. Here we show that the solitary BRC4 promotes assembly of RAD51 onto single-stranded DNA (ssDNA), but not dsDNA, to stimulate DNA strand exchange. BRC4 acts by blocking ATP hydrolysis and thereby maintaining the active ATP-bound form of the RAD51-ssDNA filament. Single-molecule visualization shows that BRC4 does not disassemble RAD51-dsDNA filaments, but rather blocks nucleation of RAD51 onto dsDNA. Furthermore, this behavior is manifest by a domain of BRCA2 comprising all eight BRC repeats. These results establish that the BRC repeats modulate RAD51-DNA interaction in two opposing, but functionally reinforcing ways: targeting active RAD51 to ssDNA and prohibiting RAD51 nucleation onto dsDNA. Thus, BRCA2 recruits RAD51 to DNA breaks and, we propose, the BRC repeats regulate DNA binding selectivity.
INTRODUCTION
The breast cancer susceptibility gene, BRCA2, encodes a protein of 3,418 amino acids (Wooster et al., 1995). BRCA2 is needed for DNA repair by homologous recombination (Moynahan et al., 2001; Tutt et al., 2001), the predominant mechanism employed by cells to accurately repair double-strand breaks (DBSs). The inability to purify full-length BRCA2 protein has hampered analysis of its role in this essential biological process. However, crystallographic studies revealed that BRCA2 possesses both a DNA Binding Domain (DBD), with unique structural features for binding ssDNA and dsDNA (Yang et al., 2002), and BRC repeats, which bind to the core of RAD51 by mimicking the structure of an adjacent RAD51 monomer (Pellegrini et al., 2002). Electron microscopy uncovered two different loci for binding to RAD51, the N-terminal domain and the nucleotide binding core (Galkin et al., 2005). In addition, another RAD51 binding site was mapped to the C-terminus of BRCA2 (Esashi et al., 2007). Together, these results suggest that the intact BRCA2 protein might bind both DNA and RAD51, and deliver RAD51 to the sites of DNA breaks (Kowalczykowski, 2005; Pellegrini et al., 2002; Shin et al., 2003; Yang et al., 2002).
The importance of the BRC motifs to BRCA2 function in the maintenance of genome integrity is supported by observations showing that point mutations within BRC which compromise interactions with RAD51 are found in individuals predisposed to breast cancer (Breast Cancer Information Core, http://research.nhgri.nih.gov/bic/). At least one BRC repeat, in concert with the DBD of BRCA2, is required for targeting RAD51 to ssDNA (San Filippo et al., 2006). Moreover, a fusion of a BRC repeat with the large subunit of the human ssDNA-binding protein, Replication protein-A (RPA), is sufficient to suppress cellular defects found in brca2 mutant mammalian cells (Saeki et al., 2006). In fact, exogenous expression of mutants deleted for the BRC repeats or with missense mutations that give rise to familial cancers eliminate the ability of BRCA2 to restore resistance to methyl methanesulphonate (MMS) (Chen et al., 1998). However, apparently contradictory effects reported for the BRC repeats on RAD51 behavior have confounded molecular understanding of their function. For instance, BRC repeats are reported to both block assembly of the RAD51 nucleoprotein filament on dsDNA and to paradoxically stabilize the same nucleoprotein filament (Chen et al., 1999; Davies et al., 2001; Galkin et al., 2005; Shivji et al., 2006). These seemingly disparate observations may simply reflect experimental differences amongst the various studies or, instead, they may suggest an unappreciated complexity in the mechanism by which the BRC repeats regulate RAD51 function. Here we use ensemble and single-molecule methods to examine the effects of the BRC4 repeat on RAD51 nucleoprotein filament assembly on both ssDNA and dsDNA. We uncover a dichotomy with regard to the effect of BRC4 and a BRCA2 domain including the eight repeats, BRCA2BRC1-8, on these two types of DNA: BRC4 and BRCA2BRC1-8 stabilize filament formation on ssDNA, but prevent filament formation on dsDNA. The consequence of this control over DNA selection results in an enhancement of the DNA strand exchange activity of RAD51 protein by the BRC repeats that we propose is a function within the intact BRCA2 protein.
RESULTS
BRC4 and BRCA2BRC1-8 stimulate the formation of RAD51 nucleoprotein filaments on ssDNA
Because prior studies had not specifically focused on the role of BRC repeats in RAD51 nucleoprotein filament on ssDNA, we decided to first examine the effect of the well characterized BRC4 repeat (Pellegrini et al., 2002) on the assembly of RAD51 onto ssDNA. BRC4 was purified as a fusion with glutathione-S-transferase (GST); as controls, a GST-fused repeat with a deletion, Δ1524-30, and a GST-fused repeat with a point mutation, T1526A, (Figure S1) each of which fails to bind RAD51 (Chen et al., 1999; Davies et al., 2001) (confirmed using a GST-pull down assays; data not shown) were also purified. For the ssDNA, to avoid the ambiguity introduced by dsDNA that results from secondary structure forming within native ssDNA, the homopolymer, dT40, was initially used. BRC4 was incubated with RAD51, ssDNA added, and then the complexes were analyzed using electrophoretic mobility shift assays (EMSA). As expected, BRC4 alone does not bind the ssDNA (Figure 1A, lane 1). In the presence of ATP, the RAD51 nucleoprotein filaments are partially stable and only ~30% of the DNA is shifted (Figure 1A, lane 2). The addition of BRC4 unexpectedly produced a BRC4 concentration-dependent increase in RAD51-ssDNA complexes from 30% to almost 100% (lanes 3-8), whereas the control peptides, Δ1524-30 (lanes 9 and 10, and Figure 1B) or T1526A (Figure 1B), had no effect. Maximum stimulation occurred at about 12 μM of peptide, and then declined at higher concentrations of BRC4 (Figure 1A, B, C).
Figure 1. Stimulation of RAD51-ssDNA complex formation by BRC4 and BRCA2BRC1-8.
(A) Gel showing RAD51 (3 μM) incubated with either GST-BRC4 or GST-Δ1524-30 prior to incubation with 32P-labeled dT40 ssDNA (0.3 μM) for 1 h in the presence of ATP, Mg2+ and Ca2+ (B) Quantification of protein-DNA complexes with GST-BRC4 (filled circles) or GST-Δ1524-30 (open circles) obtained as in (A), using: ATP, Mg2+ + Ca2+ (●); ATP and Mg2+ ( ); ADP, Mg2+ + Ca2+ (
); ADP, Mg2+ + Ca2+ ( ), ATP and Ca2+ (
), ATP and Ca2+ ( ); AMP-PNP, Mg2+ + Ca2+ (
); AMP-PNP, Mg2+ + Ca2+ ( ); or no nucleotide cofactor, Mg2+ + Ca2+ (
); or no nucleotide cofactor, Mg2+ + Ca2+ ( ). Data obtained with the control mutant, GST-T1526A, ATP, Mg2+ + Ca2+: (
). Data obtained with the control mutant, GST-T1526A, ATP, Mg2+ + Ca2+: ( ). (C) BRC4 forms a stoichiometric complex with RAD51-ssDNA complexes. RAD51 (0.15 μM) was incubated with BRC4 (
). (C) BRC4 forms a stoichiometric complex with RAD51-ssDNA complexes. RAD51 (0.15 μM) was incubated with BRC4 ( ) or BRCA2BRC1-8 (
) or BRCA2BRC1-8 ( ), or RAD51 (0.3 μM) was incubated with BRC4 (●), prior to incubation with 32P-labeled dT40 ssDNA (15 nM) for another 15 min in the presence of ATP, Mg2+ and Ca2+. Due to limitations of the protein stock, higher BRCA2BRC1-8 concentrations could not be examined. (D) BRC4 increases the stability of RAD51-ssDNA complexes to increasing concentrations salt: complexes were formed with RAD51 (0.9 μM) alone (upper panel) or RAD51 (0.9 μM) and GST-BRC4 (3.6 μM) (lower panel), in the presence of ADP and Mg2+.
), or RAD51 (0.3 μM) was incubated with BRC4 (●), prior to incubation with 32P-labeled dT40 ssDNA (15 nM) for another 15 min in the presence of ATP, Mg2+ and Ca2+. Due to limitations of the protein stock, higher BRCA2BRC1-8 concentrations could not be examined. (D) BRC4 increases the stability of RAD51-ssDNA complexes to increasing concentrations salt: complexes were formed with RAD51 (0.9 μM) alone (upper panel) or RAD51 (0.9 μM) and GST-BRC4 (3.6 μM) (lower panel), in the presence of ADP and Mg2+.
The EMSA in Figure 1A was obtained using a mixture of Ca2+ and Mg2+, which resulted in better defined RAD51-ssDNA complexes due to their greater kinetic stability imparted by the Ca2+ (Bugreev and Mazin, 2004). However, even when Ca2+ was omitted, BRC4 stabilized the RAD51 nucleoprotein filament (Figure 1B, “ATP (Mg2+)”). BRC4 also increased RAD51-ssDNA complex formation when either dT60 or dT70 were used (data not shown). When an oligonucleotide with a random 26 nucleotide sequence was used, nucleoprotein complex formation was stimulated by BRC4 from 3% to 15%; the use of longer mixed sequence DNA resulted in complex formation with RAD51 that involved more than 75% of the DNA, precluding detection of stimulation by BRC4 (data not shown). The presence of the GST tag did not affect the results, as an untagged BRC4 peptide also stimulated binding (Figure S2). When the concentrations of RAD51 were reduced (10- and 20-fold), both the profile and optimum for stimulation by BRC4 were proportionally shifted (Figure 1C), indicating that the stimulatory effect of BRC4 is the consequence of forming a defined complex with RAD51 and not a non-specific effect (Davies et al., 2001; Galkin et al., 2005); at these lower concentrations, the binding of BRC4 to RAD51 is not stoichiometric, requiring excess amounts of BRC4 to drive binding to RAD51. To confirm that these effects were not unique to the BRC4 repeat, a domain corresponding to all eight of the BRC repeats, BRCA2BRC1-8, was examined (Shivji et al., 2006). A similar profile of stimulation was observed with BRCA2BRC1-8; due to limitations of the protein stock, higher BRCA2BRC1-8 concentrations could not be examined (Figure 1C). This finding shows that the BRC4 repeat and BRCA2BRC1-8 behave similarly.
Interestingly, either when Ca2+ replaced the Mg2+ to prevent the hydrolysis of ATP (Bugreev and Mazin, 2004), or when the non-hydrolysable analogue, AMP-PNP, was used, BRC4 did not stimulate the binding of RAD51 to ssDNA but rather inhibited filament formation (Figure 1B). These results suggest that the stabilization of nucleoprotein filaments by BRC4 requires ATP hydrolysis by the nucleoprotein filament. Finally, BRC4 appeared to have little or no effect when ATP was replaced by ADP (Figure 1B) but, because the ADP-complex is more stable than the ATP-bound form (Kim et al., 2002) (and unpublished observations), this increased stability precluded detection of a stabilizing effect. Therefore, to measure the effect of BRC4 on ADP-RAD51-ssDNA complexes, we examined the dissociation of these complexes due to elevated salt concentrations. Figure 1D shows that BRC4 increased the stability of the ADP-RAD51-ssDNA complexes against dissociation at high NaCl concentrations. Thus, BRC4 stabilizes RAD51-ssDNA filaments only when hydrolysis of ATP to ADP is permitted, or when ADP is present initially.
BRC4 prevents ssDNA dependent ATP hydrolysis by RAD51, thereby maintaining the active ATP-bound form of the filament
To understand the mechanism by which BRC4 stabilizes RAD51-ssDNA complexes, we explored the effect of BRC4 on the ssDNA-dependent ATPase activity of RAD51. BRC4 reduced ATP hydrolysis by RAD51 for all the ssDNA examined, both homopolymers and natural DNA (Figure 2A). In the absence of BRC4, the rate of ATP hydrolysis was fastest for the shorter ssDNA due, likely, to the more dynamic nature of the filaments on shorter DNA. In all cases, BRC4 reduced the hydrolysis rate to the DNA-independent rate, while the control peptide had no effect (Figure 2A). This inhibition of ATP hydrolysis was not due to dissociation of the nucleoprotein filaments, since BRC4 stabilized the complexes under these conditions (Figure 1). Finally, BRC4 also inhibited similarly the ATPase activity of RAD51 when RPA was present (Figure S3), showing that the effects of BRC4 are not altered by RPA.
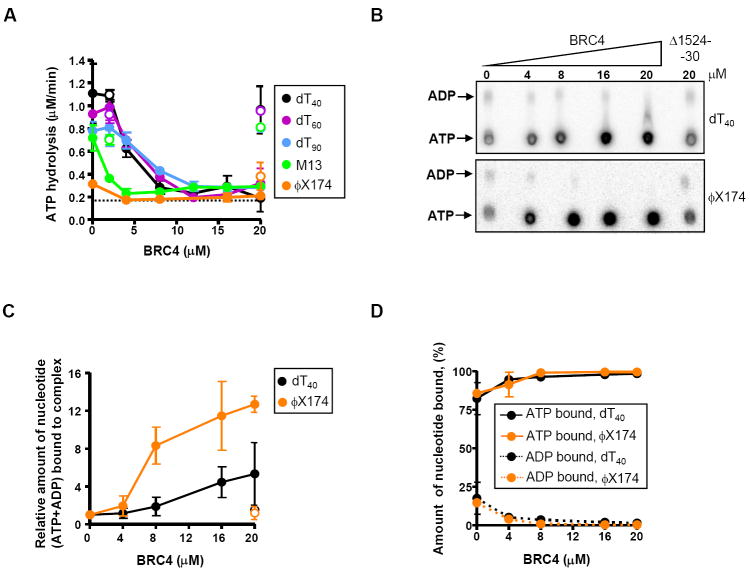
Figure 2. BRC4 inhibits ATP hydrolysis and permits accumulation of ATP-bound RAD51-ssDNA filaments.
(A) BRC4 reduces the rate of ATP hydrolysis by RAD51. RAD51 (3 μM) was incubated with increasing concentrations of GST-BRC4 (filled circles) or the control peptide GST-Δ1524-30 (open circles), as indicated, prior addition of dT40 (●), dT60 ( ), dT90 (
), dT90 ( ), M13 ssDNA (
), M13 ssDNA ( ), or ϕX174 ssDNA (
), or ϕX174 ssDNA ( ), and further incubated for 1h in the presence of 0.5 mM ATP and 4 mM Mg2+. The dashed line represents the DNA-independent rate of ATP hydrolysis by RAD51 (~0.2 μM/min). (B) Nucleotides bound to purified nucleoprotein filaments as a function of BRC4 concentration as measured by TLC. (C) Amount of total nucleotide (ATP+ADP) bound to the RAD51-ssDNA complex formed with either dT40 (black circles) or ϕX174 ssDNA (orange circles) in presence of GST-BRC4 (filled circles) or GST-Δ1524-30 (open circles), relative to the amount in the absence of BRC4. (D) Percentages of ATP (solid lines) and ADP (dashed lines) in the RAD51 nucleoproteins filaments formed on dT40 (●) or ϕX174 ssDNA (
), and further incubated for 1h in the presence of 0.5 mM ATP and 4 mM Mg2+. The dashed line represents the DNA-independent rate of ATP hydrolysis by RAD51 (~0.2 μM/min). (B) Nucleotides bound to purified nucleoprotein filaments as a function of BRC4 concentration as measured by TLC. (C) Amount of total nucleotide (ATP+ADP) bound to the RAD51-ssDNA complex formed with either dT40 (black circles) or ϕX174 ssDNA (orange circles) in presence of GST-BRC4 (filled circles) or GST-Δ1524-30 (open circles), relative to the amount in the absence of BRC4. (D) Percentages of ATP (solid lines) and ADP (dashed lines) in the RAD51 nucleoproteins filaments formed on dT40 (●) or ϕX174 ssDNA ( ).
).
Because BRC4 could prevent ATP hydrolysis at a number of different steps in the ATP hydrolysis cycle (e.g., by blocking ATP binding, ADP dissociation, or the catalytic step), we sought to determine which nucleotide was bound to the BRC4-inhibited filament. To determine whether BRC4 affected the proportion of ATP and ADP bound to the RAD51 nucleoprotein, we used a modified version of an assay described previously (Bugreev and Mazin, 2004). Nucleoprotein filaments were formed in the presence of [α-32P] ATP, isolated from free nucleotides, and then the nucleotide (ATP/ADP) composition of the filament was measured (Figure 2B, C and D). BRC4 reduced the fraction of ADP bound, and concomitantly increased the amount of ATP bound to nucleoprotein filaments formed on either dT40 (Figure 2B, upper panel) or ϕX174 ssDNA (Figure 2B, lower panel). The relative increase in amount of nucleotide-bound (for both ATP and ADP) complexes (4-fold for dT40 and more than 10-fold for ϕX174 ssDNA) correlates with the stabilization of the ssDNA complexes shown in Figure 1. More significantly, in the presence of BRC4, the resulting filaments contain nearly 100% ATP (Figure 2D). These results provide direct evidence that BRC4 permits accumulation of the active, ATP-bound form of RAD51-ssDNA filament. Moreover, since BRC4 both stimulates the assembly of RAD51 onto ssDNA only when ATP hydrolysis is occurring (Figure 1B) and stabilizes the ADP-RAD51-ssDNA complex, these findings strongly suggest that BRC4 acts by preventing dissociation of ADP-RAD51 from ssDNA, permitting exchange of ATP for ADP within the filament, and finally blocking ATP hydrolysis. The consequence of these events is formation and preservation of the active ATP-RAD51-ssDNA nucleoprotein filament.
BRC4 and BRCA2BRC1-8 prevent the formation of RAD51 nucleoprotein filaments on dsDNA
Given that RAD51 can bind both ssDNA and dsDNA (Baumann et al., 1996; Mazin et al., 2000), but only the ssDNA nucleoprotein filament is functional (Sung and Robberson, 1995), we next investigated the effect of BRC4 on the binding of RAD51 to dsDNA. RAD51 was incubated with increasing concentrations of BRC4 prior to addition of 32P-labeled ϕX174 linear dsDNA (Figure 3). As expected, BRC4 alone did not bind dsDNA (Figure 3A, lane 1). However, in the presence of ATP, increasing concentrations of BRC4 inhibited binding of RAD51 to dsDNA, which is opposite to the behavior seen with ssDNA. The control peptides, Δ1524-30 and T1526A, did not prevent RAD51-dsDNA filament formation (Figure 3A, lanes 9 and 10, and Figure 3B). As with ssDNA binding, the GST tag did not affect the results, since an untagged BRC4 peptide had the same effect (Figure S4A). The same outcome was obtained when ϕX174 dsDNA was substituted with a shorter duplex DNA, dT40·dA40 (Figure 3C), and when BRCA2BRC1-8 was employed instead of BRC4 (Figure 3D). When Ca2+ was substituted for Mg2+, BRC4 still inhibited binding of RAD51 to dsDNA (Figure S4B). But when AMP-PNP was used instead of ATP, BRC4 did not inhibit RAD51 nucleoprotein filament formation (Figure 3B). The observation that BRC4 does not block RAD51-dsDNA binding in the presence of AMP-PNP but does so in the presence of ATP and Ca2+ suggests that there is a difference in the structural conformation adopted by the BRC4-RAD51 filament that is dependent on the type of nucleotide cofactor and DNA. Previously, the amino-terminal domain of RAD51 in dsDNA filaments was found to become dynamic when ATP was used instead of AMP-PNP (Galkin et al., 2005), and this structural flexibility suggests that BRC4 acts to destabilize the RAD51-dsDNA filament through a conformation state that is accessible only when the physiological cofactor ATP is present.
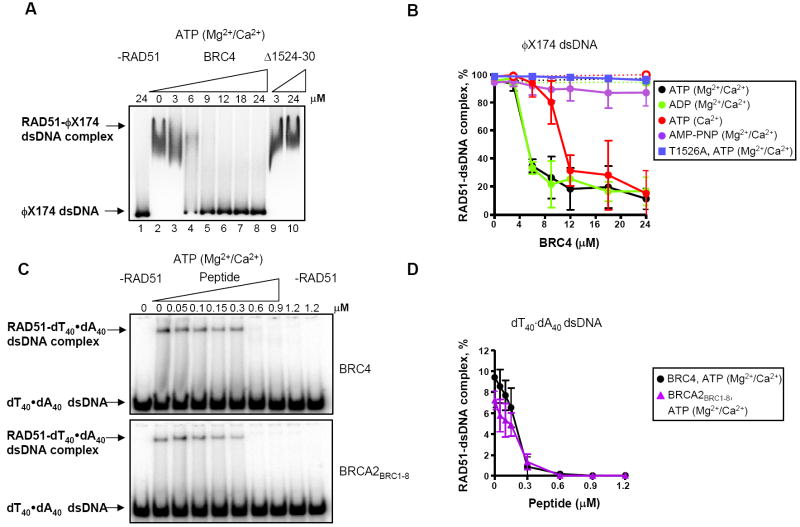
Figure 3. BRC4 and BRCA2BRC1-8 block RAD51-dsDNA complex formation.
(A) Autoradiograph of an agarose gel showing RAD51 (3 μM) incubated with either GST-BRC4 or GST-Δ1524-30 prior to incubation with 32P-labeled ϕX174 linear dsDNA (5 μM, nt) and further incubation for 1 h in the presence of ATP, Mg2+ and Ca2+. (B) Quantification of data as in (A): in the presence of ATP, Mg2+ + Ca2+ (●); ADP, Mg2+ + Ca2+ ( ); ATP and Ca2+ (
); ATP and Ca2+ ( ); or AMP-PNP, Mg2+ + Ca2+ (
); or AMP-PNP, Mg2+ + Ca2+ ( ). Filled circles and solid lines correspond to GST-BRC4; open circles and dashed lines correspond to GST-Δ1524-30. Data obtained with the control mutant, GST-T1526A, in the presence of ATP, Mg2+ + Ca2+ :(
). Filled circles and solid lines correspond to GST-BRC4; open circles and dashed lines correspond to GST-Δ1524-30. Data obtained with the control mutant, GST-T1526A, in the presence of ATP, Mg2+ + Ca2+ :( ). (C) The effect of BRC4 or BRCA2BRC1-8 on RAD51-dsDNA complex formation using dA40·dT40 dsDNA. RAD51 (0.15 μM) was incubated with GST-BRC4 or BRCA2BRC1-8, at the concentrations indicated prior to incubation with 32P-labeled dA40·dT40 dsDNA (2.4 μM, nt) and further incubation for 1 h in the presence of ATP, Mg2+ and Ca2+. Protein-DNA complexes were resolved in 6% PAGE and analyzed by autoradiography. (D) Data obtained as in (C), quantified and plotted: GST-BRC4, (●); BRCA2BRC1-8 (
). (C) The effect of BRC4 or BRCA2BRC1-8 on RAD51-dsDNA complex formation using dA40·dT40 dsDNA. RAD51 (0.15 μM) was incubated with GST-BRC4 or BRCA2BRC1-8, at the concentrations indicated prior to incubation with 32P-labeled dA40·dT40 dsDNA (2.4 μM, nt) and further incubation for 1 h in the presence of ATP, Mg2+ and Ca2+. Protein-DNA complexes were resolved in 6% PAGE and analyzed by autoradiography. (D) Data obtained as in (C), quantified and plotted: GST-BRC4, (●); BRCA2BRC1-8 ( ).
).
The BRC repeats prevent nucleation of RAD51 onto dsDNA but do not disrupt a RAD51-dsDNA filament
To understand the basis for the inhibition of RAD51-dsDNA complex formation by BRC4, we examined RAD51-dsDNA nucleoprotein filaments at the single-molecule level. A fluorescently labeled RAD51 protein (Hilario et al., in press) was used to visualize RAD51 filament formation on dsDNA following a strategy reported previously for RecA protein (Galletto et al., 2006). Briefly, a flow cell was used to generate three separate laminar flow channels (Figure 4A). In the capture channel (Step 1), a biotinylated bacteriophage λ DNA molecule, stained with the fluorescent intercalating dye, YOYO-1, and attached to a streptavidin-coated bead, was optically trapped. The bead-YOYO-1-DNA complex was then moved into the observation channel under conditions that dissociate the dye from DNA (Step 2). This dye-free DNA was subsequently moved to the protein channel containing the fluorescently modified RAD51, alone, with BRC4, or with Δ1524-30 (Step 3). After incubation for the specified times in the presence of ATP and Ca2+, the trapped DNA was moved into the observation channel (Step 4) where the bound fluorescent-RAD51 was visualized.
Figure 4. Single-molecule visualization reveals that BRC4 and BRCA2BRC1-8 inhibit nucleation of RAD51 on dsDNA.
(A) Schematic representation of the experiment. The bead-DNA-YOYO-1 complex is trapped (Step 1) and then washed to dissociate YOYO-1 (Step 2). The λ DNA is incubated in the protein-channel with fluorescently-labeled RAD51, 1 mM ATP, 2 mM Ca(OAc)2, and the indicated amount of peptide (Step 3). The resulting RAD51 filament is moved back to the observation channel for analysis (Step 4). (B) Temporal series of images showing RAD51 nucleoprotein filament assembly on one DNA molecule. From left to right: Filament formation by RAD51 alone, with GST-BRC4 and with GST-Δ1524-30, respectively, at 4:1 molar ratio (peptide:RAD51) or (C) 2:1 and 0.5:1 ratios using BRC4 or BRCA2BRC1-8. (D) Cluster appearance as a function of time on λ DNA after incubation with 250 nM RAD51 and 1 μM ( ), 0.5 μM (
), 0.5 μM ( ), 250 nM (
), 250 nM ( ), and 125 nM (
), and 125 nM ( ) GST-BRC4; or 0.5 μM (
) GST-BRC4; or 0.5 μM ( ) and 125 nM (
) and 125 nM ( ) BRCA2BRC1-8. The dashed lines represent the linear regressions of the data. (E) Rate of cluster formation vs. BRC4 or BRCA2BRC1-8 concentration; each time point is the average of 4–9 individual molecules analyzed as in D.
) BRCA2BRC1-8. The dashed lines represent the linear regressions of the data. (E) Rate of cluster formation vs. BRC4 or BRCA2BRC1-8 concentration; each time point is the average of 4–9 individual molecules analyzed as in D.
An example of a RAD51 filament formed by successively “dipping” a single dsDNA molecule in the protein channel for 20 or 30 seconds each is shown in Figure 4B, left panel: within 20 sec, RAD51 was seen along much of the dsDNA, and nucleoprotein filament formation was nearly complete in 60 sec. In marked contrast, a 4:1 molar ratio of BRC4 nearly eliminated nucleoprotein filament nucleation (Figure 4B, middle panel): a mean of 1.5 ±1.4 nucleation events were visible after 180 seconds (nucleation rate = 0.50 ± 0.46 min-1) (Figure 4D). At the same concentration, the control Δ1524-30 peptide had no effect (Figure 4B; right panel). These single-molecule results agree fully with our ensemble experiments (Figure S4B).
The RAD51 nucleation frequency was seen to depend on BRC4 concentration (Figure 4D): when reduced to 1:1 (BRC4:RAD51), nucleation was still low (1.7 ± 0.3 min-1), but upon further reduction to 0.5:1, the rate increased to 22 ± 3.8 min-1 (Figure 4C and D). At the two concentrations tested, 0.5:1 and 2:1 (BRCA2BRC1-8:RAD51), BRCA2BRC1-8 was found to affect the RAD51 nucleation rate comparably to BRC4 (Figure 4C and D), substantiating the results obtained with a single BRC repeat. In the absence of BRC4 or BRCA2BRC1-8, nucleation was too rapid to measure reliably. The effect of increasing peptide concentration on nucleus formation is summarized in Figure 4E; maximal inhibition of nucleation occurred at an approximately equimolar ratio of BRC4 or BRCA2BRC1-8 to RAD51.
We next examined whether BRC4 can disrupt preformed RAD51-dsDNA filaments. RAD51 nucleoprotein filaments were formed in the protein channel as was done for the assembly experiments, except Mg2+ was used instead of Ca2+ to permit ATP hydrolysis and subsequent dissociation. Once the filament was fully extended (~4 min in these conditions), the DNA was moved to the second channel containing both Mg2+ and ATP to commence disassembly. Within experimental error, when BRC4 was present in the second channel, the rate of disassembly of the pre-formed fluorescent RAD51-dsDNA filaments was unchanged (Figure 5A). The DNA length is proportional to the amount of RAD51 bound (Hilario et al., 2008); hence, measurement of the DNA length as a function of time permits quantification of the first order rate constant for dissociation. In the presence of BRC4, RAD51 disassembles at a rate of 0.14 (± 0.05) μm min-1; in its absence, the rate is 0.17 (± 0.09) μm min-1 (Figure 5B). These data, both the visual presence of fluorescent RAD51 and the nucleoprotein length, demonstrate that BRC4 does not accelerate disassembly of the RAD51-dsDNA filament.
Figure 5. BRC4 does not promote the disassembly of RAD51-dsDNA filaments.
(A) Temporal series of images showing disassembly of a RAD51 (250 nM) nucleoprotein filament with 2 mM Mg(OAc)2 and 1 mM ATP in all channels: left panel, RAD51; right panel, RAD51 with GST-BRC4 (1 μM). (B) Time-dependence of filament dissociation after incubation with RAD51 alone (black) or with GST-BRC4 (red). Each time point is the average of 3-4 individual molecules, and the lines represent fits of the data to a single exponential decay. The dotted line represents the starting length of the naked λ DNA.
BRC4 can stimulate RAD51-mediated DNA strand exchange
Together, the ensemble- and single-molecule experiments indicate that BRC4 stabilizes the ATP-RAD51-ssDNA filament, but blocks filament formation on dsDNA at the nucleation level. Therefore, we reasoned that BRC4 should also stimulate RAD51-mediated DNA strand exchange by both promoting filament assembly on ssDNA, and preventing inhibitory filament formation on the target dsDNA. To test this prediction, we examined DNA strand exchange activity in the presence of RPA (Figure 6A). Initially, optimal conditions employing a stoichiometric amount of RAD51 relative to ssDNA were used (Benson et al., 1994; Bugreev and Mazin, 2004; Sung and Robberson, 1995). As expected, at these conditions (Figure 6B left panel, and 6C), RAD51 promoted the formation of DNA strand exchange products, joint molecules and nicked circular dsDNA. At these conditions, BRC4 showed a negligible stimulation of product formation compared to the control Δ1524-30 peptide, although BRC4 did inhibit product formation at high concentrations (Figure S5A and B) due to inhibition of RAD51 binding to the ϕX174 ssDNA at these higher BRC4 concentrations (data not shown; see Figure 1C). However, it is well known that when the RAD51 concentration exceeds the stoichiometric optimum with regard to ssDNA, RAD51 self-inhibits DNA strand exchange in vitro by binding to the dsDNA target (Benson et al., 1994; Sung and Robberson, 1995). Indeed, increasing the RAD51 concentration to 6 μM reduced product formation (Figure S5C, first lane), and nearly completely blocked all product formation when increased further to 7.5 μM (Figure 6B, right panel, lanes labeled “0” peptide). As predicted from our binding studies, however, BRC4 restored product formation in a concentration-dependent manner (Figures 6B and C, and S5C and D). The effect was most dramatic at the highest RAD51 protein concentrations (Figure 6B and C) and, in fact, BRC4 increased the yield of the fully stranded-exchanged DNA product (the nicked circular dsDNA) to levels that were higher than even the optimal reaction (at 3.75 μM RAD51 (Figure 6 B and C); the control peptides, T1526A and Δ1524-30, had no effect in any reaction. Thus, these results reveal a previously unrecognized ability of BRC4 to stimulate RAD51-mediated DNA strand exchange.
Figure 6. BRC4 stimulates RAD51-mediated DNA strand exchange.
(A) Diagram of DNA strand exchange between circular ssDNA and homologous linear dsDNA to produce joint molecules (JM) and nicked circular dsDNA (NC). The asterisk shows the 32P-label on each strand. (B) Effect of GST-BRC4 or the control peptides GST-Δ1524-30 and GST-T1526A on DNA strand exchange between ϕX174 circular ssDNA and linear dsDNA promoted by RAD51 protein at 7.5 μM RAD51 (right panel); the uneven substrate bands resulted from a drying artifact. The left panel shows DNA strand exchange products at the optimal RAD51 concentration (3.75 μM). Size markers (kb) are shown in the left lane. (C) Quantification of the joint molecules (red) and nicked circular dsDNA (black) products using 3.75 μM RAD51 (inverted triangles); 7.5 μM RAD51 and GST-BRC4 (filled circles); 7.5 μM RAD51 and GST-Δ1524-30 (open circles); or 7.5 μM RAD51 and GST-T1526A (squares).
DISCUSSION
Taken together, our results show that both BRC4 alone and BRCA2BRC1-8 can modulate RAD51 filament formation in two mutually reinforcing ways: stimulating the assembly on ssDNA and preventing nucleation onto dsDNA. This duality with regard to control of RAD51 nucleoprotein filament formation is, in turn, responsible for the regulation of RAD51-promoted DNA strand exchange. Our results demonstrate that the interaction of BRC4 with RAD51 is sufficient to enhance RAD51 filament assembly on ssDNA (Figure 7A, step 1). This enhancement results from both an inhibition of ATP hydrolysis (Figure 7A, step 2) and a stabilization of the ADP-bound form of the nucleoprotein filament (Figure 7A, step 3). The combined consequence is to allow exchange for ATP within the filament rather than the dissociation of ADP-RAD51 from the ssDNA. The net effect is maintenance of a protein filament assembled on ssDNA in its ATP-bound form, which is the form that is required for DNA pairing function. In addition, BRC4 prevents a competing inhibitory reaction: namely, the nonfunctional assembly of RAD51 on dsDNA (Figure 7B, step 4). BRC4 achieves this function by blocking the rate-limiting nucleation of filaments on dsDNA. The similar results obtained with a domain comprising the eight BRC repeats, BRCA2BRC1-8, previously shown to stimulate RAD51-mediated DNA strand exchange (Shivji et al., 2006), support the conclusions obtained with the single BRC4 repeat, and they further suggest that the entire BRCA2BRC1-8 domain behaves as a regulator of DNA binding much as BRC4.
Figure 7. Model showing regulation of RAD51-DNA interactions by the BRC repeats.
(A) The BRC repeats promote formation of the active ATP-RAD51-ssDNA filament at several steps: (1) In the presence of Mg2+-ATP, the BRC repeats promote binding of RAD51 to ssDNA. (2) By decreasing the rate of ATP hydrolysis, they permit accumulation of the active ATP-bound form of the filament. (3) The BRC repeats also stabilize the ADP-RAD51-ssDNA complex, permitting exchange of ADP for ATP and further increasing accumulation of an ATP-bound nucleoprotein filament. This filament is the active species in DNA strand exchange. (B) The BRC repeats prevent assembly of the non-productive ATP-RAD51-dsDNA filament: (4) The BRC repeats block nucleation of RAD51 onto dsDNA. (C) Proposed function of the intact BRCA2 protein in DSB repair (see text for details). A break in dsDNA is resected to produce ssDNA. RPA binds to the ssDNA, hindering RAD51 filament assembly. BRCA2 binds to the dsDNA-ssDNA junction, and loads RAD51 onto the ssDNA but blocks assembly onto the dsDNA. RAD51 filament growth extends the BRCA2-stablized nucleus, to form an ATP-bound nucleoprotein filament capable of homologous DNA pairing. Blue spheres represent a RAD51 monomer, T represents ATP, and D represents ADP; RPA is in orange, BRCA2 is brown, and the BRC repeats are represented in purple.
Crystallographic analysis defined an interaction interface between BRC4 and RAD51 that involves the ATPase core of RAD51 and that mimics the intermonomer interactions within the filament (Pellegrini et al., 2002). It was found that the nucleotide-binding site of RAD51adopts a more closed conformation when bound to BRC4, and the authors suggested that this conformation might, in turn, alter ATP occupancy. This interpretation could provide a structural explanation for the inhibition of the ATPase activity of RAD51 by BRC4 and the resultant stabilization of RAD51- ssDNA filaments if the ATP binding pocket closed with ATP bound and BRC4 prevented the conformational changes needed for ATP hydrolysis.
Paradoxically, BRC repeats were also shown both to disrupt RAD51 nucleoprotein filament formation on dsDNA and to bind to these nucleoprotein filaments. This seemingly contradictory outcome is likely a consequence of the two different binding modes reported for the BRC repeats: a mode that binds RAD51 in a monomeric state and another that binds the nucleoprotein filament, respectively (Davies et al., 2001; Galkin et al., 2005; Pellegrini et al., 2002); a third interaction mode, described for the C-terminal region of BRCA2, reportedly involves oligomeric RAD51 binding (Davies and Pellegrini, 2007; Esashi et al., 2007). Some prior studies are seemingly contradictory to some of the observations reported here, but our studies readily rationalize those findings. We showed that BRC4 will not inhibit RAD51 filament formation on dsDNA, unless the RAD51 is allowed to dissociate via ATP hydrolysis; once dissociated, however, our studies also show that reassembly of the dissociated RAD51 protein will be blocked by BRC4, leading to an apparent disruption of the ATP-nucleoprotein filaments. For this reason, nucleoprotein filaments formed on dsDNA in the presence of the non-hydrolysable analog, AMP-PNP, are much less affected by BRC repeats, whereas those assembled in the presence of ATP are apparently disrupted; however, these latter complexes are not, in fact, disrupted by the BRC repeat but, instead, they dissociate normally and their reassembly is blocked on a steady-state kinetic basis. In addition, BRC4 was found to inhibit the binding of RAD51 to longer ssDNA (e.g., poly(dT)) (Davies and Pellegrini, 2007). However, here we discovered that there are two distinct regimes of BRC4 behavior with regard to RAD51 binding to ssDNA: stimulation at low concentrations of BRC4, and inhibition at high concentrations. Because RAD51 forms unstable complexes with short oligonucleotides, such as dT40 or dT60, we can detect both the stimulatory regime and the inhibitory regime (Figure 1). However, for longer ssDNA, the binding of RAD51 is stable (close to 100% binding); thus, the stimulatory behavior cannot be detected, and only the inhibitory behavior is seen. This dichotomous behavior resolves the apparent discrepancy with studies using ssDNA that forms stable complexes with RAD51.
It was recently reported that a peptide derived from the BRC repeat of BRCA2 can inhibit the binding of RAD51 to the fluorescently-modified ssDNA, poly(etheno-dA) (Nomme et al., 2008); in agreement, we observed a similar behavior with M13 etheno-ssDNA (unpublished observations). We conclude that the fluorescent modification of these ssDNA substrates precludes the stimulation of binding seen for BRC4 with the unmodified ssDNA. Our results show that these distinct modes of action may have disparate consequences which depend on the type of DNA, single- or double-stranded, involved.
When considered in conjunction with previous work which showed that BRC repeats are necessary to target RAD51 to ssDNA (San Filippo et al., 2006), our findings suggest that the dominant negative phenotype resulting from expression of the single BRC4 in vivo (Chen et al., 1999) arises either from the inability of an isolated repeat, without the DNA binding domains of BRCA2, to efficiently deliver RAD51 to the site of a processed dsDNA break or from an inhibitory effect of excessive BRC4 expression. Consistent with the former interpretation, expression of a fusion of BRC4 and the ssDNA binding domain of a heterologous protein, RPA, restores recombinational DNA repair in mammalian cells to nearly wild-type levels (Saeki et al., 2006). In agreement with the latter possibility, high concentrations of BRC4 block DNA strand exchange in vitro (Figure S5A and B).
The BRCA2-homologue from C. elegans, CeBRC2, which possesses a single BRC repeat, was reported to reduce the rate of ATP hydrolysis by CeRAD51 in vitro and to promote persistent CeRAD51 nucleoprotein focus formation in vivo (Petalcorin et al., 2007). In agreement, here we showed that a single BRC repeat from the human BRCA2 is sufficient to stabilize the human RAD51-ssDNA nucleoprotein complex by decreasing ATP hydrolysis and by both stabilizing and increasing the fraction of active ATP-bound nucleoprotein filaments on ssDNA (Figure 2B and C). Collectively, the published reports and our results support the conclusion that BRC repeats enhance the DNA pairing activity of RAD51 by slowing ATP hydrolysis. Interestingly, a similar mechanism was invoked to explain the Ca2+-dependent stabilization of the RAD51-ssDNA filament and the consequent stimulation of DNA strand exchange (Bugreev and Mazin, 2004). The accumulation of ATP-bound form of the filament by either Ca2+ or BRC4 promotes formation of an active nucleoprotein filament on ssDNA that is necessary for DNA strand exchange. The interaction with BRC4 goes one step further: it limits the non-productive formation of filaments on dsDNA. Thus, BRC4 shows two different modes of interaction: one that stabilizes RAD51-ssDNA filaments by modulating ATPase activity and another that prevents nucleation of filaments on dsDNA (Figure 7A and B).
Further research will be required to understand how the BRC repeats and the C-terminal region orchestrate RAD51 nucleoprotein formation in the context of entire BRCA2 protein and throughout the cell cycle. However, our observations have revealed an unexpected dichotomy of the seemingly simple BRC repeats in controlling RAD51 nucleoprotein filament assembly that might be crucial for understanding the role of BRCA2 in the DNA damage response and repair. In Figure 7C, we illustrate the likely function of the intact BRCA2 protein. Duplex DNA that suffers a DSB is resected to produce ssDNA. The ssDNA is initially bound by RPA, precluding the binding of RAD51 and necessitating a catalyst to facilitate assembly of RAD51 on the ssDNA (Sugiyama and Kowalczykowski, 2002). BRCA2 is proposed to bind to the ssDNA-dsDNA junction by virtue of its DNA binding domain, which comprises both ssDNA and dsDNA binding domains (Yang et al., 2002; Yang et al., 2005). The BRC repeats recruit RAD51 to this junction, and load RAD51 onto the ssDNA, but they block its non-productive assembly onto dsDNA. The major role of BRCA2 is seen as nucleating the nucleoprotein filament near the junction by preventing turnover of the nascent nucleus of the ATP-bound RAD51 filament. Once a nascent filament forms, growth and RPA-displacement ensues, likely aided by other mediators such as RAD52 (Benson et al., 1998; New et al., 1998; Shinohara and Ogawa, 1998; Sung, 1997; Sung et al., 2003).
EXPERIMENTAL PROCEDURES
Protein expression and purification
See supplemental data.
Electrophoretic mobility shift assay
The procedure was essentially as described (Galkin et al., 2005). RAD51 was pre-incubated with GST-tagged BRC4, BRCA2BRC1-8, Δ1524-30, or T1526A at the indicated concentrations for 15 min, followed by addition of ssDNA (dT40, labeled with 32P at the 5’-end) or dsDNA (XhoI-linearized ϕX174 dsDNA, 32P-labelled at the 5’ end or 32P-5’end labelled duplex dT40·dA40 prepared by annealing), at the concentrations indicated, in buffer containing 20 mM Tris-HCl (pH 7.5), 10 mM Mg(OAc)2, 2 mM CaCl2, 2 mM ATP, 100 μg/ ml BSA. The mixture was incubated for 15 min or 60 min at 37 °C, as indicated. The reaction products were resolved by 6% PAGE at 4°C in 1 × TAE, (40 mM Tris-acetate, pH 7.5, 0.5 mM EDTA) or using 0.5 % agarose gels at 40 V o/n (in the case of ϕX174 dsDNA) in 1 × TAE. Where indicated, ATP was replaced with ADP or AMP-PNP (2 mM), and Mg2+ and Ca2+ were replaced by either Mg2+ (10 mM) or Ca2+ (2 mM). The agarose gels were dried and analyzed on a Molecular Dynamics Storm 840 PhosphorImager using ImageQuant software. The percentage of protein-DNA complexes was quantified as the free radiolabeled DNA remaining in a given lane relative to the protein-free lane, which defined the value of 0% complex (100% free DNA).
ATP hydrolysis assay
RAD51 (3 μM) was pre-incubated with the GST-tagged BRC4, or Δ1524-30 at 37°C for 15 min, followed by addition of ssDNA (9 μM nucleotides, nt) in 10 μl of buffer containing 20 mM TrisHCl (pH 7.5), 4 mM MgCl2, 1 mM DTT, 0.5 mM ATP, and 20 μCi/ml [γ32P]ATP, and further incubated at 37 °C for 1h. In figure S3, RPA (1 μM) or RPA storage buffer was pre-incubated with dT40 (9 μM nt) for 5 min at 37 °C, followed by the addition of RAD51 (3 μM) and the concentrations specified of GST-tagged BRC4 in the same buffer as above, and incubated at 37 °C for 1h. Aliquots (1 μl) were spotted onto a polyethyleneimine (PEI) thin layer chromatography (TLC) plate (EMD Chemicals). The spots were air dried and the plates were developed in 1 M formic acid and 0.5 M LiCl. The amount of ATP hydrolyzed was determined from dried plates using a Molecular Dynamics Storm 840 PhosphorImager. The amount of 32Pi and [γ32P] ATP was quantified using ImageQuant software.
Isolation and analysis of the nucleotide composition of RAD51 nucleoprotein complexes
RAD51 (3 μM) was pre-incubated with the GST-tagged BRC4, or Δ1524-30, at 37°C for 15 min, followed by addition of ssDNA (9 μM nt), dT40 or linear ϕX174 ssDNA. (The linear ϕX174 ssDNA was prepared by annealing an oligonucleotide (80-mer) complementary to the AluI restriction site contained within the sequence (5207-5286) of ϕX174 ssDNA, subsequently cutting with AluI, and then purified using a MicroSpin S-400 HR column, (Amersham)). The protein-DNA mixture was incubated for 1 h at 37 °C in 20 μl of buffer containing 20 mM TrisHCl (pH 7.5), 4 mM MgCl2, 1mM DTT, 0.5 mM ATP, and 6 μCi [α32P] ATP. Aliquots of 1 μl were spotted on PEI plates as described above. Aliquots (19 μl) were loaded onto Micro Bio-Spin P30 columns (Bio-Rad) pre-equilibrated in Tris-EDTA (TE (10 mM TrisHCl (pH 7.5), 1 mM EDTA), and 10 μl of the eluate was spotted on PEI plates, air dried, and analyzed as described above. The amount of nucleotide bound to the isolated protein-DNA complexes was calculated from the [α32P] ATP + [α32P] ADP content of each lane and expressed relative to the amount of [α32P] ATP + [α32P] ADP bound when BRC4 was absent. The ATP and ADP bound to free RAD51 was determined in parallel by doing the same experiment in the absence of DNA and was subtracted from each lane of the experiment done in the presence of DNA.
Single-molecule visualization
The experimental protocol was similar to that reported previously (Bianco et al., 2001; Galletto et al., 2006). A three-channel flow cell (4.5 mm wide and 70 μm deep) generated three 1.5 mm laminar flow paths. Bacteriophage λ DNA, biotinylated at one end, was attached to a 1 μM streptavidin-coated polystyrene bead (Bangs Laboratories). The bead-DNA complex in buffer T (50 mM TrisOAc, pH 8.3, 15% sucrose, and 30 mM DTT), containing 10-20 nM YOYO-1, was trapped at a linear flow rate of 180 μm s -1 at 25°C. The trapped bead-λ DNA complex was then moved to the observation channel which contained buffer T and 5 mM Ca(OAc)2, where the YOYO-1 dissociated from the DNA in less than one min. The YOYO-1 allowed measurement of DNA length and verification that a single DNA molecule was captured. The bead-λ DNA complex was then moved to the protein channel containing the fluorescent RAD51 (using carboxyfluorescein succinimidyl ester coupling to the N-terminal amino group at pH 7.0 (Galletto et al., 2006)) in buffer T containing 2 mM Ca(OAc)2 with 1 mM ATP and GST-tagged BRC4, Δ1524-30, or BRCA2BRC1-8, at the concentrations specified. After each incubation time, the protein-DNA-bead complex was moved back to the second channel for direct observation.
The dissociation experiment was conducted by forming the RAD51 nucleoprotein filament in the protein channel in buffer T containing 1 mM ATP, 2 mM Mg(OAc)2, and 250 nM RAD51. After 4 min of incubation, the filament was moved to the observation channel in buffer T containing 2 mM ATP and 10 mM Mg(OAc)2, with or without 1 μM GST-BRC4. The length of the λ DNA was calculated from 11 molecules, corrected for the radius of the bead and for extension due to the YOYO-1 dye. All the experiments were conducted using illumination from a mercury lamp, and fluorescence was observed using a FITC filter (Chroma Technology Corp., number 41001). Data were collected on S-VHS tapes, digitized, and analyzed with Image J software and associated plug-ins.
DNA strand exchange assay
Reactions (20 μl) were as described (Bugreev and Mazin, 2004). RAD51 and GST-tagged BRC4 or the control peptides Δ1524-30 and T1526A, at the concentrations indicated, were incubated with ϕX174 ssDNA (15 μM, nt) for 5 min at 37 °C in buffer containing: 25 mM TrisOAc, pH 7.5; 250 mM NaCl, 2 mM ATP, 1 mM DTT, 1 mM MgCl2, 2 mM CaCl2. RPA (1 μM) was then added, and incubation continued for 5 min at 37 °C. The reaction was started by the addition of XhoI- linearized 32P-labelled ϕX174 duplex DNA (15 μM, nt). After 2 h at 37 °C, the samples were treated with Proteinase K (Roche) for 15 min at 37 °C. Products were resolved by electrophoresis on a 1% agarose gel (1 × TAE) at 40 V over night. The gels were dried and analyzed on a Molecular Dynamics Storm 840 PhosphorImager using ImageQuant software. The amount of DNA strand exchange product at each BRC4 concentration was calculated as a percentage of the joint molecules (JM) or nicked circular DNA (NC) products relative to the total amount of DNA in the same lane.
Analysis
In all graphs, error bars represent the standard deviation derived from at least three independent experiments and, in some cases, error bars are smaller than the symbol; all analyses used GraphPad Prism (version 5.01).
Acknowledgments
We are grateful to Dr. Ryan Jensen for providing us with full length BRCA2 construct and to all the members of the Kowalczykowski lab (Jason Bell, Petr Cejka, Anthony Forget, Ryan Jensen, Taeho Kim, Edgar Valencia-Morales, Amitabh Nimonkar, Jody Plank, Katsumi Morimatsu, Behzad Rad, Jason Wong, Rebecca Wright, and Liang Yang) for insightful comments on the manuscript. This work was supported by NIH grants (GM-62653 and GM-64745) to S.C.K., a postdoctoral fellowship from Ministerio de Educación y Ciencia (Spain) to A.C., a Susan G. Komen postdoctoral fellowship (PDF0707927) to J.H., and funds from the U.K. Medical Research Council to M.K.K.S. and A.R.V. We thank Owen Davies and Luca Pellegrini for BRCA2BRC1-8 purification.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, Baskin RJ. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci U S A. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CF, Chen PL, Zhong Q, Sharp ZD, Lee WH. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J Biol Chem. 1999;274:32931–32935. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- Chen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci U S A. 1998;95:5287–5292. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat Struct Mol Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Esashi F, Yu X, Yang S, West SC, Egelman EH. BRCA2 BRC motifs bind RAD51-DNA filaments. Proc Natl Acad Sci U S A. 2005;102:8537–8542. doi: 10.1073/pnas.0407266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- Hilario J, Amitani I, Baskin R, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0811965106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Morimatsu K, Norden B, Ardhammar M, Takahashi M. ADP stabilizes the human Rad51-single stranded DNA complex and promotes its DNA annealing activity. Genes Cells. 2002;7:1125–1134. doi: 10.1046/j.1365-2443.2002.00588.x. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC. Cancer: catalyst of a catalyst. Nature. 2005;433:591–592. doi: 10.1038/433591a. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc Natl Acad Sci U S A. 2001;98:8461–8468. doi: 10.1073/pnas.151260698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin AV, Zaitseva E, Sung P, Kowalczykowski SC. Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 2000;19:1148–1156. doi: 10.1093/emboj/19.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- Nomme J, Takizawa Y, Martinez SF, Renodon-Corniere A, Fleury F, Weigel P, Yamamoto K, Kurumizaka H, Takahashi M. Inhibition of filament formation of human Rad51 protein by a small peptide derived from the BRC-motif of the BRCA2 protein. Genes Cells. 2008;13:471–481. doi: 10.1111/j.1365-2443.2008.01180.x. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- Petalcorin MI, Galkin VE, Yu X, Egelman EH, Boulton SJ. Stabilization of RAD-51-DNA filaments via an interaction domain in Caenorhabditis elegans BRCA2. Proc Natl Acad Sci U S A. 2007;104:8299–8304. doi: 10.1073/pnas.0702805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki H, Siaud N, Christ N, Wiegant WW, van Buul PP, Han M, Zdzienicka MZ, Stark JM, Jasin M. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc Natl Acad Sci U S A. 2006;103:8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Chi P, Sehorn MG, Etchin J, Krejci L, Sung P. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J Biol Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Shivji MK, Davies OR, Savill JM, Bates DL, Pellegrini L, Venkitaraman AR. A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange. Nucleic Acids Res. 2006;34:4000–4011. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J Biol Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, Griffin C, Thacker J, Ashworth A. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]