Abstract
Dendritic cells (DCs) are highly potent antigen-presenting cells (APCs) and play a vital role in stimulating naïve T cells. Treatment of human blood monocytes with the cytokines granulocyte–macrophage colony-stimulating factor (GM-CSF) and interleukin (IL)-4 stimulates them to develop into immature dendritic cells (iDCs) in vitro. DCs generated by this pathway have a high capacity to prime and activate resting T cells and prominently express CD1 antigen-presenting molecules on the cell surface. The presence of human serum during the differentiation of iDCs from monocytes inhibits the expression of CD1a, CD1b and CD1c, but not CD1d. Correspondingly, T cells that are restricted by CD1c showed poor responses to DCs that were generated in the presence of human serum, while the responses of CD1d-restricted T cells were enhanced. We chemically fractionated human serum to isolate the bioactive factors that modulate surface expression of CD1 proteins during monocyte to DC differentiation. The human serum components that affected CD1 expression partitioned with polar organic soluble fractions. Lysophosphatidic acid and cardiolipin were identified as lipids present in normal human serum that potently modulate CD1 expression. Control of CD1 expression was mediated at the level of gene transcription and correlated with activation of the peroxisome proliferator-activated receptor (PPAR) nuclear hormone receptors. These findings indicate that the ability of human DCs to present lipid antigens to T cells through expression of CD1 molecules is sensitively regulated by lysophosphatidic acid and cardiolipin in serum, which are ligands that can activate PPAR transcription factors.
Keywords: cell surface molecules, dendritic cells, human, lipid mediators, T cells
Introduction
Dendritic cells (DCs) and other professional antigen-presenting cells (APCs) express the lipid antigen-presenting molecules CD1a, CD1b and CD1c (group 1), as well as CD1d (group 2).1,2 For example, Langerhans cells express CD1a and CD1c,3 and lymph node interdigitating cells may express CD1a, CD1b and CD1c as do DCs that infiltrate infectious lesions4 and tumours.5In vitro, CD14+ monocyte precursors cultured in the presence of interleukin (IL)-4 and granulocyte–macrophage colony-stimulating factor (GM-CSF) differentiate into immature myeloid dendritic cells (iDCs) that express high levels of CD1a, CD1b and CD1c cell surface molecules6 while displaying reduced levels of CD1d.7 Previous studies have identified a variety of microbial lipids as antigens presented by group 1 CD1 molecules to specific T cells.8–10 In general, T cells with reactivity against these antigens have a pro-inflammatory phenotype displaying cytolytic potential and secretion of T helper type 1 (Th1) cytokines, suggesting that they play a role in host defence against microbial infection.1,11 Additionally, group 1 self-reactive T cells that recognize CD1 molecules can mediate the maturation of pro-inflammatory myeloid DCs in a CD1-dependent manner.12,13 T-cell recognition of self-lipids presented by group 1 CD1 molecules has also been implicated in human inflammatory diseases including systemic lupus erythematosus (SLE),14 multiple sclerosis (MS),15 and autoimmune thyroiditis.16 As group 1 CD1-restricted T cells have been implicated in pro-inflammatory responses relating to both host defence and autoimmunity, the regulation of cell surface CD1 expression on DCs is of interest as a mechanism that controls their activation.
Recent studies have shown that the expression of CD1 molecules on DCs is modulated by activation of peroxisome proliferator-activated receptor-γ (PPARγ), which can occur as a result of exposure to synthetic ligands or serum.17–19 PPARγ is a transcription factor that regulates expression of genes relating to lipid metabolism and uptake, as well as cellular glucose metabolism and energy expenditure.20 As a member of the nuclear hormone receptor superfamily, PPARγ resides in the cytoplasm in an inactivated state until it binds a specific ligand (e.g. prostaglandins, oxidized fatty acids, and thiazolidinedione drugs). Upon ligand binding, PPARγ is activated by de-phosphorylation of serine 82 by mitogen-activated protein (MAP) kinases, heterodimerizes with the retinoid X receptor (RXR), and translocates to the nucleus, where it binds specific PPAR-response elements (PPREs) in the promoters of target genes.21 Szatmari and coworkers have demonstrated that CD1d expression is up-regulated on DCs by the addition of synthetic PPARγ agonists such as Rosiglitazone, and the resulting increase in CD1d expression on the cell surface correlated with enhanced natural killer T (NKT) cell activity.17,18 PPARγ can also be activated by endogenous lipid molecules that are present in serum; however, the identity of the activating ligands remains unclear.22–24
Here, we observed that the phospholipids cardiolipin and lysophosphatidic acid, which are present in micromolar concentrations in normal human serum, can inhibit expression of CD1a, CD1b and CD1c (group 1) molecules by DCs that differentiate from human blood monocytes, resulting in a reduced capacity to stimulate group 1 CD1-restricted T cells. The down-regulation of group 1 CD1 expression by serum lipids is controlled at the level of transcription and was associated with activation of multiple members of the PPAR nuclear hormone receptor family. Importantly, down-modulation of CD1 expression on DCs by serum lipids was completely reversible by removal of the inhibitory serum component. These data indicate that the presence of specific lipid species within the cellular microenvironment can negatively regulate pro-inflammatory DC function by inhibiting expression of group 1 CD1 molecules and thus preventing efficient activation of group 1 CD1-restricted T cells.
Materials and methods
Antibodies and flow cytometry
The following monoclonal antibodies (mAbs) were used for flow cytometry: immunoglobulin G1-phycoerythrin (IgG1-PE), IgG2a-PE, CD1a-PE, CD1d-PE, IgG1-fluorescein isothiocyanate (FITC), CD83-FITC, CD86-PE (anti-B7.2), CD14-PE, CD209-PE and human leucocyte antigen (HLA)-DR-PE (all from BD-Pharmingen, San Diego, CA), and CD1b-PE and CD1c-PE (Ancell, Bayport, MN). Antibodies were used at saturating concentrations per the manufacturer’s instructions and staining was performed in 96-well plates. All antibody staining was carried out for 30 minutes at 4° in the presence of 0·02% NaN3. Samples were then analysed utilizing a FACSort flow cytometer (Becton Dickinson, Franklin Lakes, NJ).
T-cell lines and clones
The CD1c-reactive clone Ye2.3 was derived by limited dilution cloning from normal donor peripheral blood as previously described.12 CD1d-reactive clone J3N.5 expressing an invariant Vα24/JαQ T-cell receptor (TCR) was derived by staining human peripheral blood mononuclear cells (PBMC) with fluorescently labelled CD1d tetramers loaded with α-galactosyl ceramide (α-GalCer) followed by sorting by flow cytometry and limiting dilution cloning as previously described.25
Differentiation of monocytes to dendritic cells
Monocyte-derived DCs were generated from human blood monocytes that were isolated by CD14 magnetic antibody cell sorting (CD14-MACS) selection (Miltenyi Biotec, Auburn, CA) from the byproducts of platelet pheresis and induced to differentiate and express CD1a, CD1b and CD1c by incubation with GM-CSF and IL-4 as described previously.26 All protocols involving human samples were performed with approval and in accordance with institutional guidelines. Monocytes were cultured in 24-well tissue culture plates at a concentration of 1 × 106 cells/ml for 3 days in RPMI complete medium containing l-glutamine, penicillin-streptomycin and 10% fetal bovine serum (FBS) in the absence (FBS-DCs) or presence [human serum (HS)-DCs] of normal pooled male human AB serum (1–10%) (Gemini Bioproducts, West Sacramento, CA). For experiments utilizing the addition of specific lipids the following compounds were used: phosphatidic acid, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol (all synthetic 18:2), cholesterol, cholesteryl stearate, cardiolipin (CL; bovine heart) (Avanti Polar Lipids, Alabaster, AL) and lysophosphatidic acid (LPA) 20:4 (Echelon Biosciences, Salt Lake City, UT). Studies of PPARγ activation utilized the PPARγ agonist troglitazone at a concentration of 20 μm or the PPARγ antagonist GW9662 at a concentration of 10 μm (Tocris, Ellisville, MS) during the differentiation of monocytes to DCs.
Organic phase separation and thin-layer chromatography (TLC)
Human pooled male AB serum (Gemini Bioproducts) was separated into organic, interphase and aqueous phases using the method of Folch as previously described.27 In brief, 20 ml of serum was added to 80 ml of chloroform and 40 ml of methanol (4 : 2 : 1 chloroform:acetone:methanol) and phase separation allowed to occur. Individual fractions were removed, dried down under a nitrogen gas stream, and weighed for use in the DC differentiation experiments. Further separation of the organic soluble fraction was performed by loading the total material onto an open silica column consisting of 32–63-μm silica beads (Selecto Scientific, Suwanee, GA). Fractions were eluted in progressively polar solvents: chloroform, acetone, and then methanol [high-performance liquid chromatography (HPLC) grade; Fisher Scientific, Waltham, MA]. These fractions were then dried, weighed, and utilized in the biological assays described.
CD1-restricted T-cell/DC co-cultures
The CD1-restricted T-cell clones YE2.312 and J3N.525 were utilized for all studies. FBS-DCs or HS-DCs were generated as described above. T-cell proliferation in response to these DCs was assessed by culturing 5 × 104 T cells with 5 × 104γ-irradiated (5000 rads) FBS-DCs or HS-DCs per well for 3 days in 96-well plates. For experiments using CD1d-reactive T-cell clone J3N.5, α-GalCer was utilized at a final concentration of 100 ng/ml. During the final 6 hr of culture, cells were pulsed with 1 μCi [3H]-thymidine (2 Ci/mmol; New England Nuclear, Waltham, MA) harvested using a Tomtec harvester (Hamden, CT) and the mean count per minute ± standard error of the mean (c.p.m. ± SEM) was determined using a beta plate counter (Wallac, Waltham, MA). For determination of T-cell interferon (IFN)-γ release, culture supernatants were collected at 24 hr and assayed for IFN-γ using sandwich enzyme-linked immunosorbent assay (ELISA) mAbs (Pierce Endogen, Rockford, IL) and analysed as described above. ELISA results are expressed in pg/ml ± SEM.
Allogeneic mixed lymphocyte reaction
FBS-DCs or HS-DCs were generated as described above. These DCs were then cultured for three additional days in the same medium or in the presence of 100 ng/ml Escherichia coli lipopolysaccharide (LPS) (Sigma, St Louis, MO). DCs were washed and γ-irradiated (5000 rads), and then these cells were cultured with 1 × 105 allogeneic CD4+ T cells (isolated by immunomagnetic selection; Miltenyi Biotec) per well at DC:T-cell ratios of 1 : 200 to 1 : 2000 for 5 days. During the final 6 hr of culture, cells were pulsed with 1 μCi [3H]-thymidine (2 Ci/mmol; New England Nuclear) and harvested using a Tomtec harvester. The c.p.m. was determined using a beta-plate scintillation counter (Wallac). Results represent triplicate samples ± SEM.
Measurement of cytokines by ELISA
FBS-DCs or HS-DCs were generated as described above. Following additional culture with E. coli LPS (1 μg/ml; Sigma) or control medium for 24 hr, supernatants were harvested and analysed for IL-12p70 by sandwich ELISA assay using antibody pairs purchased from Pierce Endogen. Samples were analysed using a Molecular Devices (Sunnyvale, CA) plate reader and SoftMax Pro software (Sunnyvale, CA). Results were expressed as pg/ml ± SEM.
Confocal microscopy
Following culture for 72 hr in the presence of medium alone or medium containing 10% human AB serum, DCs were adhered to fibronectin-coated coverslips, fixed and permeabilized as previously described.28 The cells were then labelled with mAb specific for CD1a, CD1b, CD1c and CD1d.29 Alexa 488-goat anti-mouse IgG (Jackson Immunologic, West Grove, PA) was utilized as a secondary reagent. Cells were counterstained with mAb against the lysosomal marker lysosome-associated membrane glycoprotein 1 (LAMP-1) (BD Pharmingen) conjugated to Alexa 546 (Molecular Probes, Carlsbad, CA). Labelled cells were then examined using a Nikon C1 confocal laser scanning microscope (Melville, NY) fitted with krypton and argon lasers.
Real-time polymerase chain reaction (PCR)
Following differentiation of immature DCs by 3-day culture of CD14+ cells in IL-4 and GM-CSF in the absence or presence of human AB serum, total RNA was extracted using an Rneasy kit (Qiagen, Valencia, CA). After retrotranscription using Superscript II H-Reverse Transcriptase (Invitrogen, Carlsbad, CA), the relative CD1 expression levels were analysed by real-time PCR using SYBR Green amplification (BioRad, Hercules, CA) and the ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA). Primers specific for each CD1 isoform were generated as described previously.30 The relative CD1 mRNA levels were determined by the comparative threshold cycle (Ct) method relative to the expression of the housekeeping gene GAPDH and data expressed as fold change in HS-DCs compared with FBS-DCs.
PPAR gene expression
Total cellular RNA from monocytes and immature DCs (2 × 106 cells) was extracted using TRIzol (Invitrogen). Reverse transcription was performed on 500 ng of total RNA using an oligo-(dT)18 primer with the first-strand cDNA synthesis kit (Roche, Indianapolis, IN). Primers were as follows: GAPDH forward, 5′-ACTCCACGACGTACTCAGCG-3′ and reverse, 5′-GGTCGGAGTCAACGGATTTG-3′; PPARα forward, 5′-CAAGTGCCTTTCTGTCGGGA-3′ and reverse, 5′-TGGCAGCAGTGAAAGATGCG-3′; PPARβ/δ forward, 5′-TCTCCAAGCACATCTACAATGCC-3′ and reverse, 5′-CAGGTCACTGTCATCAAGTTCCAG-3′; PPARγ forward, 5′-TCATAATGCCATCAGGTTTGG-3′ and reverse, 5′-CTGGGCGGTCTCCACTG-3′. PCR amplification of the specific PPAR isoforms (α, β/δ, and γ) and GAPDH was performed and PCR products were separated by agarose gel electrophoresis and a digitized image generated. Quantification of PCR products was carried out using ImageJ software (http://rsb.info.nih.gov/ij/) and normalized to the GAPDH band intensities.
PPARγ reporter construct assay
To test for transcriptional activity of PPARγ, we utilized a PPRE-luciferase construct described previously.31 Briefly, 1 × 106 HCT-116 colon cancer cells (American Type Culture Collection, Rockville, MD) grown in 24-well cell plates in McCoy’s medium were transfected with 100 ng of a plasmid encoding a PPRE DR-1 firefly luciferase reporter construct and 100 ng of pRL-TK (Promega, Madison, WI) vector encoding the Renilla lucifierase under a constitutive promoter utilizing FuGene-6 (Roche) according to the manufacturer’s instructions. For some assays, an additional plasmid construct expressing full-length cDNA of human PPARγ was cotransfected. Cells were incubated for 24 hr and test compounds were then added, and the cells were incubated for an additional 18 hr. Cells were then lysed and assayed for luciferase activities using a dual-luciferase reporter assay system (Promega) and a luminometer. Results were calculated as the ratio of specific PPRE-luciferase to Renilla lucifierase activity and normalized to the medium control. Results were expressed as fold induction ± SEM.
Results
Human serum modulates DC expression of CD1 cell surface molecules
Myeloid dendritic cells are known to express all group 1 (CD1a, CD1b and CD1c) as well as group 2 (CD1d) isoforms.32 As these DCs expressing CD1 are important APCs for lipid recognition by CD1-reactive T cells, we hypothesized that regulatory control of CD1 cell surface expression might play a critical role in regulating the activation of CD1-restricted T cells. In an initial series of experiments, we found an inverse correlation between the quantity of human serum present in medium during in vitro monocyte differentiation to DCs and the cell surface expression CD1 molecules. Fresh blood monocytes were cultured with GM-CSF and IL-4 in complete medium containing 10% FBS for 3 days to induce the differentiation into myeloid DCs (FBS-DCs) or the same FBS medium supplemented with an additional 10% human AB serum (HS-DCs). The DCs were then stained with mAbs against CD1a, CD1b, CD1c, CD1d, the monocyte marker CD14, the DC marker CD209, or HLA-DR and analysed by flow cytometry (Fig. 1). The presence of human serum during differentiation strongly inhibited the cell surface levels of CD1a [HS-DC mean fluorescence intensity (MFI) = 4·5 versus FBS-DC MFI = 712], CD1b (MFI = 27·5 versus 317), and CD1c (MFI = 13·5 versus 153). In contrast, CD1d expression was slightly increased in the presence of human serum (HS-DC MFI = 12·4 versus FBS-DC MFI = 3·7). No significant change in cell surface levels of major histocompatibility complex class II (MHC II) was observed. Like the control DCs, the human serum-treated cells had a CD14− and CD209+ DC phenotype, suggesting that the effect of human serum was not a result of a generalized blockade of blood monocyte differentiation. The serum-mediated effect on CD1 expression was also observed using serum samples from multiple healthy individual donors, suggesting that it was a general effect. Additionally, increasing the concentration of FBS (up to 20%) had no effect upon CD1 expression, demonstrating that the observed effects were not attributable to the absolute concentration of serum in the media (data not shown). These data suggest that factors present in normal human serum have the capacity to inhibit the expression of cell surface CD1a, CD1b and CD1c on DCs.
Figure 1.
Human serum inhibits group 1 CD1 expression by dendritic cells (DCs). CD14+ monocytes were cultured for 72 hr in interleukin (IL)-4 and granulocyte–macrophage colony-stimulating factor (GM-CSF) in fetal bovine serum (FBS)-containing medium (FBS-DCs), or similar medium containing additional 10% human pooled AB serum [human serum (HS)-DCs] to derive immature monocyte-derived DCs. Cell surface expression of CD1a, CD1b, CD1c, CD1d, CD14, CD209, and human leucocyte antigen (HLA)-DR were assessed by monoclonal antibody (mAb) staining and flow cytometry. The presence of human serum (HS-DCs) inhibited expression of CD1a, CD1b and CD1c and promoted up-regulation of CD1d. HS-DCs also lacked expression of CD14 and expressed CD209, consistent with a DC phenotype. These results are representative of five independent experiments using different DC donors.
DC maturation is altered by the addition of human serum
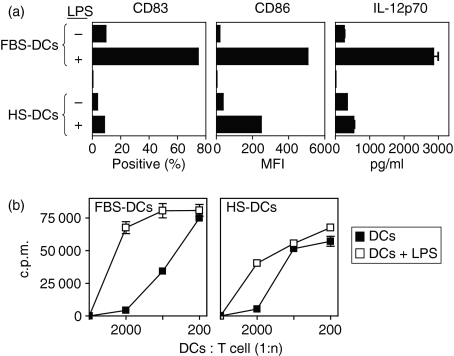
To further characterize the HS-DCs, we examined the expression of the maturation marker CD83 and the costimulatory molecule CD86 (B7.2) in response to exposure to the Toll-like receptor 4 (TLR4) ligand LPS. Immature DCs lack expression of CD83 and express low levels of CD86 but up-regulate these cell surface molecules following exposure to maturation stimuli such as LPS. Figure 2(a) shows that, following a 3-day culture in the presence of 100 ng/ml LPS, fewer HS-DCs expressed CD83 (8·2%) when compared with control FBS-DCs (74·3%). Additionally, although immature HS-DCs had slightly higher levels of CD86 (MFI = 34·3) than did FBS-DCs (MFI = 16·4), these HS-DCs up-regulated this molecule to a lesser extent (MFI = 245) than did FBS-DCs (MFI = 503·4) in the presence of LPS (Fig. 2a). These data suggest that DCs exposed to factors present in human serum are refractory to maturation stimuli such as LPS.
Figure 2.
Human serum dendritic cells (DCs) are refractory to lipopolysaccharide (LPS)-induced maturation. (a) Human serum (HS)-derived DCs fail to express the maturation marker CD83, up-regulate the costimulatory molecule CD86, or produce interleukin (IL)-12p70 in response to LPS. Fetal bovine serum (FBS)-DCs or HS-DCs were generated as described and were further cultured in the presence of LPS (1 μg/ml) for 3 days to induce maturation. DCs were then assessed by monoclonal antibody (mAb) staining and flow cytometry or enzyme-linked immunosorbent assay (ELISA) to assess IL-12p70 production. Note that HS-DCs express less CD83 and CD86 and fail to produce IL-12p70 in response to LPS as a maturation stimulus. Error bars on the IL-12p70 ELISA represent the standard error of the mean (SEM) from triplicate samples. (b) HS-DCs are less effective at supporting an allogeneic mixed lymphocyte reaction (MLR). Immature FBS-DCs or HS-DCs were derived as described and cultured for an additional 72 hr in the presence of medium alone or LPS (100 ng/ml) as a maturation signal. DCs were then irradiated and used to stimulate allogeneic CD4+ T cells. Note that maximum allogeneic stimulatory capacity was decreased for immature HS-DCs (right panel; closed squares) when compared with immature FBS-DCs (left panel; closed squares). Additionally, LPS (open squares) had a lesser effect upon increasing the ability of HS-DCs to stimulate allogeneic T cells than upon the ability of FBS-DCs to stimulate these T cells. These results are representative of three independent experiments using different DC donors. Results represent triplicate samples ± SEM. c.p.m., counts per minute; MFI, mean fluorescence intensity.
One of the important hallmarks of mature DCs is their ability to produce bioactive IL-12p70, which is essential for the subsequent Th1 polarization of naïve T cells encountered in secondary lymphoid tissues.33 DCs exposed to high levels of TLR agonists (e.g. LPS) in vitro are capable of transiently producing IL-12p70. Therefore, we next cultured FBS-DCs or HS-DCs in the presence of LPS for 24 hr and assayed production of IL-12p70 by ELISA. In contrast to FBS-DCs, cells derived in human serum produced markedly less IL-12p70 (FBS-DCs, 2838 ± 192 pg/ml versus HS-DCs, 521 ± 54 pg/ml) (Fig. 2a). Recent evidence suggests that both microbial signals, mediated via pattern recognition receptors such as TLR, and host T-cell signals are required for the generation of mature DCs capable of producing IL-12p70. We previously showed that CD1a-, CD1b- and CD1c-restricted T cells can induce DC secretion of IL-12p70 in the presence of very low levels of LPS.12,13 In our current studies we observed that DCs differentiated in the presence of human serum failed to produce IL-12p70 when cultured with CD1-restricted T cells and LPS (data not shown), suggesting that anti-inflammatory signals from the human serum override the pro-inflammatory signals from the T cells and LPS.
We next performed allogeneic mixed lymphocyte reactions (MLRs) to compare the functional capacities of HS-DCs and FBS-DCs to activate T cells. Purified CD4+ T cells were co-cultured for 5 days with irradiated allogeneic HS-DCs or FBS-DCs. Proliferation to alloantigens using the MLR assay was similar when T cells were stimulated with either FBS-DCs or HS-DCs, although maximum allogeneic T-cell proliferation was somewhat lower when HS-DCs were used as APCs. To determine if the maturation state of the DCs affected their ability to function as APCs, FBS-DCs and HS-DCs were also cultured in the presence of the LPS as a maturation signal. When DCs were cultured in the presence of LPS for an additional 3 days prior to being used as APCs, HS-DCs (37 080 ± 1120 c.p.m.) were again less effective at supporting the proliferation of allogeneic T cells than were FBS-DCs (69 540 ± 2320 c.p.m.) at a DC:T-cell ratio of 1 : 2000 (Fig. 2b). Thus, HS-DCs are less efficient APCs in the MLR assay when compared with FBS-DCs, suggesting an attenuated functional capacity for T-cell stimulation. Taken together, these data suggest that, although monocyte differentiation to immature DCs occurs after the addition of human serum, subsequent DC maturation and function are inhibited.
Activation of CD1-restricted T cells is modulated by human serum effects on DCs
As our initial experiments demonstrated that cell surface expression of group 1 CD1a, CD1b and CD1c molecules is decreased by the addition of human serum while that of group 2 CD1d molecules is slightly increased (Fig. 1), we next determined if these changes in DC CD1 expression alter the activation of CD1-restricted T cells. As an example of a group 1 CD1-restricted T cell, we used the previously described T-cell clone Ye2.3 which recognizes CD1c in the absence of exogenous lipid antigen.12 To assess the interaction between DCs and group 2 CD1d-restricted T cells, we utilized previously described J3N.5, an NKT cell clone that expresses an invariant Vα24/JαQ TCR and recognizes the model lipid antigen α-GalCer in the context of CD1d.25 The CD1c-restricted T-cell clone Ye2.3 or CD1d-restricted T-cell clone J3N.5 was co-cultured for 3 days with irradiated FBS-DCs or HS-DCs and T-cell activation was measured in a proliferation assay. For J3N.5, α-GalCer (100 ng/ml) was utilized as an exogenous lipid antigen. Both CD1c-restricted Ye2.3 T cells and CD1d-restricted J3N.5 NKT cells proliferated in the presence of FBS-DCs (or FBS-DCs + α-GalCer in the case of J3N.5 T cells). In contrast, the CD1c-restricted Ye2.3 T cells failed to proliferate when HS-DCs were used as APCs while the proliferation of the CD1d-restricted clone J3N.5 was similar with either FBS-DCs or HS-DCs (Fig. 3a). As a second measure of CD1-restricted T-cell function, we examined the production of IFN-γ by these T cells in response to co-culture in the presence of FBS-DCs or HS-DCs. FBS-DCs supported IFN-γ production by both CD1c-restricted Ye2.3 (1346 ± 102 pg/ml) and CD1d-restricted J3N.5 (403 ± 1·2 pg/ml) T cells. In contrast, the Ye2.3 T cells showed reduced IFN-γ (31 ± 1·8 pg/ml) when HS-DCs were used as APCs. Production of IFN-γ by J3N.5 (501 ± 31 pg/ml) was only slightly higher using HS-DCs as APCs (Fig. 3b). These results indicate that factors present in human serum that modulate the expression of CD1 molecules on DCs can lead to the differential activation of group 1 and group 2 CD1-restricted T cells.
Figure 3.
Human serum regulates dendritic cell (DC) activation of CD1-restricted T cells. (a) CD1c-restricted T cells do not proliferate in response to co-culture with human serum (HS)-DCs. CD1c-restricted autoreactive Ye2.3 (left panel) or CD1d-restricted, α-galactosyl ceramide (α-GalCer)-reactive J3N.5 T cells (right panel) were incubated with fetal bovine serum (FBS)-DCs or HS-DC for 72 hr and proliferation was measured. For J3N.5, α-GalCer was used at a final concentration of 100 ng/ml. FBS-DCs, expressing both CD1c and CD1d, produced proliferation of both CD1c-reactive Ye2.3 and CD1d-reactive J3N.5 T cells. Note that HS-DCs expressing lower cell surface levels of CD1c failed to activate CD1c-restricted Ye2.3. (b) CD1c-restricted T cells do not produce interferon (IFN)-γ in response to HS-DCs. CD1-restricted T cells were cultured with DCs as described above. Note that CD1c-restricted Ye2.3 T cells do not make IFN-γ in response to culture with HS-DCs. Results represent triplicate samples ± standard error of the mean (SEM). These data are representative of three independent experiments using different DC donors.
Human serum factors inhibit DC expression of CD1 at the level of transcription
We first performed confocal microscopy to determine if the effect of human serum in modulating CD1 expression at the cell surface was a result of altered cellular distribution of these molecules. HS-DCs or FBS-DCs were fixed and permeabilized and then stained with mAbs against CD1a, CD1b, CD1c and CD1d (green staining). The lysosomal marker LAMP-1 was used as a marker in a second colour (red staining). FBS-DCs expressed high levels of CD1a, CD1b and CD1c molecules but only very low levels of CD1d. In contrast, HS-DCs lacked detectable intracellular or cell surface staining of group 1 CD1a, CD1b or CD1c molecules while staining for CD1d was slightly increased (Fig. 4a). As seen in prior studies of the influence of TLR-2 on CD1 expression,30 these results indicated that the effect of human serum on CD1 is not attributable to cellular redistribution, but rather to altered synthesis of CD1 proteins.
Figure 4.
Human serum regulates dendritic cell (DC) expression of CD1 at the level of transcription. (a) Cell surface expression of CD1 is not a result of altered cellular distribution of CD1. Fetal bovine serum (FBS)-DCs (upper panels) or human serum (HS)-DCs (lower panels) were generated as described and stained with monoclonal antibodies (mAbs) against CD1 molecules (green staining) and the lysosomal marker lysosome-associated membrane glycoprotein 1 (LAMP-1) (red staining). Note that, in contrast to FBS-DCs, HS-DCs express no group 1 CD1a, CD1b or CD1c but express increased levels of CD1d. (b) Alteration of DC CD1 expression is regulated at the level of transcription. CD1 mRNA expression by FBS-DCs and HS-DCs was assessed using quantitative real-time polymerase chain reaction. Results are expressed as relative fold change in HS-DCs compared with FBS-DCs. Note that, while HS-DCs have decreased transcription of CD1a, CD1b and CD1c, CD1d is increased when compared with FBS-DCs. (c) Inhibition of DC CD1a expression is reversible. Left panel: FBS-DCs were differentiated as described and at day 3 (red arrow) 10% human serum (red triangles, dashed line) was added to cultures and inhibited further acquisition of CD1a. In contrast, cells continued in culture without the addition of human serum continued to increase cell surface expression of CD1a (squares, solid line). Right panel: HS-DCs were generated over a 3-day culture period and at day 3 (red arrow) human serum was removed from cultures and CD1a cell surface expression increased over the next 48 hr (red triangles, dashed line) while cultures in which human serum remained failed to express CD1a (squares, solid line). These results are representative of three independent experiments using different DC donors. MFI, mean fluorescence intensity.
To determine if factors in serum affect CD1 expression at the level of transcription, we performed real-time PCR to assess the levels of CD1 mRNA. FBS-DCs or HS-DCs were generated as described above and RNA was isolated. The levels of each CD1 isoform mRNA were assessed using CD1-specific primers and normalized to GAPDH. When compared with FBS-DCs, HS-DCs expressed significantly less mRNA for group 1 CD1a (47-fold decrease), CD1b (12·9-fold decrease), and CD1c (sixfold decrease) but displayed increased levels of group 2 CD1d (13·2-fold increase) (Fig. 4b). Taken together, these data strongly support the theory that the altered cell surface expression of CD1 proteins by DCs differentiated in the presence of human serum is a result of changes at the level of CD1 gene transcription.
The cellular microenvironment in which DCs reside at any given time can be altered by changes in the location of the DCs (i.e. migration and trafficking) or by local changes in the surrounding tissues in which DCs may be present (i.e. disease or tissue injury). Given the dynamic nature of DC trafficking, it would seem critical that any regulatory mechanisms that control expression of important antigen-presenting molecules such as CD1 retain the capacity to respond to feedback signals from the local cellular environment. Therefore, we determined if the inhibition of DC CD1a, CD1b and CD1c expression by human serum is a dynamic and potentially reversible process. We first derived FBS-DCs or HS-DCs using a 3-day culture period as described above. Cells were then washed and re-plated in medium in the presence or absence of 10% human serum for an additional 3 days. These cells were analysed by flow cytometry for CD1a expression on each day during the culture period. Addition of human serum at day 3 to FBS-DCs that already expressed CD1a completely inhibited further up-regulation of this molecule (Fig. 4c, left panel). Conversely, DCs that had the inhibitory human serum removed at day 3 up-regulated CD1a at the cell surface to levels approaching those of the medium control, while DCs maintained in human serum failed to express CD1 during this culture period (Fig. 4c, right panel). Although the effect was greatest for CD1a, a similar effect was seen for CD1b and CD1c, while group 2 CD1d levels were low and remained unchanged throughout the culture period (data not shown). These data demonstrate that the inhibition of DC group 1 CD1 expression by factors present in human serum is a reversible process.
Lipid molecules mediate inhibition of DC CD1 expression
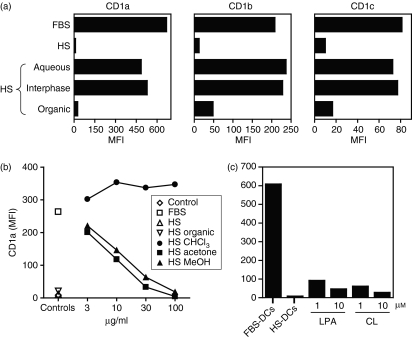
We next performed a series of experiments to characterize the active compounds in human serum responsible for CD1 inhibition. Utilizing phase separation by Folch extraction, we isolated organic, interphase and aqueous soluble fractions from normal human serum.27 This technique allows separation of organic solvent soluble molecules such as lipids from aqueous solvent soluble molecules such as proteins. DCs were then differentiated in IL-4 and GM-CSF in the presence of FBS-containing medium or the same medium containing either the organic, interphase or aqueous extracts from human serum. Cells were cultured for 3 days and assessed for cell surface expression of CD1a, CD1b and CD1c by flow cytometry. The CD1 inhibitory activity in human serum appeared to completely segregate to the organic fraction (Fig. 5a). These results provide strong evidence that the inhibitory molecules in human serum are hydrophobic species such as lipids.
Figure 5.
Polar human serum (HS) lipids inhibit dendritic cell (DC) expression of group 1 CD1 molecules. (a) Inhibition of DC group 1 CD1 expression is mediated by organic soluble molecules. Human serum was separated into aqueous, interphase and organic fractions and added to CD14+ cells differentiated to DCs in the presence of interleukin (IL)-4 and granulocyte–macrophage colony-stimulating factor (GM-CSF) in fetal bovine serum (FBS)-containing medium. DCs were also derived in the presence of similar medium containing 10% HS as a positive control. Cells were analysed for CD1a, CD1b and CD1c cell surface expression by flow cytometry. Note that the organic phase of human serum contained the CD1 inhibitory activity. (b) Polar serum lipid fractions inhibit CD1a. Total human serum organic extract was separated using silica column elution with progressively polar solvents (chloroform, followed by acetone, and then methanol). CD14+ cells were cultured in medium alone without cytokines (open diamond) or differentiated to DCs in IL-4 and GM-CSF in medium containing FBS (open square), similar medium plus 10% HS (open triangle), total HS organic extract (inverted open triangles), HS chloroform extract (solid circles), HS acetone extract (solid squares), or HS methanol extract (solid triangles). Cells were then assessed for CD1a expression by monoclonal antibody (mAb) staining and flow cytometry. Note that the more polar acetone and methanol fractions inhibited DC expression of CD1a. (c) Lysophosphatidic acid (LPA) and cardiolipin (CL) inhibit CD1a expression. FBS-DCs or HS-DCs were generated as described. Additionally, additional FBS-DCs were cultured in the presence of LPA or CL at concentrations of 1 and 10 μm. DCs were then assessed for CD1a cell surface expression by mAb staining and flow cytometry. Note that LPA and CL inhibited CD1a expression at both 1 and 10 μm. These results are representative of four independent experiments using different DC donors. MFI, mean fluorescence intensity.
To further characterize the inhibitory activity within the organic solvent soluble fraction, we employed a strategy of open silica column chromatography of the total human serum organic extract and eluted with progressively polar solvents: chloroform, then acetone, and finally methanol. Each fraction was then subjected to analytical TLC to assess content and compared with known lipid standards (data not shown). The eluted fractions were weighed, dried under N2, and reconstituted in IL-4- and GM-CSF-containing medium and cultured with CD14+ monocytes to derive DCs. Analysis by flow cytometry revealed that CD1a, CD1b and CD1c expression was inhibited in a dose-dependent manner by the fractions that had been eluted in the more polar acetone and methanol solvents, while the less polar chloroform fraction had no effect on the inhibition of CD1 expression (CD1a shown, Fig. 5b). These results suggest that the inhibition of DC group 1 CD1 expression by human serum is probably attributable to polar lipids.
As phospholipids are polar lipids that are known to be present in significant quantities in human serum, we screened a panel of individual lipids for their ability to inhibit expression of CD1 by DCs during the 3-day culture in IL-4 and GM-CSF. Two of the phospholipids tested, CL and LPA, markedly inhibited group 1 CD1a expression in the micromolar range (CL = 96% decrease in MFI at 1 μm; LPA = 87% decrease in MFI at 1 μm) (Fig. 5c). In contrast, other prominent serum lipids such as cholesterol and cholesteryl stearate had no inhibitory effect on CD1 expression (data not shown).
Modulation of CD1 expression by DCs is associated with PPAR activation
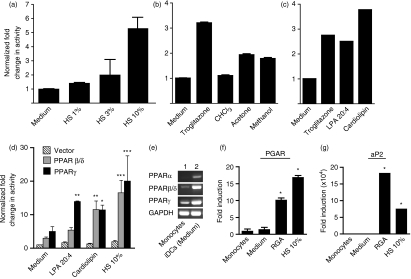
As CD1 gene expression has previously been reported to be influenced by PPARγ,17,18,34 and this nuclear hormone receptor is known to be activated by fatty acyl compounds,35 we investigated whether the bioactivity of the lipid molecules we identified above was linked to activation of members of the PPAR transcription factor family. Therefore, we tested whether addition of the specific lipids we identified results in activation of the PPAR. In initial studies we tested lipids using a cell-based PPRE luciferase reporter (DR1) which measures PPAR-dependent promoter activity.31Figure 6(a) shows that human serum is capable of stimulating PPRE promoter activity in dose-dependent manner. We also tested serum lipid fractions and individual serum lipid components in the same reporter assay system. Consistent with the modulation of CD1 expression shown in Fig. 5b, the methanol and acetone fractions were able to induce PPRE-dependent activity relative to the medium control using the DR1 reporter (Fig. 6b). The positive control in these assays was troglitazone; an agonist of PPARγ that has been shown previously to modulate CD1 expression on DCs.17,18 Rosiglitazone, another compound in the thiazolidinedione class, was also shown to have a similar effect to troglitazone (data not shown). We next tested the lipids LPA and CL using the same assay system. Figure 6(c) shows that both LPA and CL were indeed able to activate the PPRE reporter system, indicative of PPAR activity. To determine whether PPARγ was contributing to the activity, the PPRE reporter plasmid was cotransfected with a second plasmid expressing the PPARγ protein and re-tested with the self-lipids. Figure 6(d) shows that PPARγ exhibited significantly higher reporter activity in response to serum or individual lipids than medium alone. Surprisingly, we also detected similar activity when PPARβ/δ was cotransfected. However, because of the high level of endogenous PPARα activity in the cell line used for this assay, we could not determine its role in lipid-dependent activation. Taken together, these data suggest that at least two PPAR isoforms can contribute to the response to serum or serum lipid components.
Figure 6.
Human serum lipids activate peroxisome proliferator-activated receptor γ (PPARγ). (a) Human serum (HS) activates PPARγ. HCT-116 colon cancer cells were transiently transfected with a DR1- PPAR-response element (PPRE)-luciferase reporter construct and cultured for 18 hr in the presence of medium alone or in the presence of increasing concentrations of human serum (1–10%) to assess activation of PPARγ. Cells were then lysed and luciferase activity was measured. Fold induction was calculated by comparing the ratio of activity in human serum-treated cells compared with medium control cells. Note that increasing the percentage of human serum (from 1 to 10%) led to increased PPARγ activity. (b) Human serum polar lipid fractions activate PPARγ. HCT-116 cells transfected with the PPRE-luciferase construct were cultured as described above in the presence of medium (negative control), the non-polar human serum chloroform fraction, more polar human serum acetone and methanol fractions, or the PPARγ agonist troglitazone. Cells were then assayed for luciferase activity. Note that the more polar human serum methanol and acetone eluted fractions caused approximately a twofold induction of luciferase activity while the non-polar chloroform-eluted human serum had no effect upon activation of PPARγ. (c) Lysophosphatidic acid (LPA) and cardiolipin (CL) activate PPARγ. LPA and CL, which inhibit DC expression of group 1 CD1 molecules, were assayed for PPARγ activity as described above. Troglitazone was used as a positive control. Note that both CL and LPA led to increased induction of PPARγ activity. Results represent triplicate samples ± standard error of the mean (SEM). (d) Cotransfection of the PPRE luciferase reporter with plasmids expressing PPARβ/δ, PPARγ, or empty vector. Transfected cells were treated with the indicated compounds for 24 hr prior to luciferase assays. Error bars are the SEM for triplicate samples. A two-way analysis of variance (ANOVA) was performed comparing PPAR-expressing plasmids with the vector controls for each treatment group. The statistical significance is indicated: *, P < 0·05; **, P < 0·01; ***, P < 0·001. (e) Agarose gel showing specific bands amplified from mRNA derived from whole blood monocytes (lane 1) and the same cell preparation after 3 days in culture using standard media containing fetal bovine serum (FBS) with granulocyte–macrophage colony-stimulating factor (GM-CSF) and interleukin (IL)-4 (lane 2). (f, g) Real-time polymerase chain reaction of human DC mRNA using probes for (f) human PPARγ angiopoietin related (PGAR) and (g) human fatty acid binding protein 4 (aP2). For panels (f) and (g), the * symbol indicates P < 0·05 using a one-way analysis of variance (ANOVA) test for each treatment compared to the value for each gene in monocytes. RGA, rosiglitazone.
We next demonstrated that all of the PPAR genes (PPARα, PPARβ/δ and PPARγ) were up-regulated in iDCs following the standard 3-day culture of CD14+ monocytes in FBS medium with GM-CSF and IL-4 (Fig. 6e). In addition, co-incubation of cells with human serum or rosiglitazone did not significantly alter PPAR mRNA levels (data not shown). These data confirm the expression of PPAR under the culture conditions used in this study for in vitro DC differentiation and are consistent with several other previously published reports.17–19 To determine if PPARγ was active in the iDCs, we determined the relative levels of mRNA using real-time PCR for two prominent target genes [PPARγ angiopoietin related (PGAR) and fatty acid binding protein 4 (aP2)] which are known to be up-regulated in response to PPARγ activation in iDCs. Figure 6(f and g) shows a significant increase in the mRNA levels for both genes, particularly for the aP2 gene, which codes for a fatty acid binding protein. These data support the findings of the PPRE reporter experiments above (Fig. 6a–d) indicating that PPARγ is activated and exhibits its normal functional activity by up-regulating known PPARγ target genes.
To confirm that the modulation of CD1 expression by DCs in the presence of human serum is dependent upon activation of PPARγ, we utilized the PPARγ antagonist GW9662, which predominantly blocks the activity of PPARγ. FBS-DCs and HS-DCs were derived in the absence or presence of GW9662 during 3-day culture in IL-4 and GM-CSF. Cell surface expression of CD1 was then assessed by flow cytometry. As observed previously, HS-DCs expressed little CD1a, CD1b or CD1c at the cell surface when compared with FBS-DCs, while the level of CD1d was slightly up-regulated in the presence of human serum. Importantly, the presence of GW9662 partially restored expression of group 1 CD1 on HS-DCs (Fig. 7a; CD1a is shown), and decreased CD1d expression on HS-DCs to control levels (Fig. 7b). The inability of GW9662 to fully restore CD1a expression levels may reflect the contribution of other PPARs (Fig. 6d) or non-PPAR-dependent pathways. These data support the theory that CD1 expression on iDCs is modulated by prominent serum lipid components, and the presence of these lipids during cytokine-induced DC differentiation correlates with the activation of PPAR transcription factors.
Figure 7.
The peroxisome proliferator-activated receptor γ (PPARγ) antagonist GW9662 partially reverses the effect of human serum upon dendritic cell (DC) CD1 expression. Fetal bovine serum (FBS)-DCs or human serum (HS)-DCs were generated as previously described in the presence or absence of the PPARγ antagonist GW9662 (10 μm) for 3 days. Cells were then stained with monoclonal antibody (mAb) against CD1a and CD1d and analysed by flow cytometry. (a) The presence of GW9662 had no effect upon FBS-DC CD1a expression but partially reversed the inhibition of CD1a in HS-DCs. (b) GW9662 had no effect upon FBS-DC CD1d expression but reversed the up-regulation of CD1d in HS-DCs. These results are representative of three independent experiments using different DC donors.
Discussion
The links between lipid metabolism and the immune system are becoming an area of increasing research interest.36 This interest is a result of an increased appreciation of the role that chronic inflammation appears to play in the aetiology of a number of important human diseases including diabetes, atherosclerosis and cancer. In this study, we have described the ability of the serum lipids LPA and CL to modulate human DC CD1 expression and function. The presence of these endogenous lipids inhibits the transcription and subsequent cell surface expression of group 1 CD1a, CD1b and CD1c lipid antigen-presenting molecules by DCs, which reduces the ability of the DCs to activate CD1-restricted T cells. A range of molecules present in normal serum are likely to contribute to the modulation of DC function. However, of the molecules we tested, CL and LPA had the most potent bioactivity. Although CL is primarily known as an intracellular lipid enriched in mitochondrial membranes, it is also present in normal human serum at a concentration of approximately 15 μm.37 LPA is also present in the micromolar range in normal serum, and has been well characterized as a bioactive lipid that promotes cell growth and activation via interaction with specific LPA receptors.38 Our results suggest that these lipids may also have anti-inflammatory effects by controlling the expression of group 1 CD1 molecules on DCs.
LPA has recently been shown to act as an agonist of the transcriptional regulator PPARγ.24,39 A number of detailed studies have implicated PPARγ in the regulation of CD1 expression and, more broadly, in the modulation of the DC phenotype.17–19 This effect on CD1 expression was demonstrated previously by Szatmari and coworkers, who showed that DCs derived in the presence of the PPARγ agonist rosiglitazone expressed increased cell surface levels of CD1d and promoted expansion of human invariant NKT cells.17 A more recent study by the same authors has implicated the activation of retinoic acid (RA) synthesis genes by the PPARγ transcription factor with downstream activation of RARα by RA as the regulatory cascade that subsequently modulates CD1d expression.18 A direct interaction of the RXR/RARα receptor dimer with the CD1 promoter regions has not been described. However, activation of PPARγ by chemical agonists such as thiazolidinediones or human serum appears to be an early step in regulating DC function in these in vitro differentiation systems.
The data presented here indicate that PPARγ is activated in response to the lipid molecules CL and LPA in a manner analogous to its activation by PPARγ synthetic ligands.40 However, we have not determined whether these lipids interact directly with the ligand binding domain (LBD) of the PPARs. Thus, it remains unclear whether there is a direct interaction of the PPAR receptor with the lipid ligand or whether the effect is an indirect one resulting from, for example, a secondary signal derived from the activation of an upstream pathway. In support of this possibility, it is well known that LPA has a broad range of effects on cellular function and metabolism.41 Hence, cross-talk between PPAR and other signalling pathways is possible. However, previous biochemical data have shown that LPA can bind to the LBD of PPARγ and can function as an agonist.24 Interestingly, a single cleavage of CL can result in the generation of LPA, which may explain the equally potent activity of the two molecules. Therefore, a direct interaction between LPA and PPARγ would also be consistent with the current data.
Our studies provide insights into the regulatory control of CD1a, CD1b and CD1c expression by DCs. Modulation of CD1 expression and DC functional phenotype by self-lipids may provide the host with a mechanism by which to tightly regulate the steady-state levels of CD1 by DCs. This, in turn, directly influences the activation of CD1-restricted T cells. The regulation of CD1 expression and function may have implications in the treatment of human inflammatory diseases in which CD1 responses play a pathogenic role. The use of serum lipid profiles as risk factors for cardiovascular disease and diabetes is an area of active research. Moreover, there is mounting evidence of the role of chronic inflammation and the immune system in the pathogenesis of these diseases. Our findings suggest that specific endogenous lipid molecules can reversibly modulate the activation state of DCs and alter the subsequent inflammatory response. Translation of these findings to the in vivo setting may point to novel therapeutic strategies that could alter DC function for treatment of chronic inflammatory diseases.
Acknowledgments
This research was supported by the Arthritis National Research Foundation (D.S.L.), an ARA Award to D.S.L. sponsored by Pfizer, Inc., the National Institutes of Health (K08AR02171 to D.S.L.), the Medical Research Council (G9901077 and G0400421 to G.S.B.), and the Wellcome Trust (07221/Z/03/Z to G.S.B.).
References
- 1.Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–90. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 2.Barral DC, Brenner MB. CD1 antigen presentation: how it works. Nat Rev Immunol. 2007;7:929–41. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
- 3.Sugita M, van Der Wel N, Rogers RA, Peters PJ, Brenner MB. CD1c molecules broadly survey the endocytic system. Proc Natl Acad Sci USA. 2000;97:8445–50. doi: 10.1073/pnas.150236797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sieling PA, Jullien D, Dahlem M, Tedder TF, Rea TH, Modlin RL, Porcelli SA. CD1 expression by dendritic cells in human leprosy lesions: correlation with effective host immunity. J Immunol. 1999;162:1851–8. [PubMed] [Google Scholar]
- 5.Garcia-Plata D, Lessana-Leibowitch M, Palangie A, Gulliemette J, Sedel D, Mendez L, Mozos E. Immunophenotype analysis of dendritic cells and lymphocytes associated with cutaneous malignant melanomas. Invasion Metastasis. 1995;15:125–34. [PubMed] [Google Scholar]
- 6.Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4– CD8– cytolytic T lymphocytes. Nature. 1989;341:447–50. doi: 10.1038/341447a0. [DOI] [PubMed] [Google Scholar]
- 7.Spada FM, Borriello F, Sugita M, Watts GF, Koezuka Y, Porcelli SA. Low expression level but potent antigen presenting function of CD1d on monocyte lineage cells. Eur J Immunol. 2000;30:3468–77. doi: 10.1002/1521-4141(2000012)30:12<3468::AID-IMMU3468>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 8.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. 1994;372:691–4. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 9.Beckman EM, Melian A, Behar SM, et al. CD1c restricts responses of mycobacteria-specific T cells. Evidence for antigen presentation by a second member of the human CD1 family. J Immunol. 1996;157:2795–803. [PubMed] [Google Scholar]
- 10.Moody DB, Reinhold BB, Guy MR, et al. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science. 1997;278:283–6. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- 11.Dascher CC, Brenner MB. CD1 antigen presentation and infectious disease. In: Herwald H, editor. Host Response Mechanisms in Infectious Disease. Vol. 10. Basel: Karger; 2003. pp. 164–82. [DOI] [PubMed] [Google Scholar]
- 12.Vincent MS, Leslie DS, Gumperz JE, Xiong X, Grant EP, Brenner MB. CD1-dependent dendritic cell instruction. Nat Immunol. 2002;3:1163–8. doi: 10.1038/ni851. [DOI] [PubMed] [Google Scholar]
- 13.Leslie DS, Vincent MS, Spada FM, Das H, Sugita M, Morita CT, Brenner MB. CD1-mediated gamma/delta T cell maturation of dendritic cells. J Exp Med. 2002;196:1575–84. doi: 10.1084/jem.20021515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sieling PA, Porcelli SA, Duong BT, Spada F, Bloom BR, Diamond B, Hahn BH. Human double-negative T cells in systemic lupus erythematosus provide help for IgG and are restricted by CD1c. J Immunol. 2000;165:5338–44. doi: 10.4049/jimmunol.165.9.5338. [DOI] [PubMed] [Google Scholar]
- 15.De Libero G, Donda A, Gober HJ, Manolova V, Mazorra Z, Shamshiev A, Mori L. A new aspect in glycolipid biology: glycosphingolipids as antigens recognized by T lymphocytes. Neurochem Res. 2002;27:675–85. doi: 10.1023/a:1020280201809. [DOI] [PubMed] [Google Scholar]
- 16.Roura-Mir C, Catalfamo M, Cheng TY, Marqusee E, Besra GS, Jaraquemada D, Moody DB. CD1a and CD1c activate intrathyroidal T cells during Graves’ disease and Hashimoto’s thyroiditis. J Immunol. 2005;174:3773–80. doi: 10.4049/jimmunol.174.6.3773. [DOI] [PubMed] [Google Scholar]
- 17.Szatmari I, Gogolak P, Im JS, Dezso B, Rajnavolgyi E, Nagy L. Activation of PPARgamma specifies a dendritic cell subtype capable of enhanced induction of iNKT cell expansion. Immunity. 2004;21:95–106. doi: 10.1016/j.immuni.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Szatmari I, Pap A, Ruhl R, et al. PPARgamma controls CD1d expression by turning on retinoic acid synthesis in developing human dendritic cells. J Exp Med. 2006;203:2351–62. doi: 10.1084/jem.20060141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szatmari I, Torocsik D, Agostini M, Nagy T, Gurnell M, Barta E, Chatterjee K, Nagy L. PPARγ regulates the function of human dendritic cells primarily by altering lipid metabolism. Blood. 2007;110:3271–80. doi: 10.1182/blood-2007-06-096222. [DOI] [PubMed] [Google Scholar]
- 20.Feige JN, Gelman L, Michalik L, Desvergne B, Wahli W. From molecular action to physiological outputs: peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Prog Lipid Res. 2006;45:120–59. doi: 10.1016/j.plipres.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Camp HS, Tafuri SR, Leff T. c-Jun N-terminal kinase phosphorylates peroxisome proliferator-activated receptor-gamma1 and negatively regulates its transcriptional activity. Endocrinology. 1999;140:392–7. doi: 10.1210/endo.140.1.6457. [DOI] [PubMed] [Google Scholar]
- 22.Straus DS, Glass CK. Anti-inflammatory actions of PPAR ligands: new insights on cellular and molecular mechanisms. Trends Immunol. 2007;28:551–8. doi: 10.1016/j.it.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, Baker DL, Yasuda S, et al. Lysophosphatidic acid induces neointima formation through PPARgamma activation. J Exp Med. 2004;199:763–74. doi: 10.1084/jem.20031619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McIntyre TM, Pontsler AV, Silva AR, et al. Identification of an intracellular receptor for lysophosphatidic acid (LPA): LPA is a transcellular PPARgamma agonist. Proc Natl Acad Sci USA. 2003;100:131–6. doi: 10.1073/pnas.0135855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med. 2002;195:625–36. doi: 10.1084/jem.20011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porcelli S, Morita CT, Brenner MB. CD1b restricts the response of human CD4-8-T lymphocytes to a microbial antigen. Nature. 1992;360:593–7. doi: 10.1038/360593a0. [DOI] [PubMed] [Google Scholar]
- 27.Folch J, Lebaron FN. The chemistry of the phosphoinositides. Can J Biochem Physiol. 1956;34:305–19. [PubMed] [Google Scholar]
- 28.Sugita M, Grant EP, van Donselaar E, Hsu VW, Rogers RA, Peters PJ, Brenner MB. Separate pathways for antigen presentation by CD1 molecules. Immunity. 1999;11:743–52. doi: 10.1016/s1074-7613(00)80148-x. [DOI] [PubMed] [Google Scholar]
- 29.Cao X, Sugita M, Van Der Wel N, Lai J, Rogers RA, Peters PJ, Brenner MB. CD1 molecules efficiently present antigen in immature dendritic cells and traffic independently of MHC class II during dendritic cell maturation. J Immunol. 2002;169:4770–7. doi: 10.4049/jimmunol.169.9.4770. [DOI] [PubMed] [Google Scholar]
- 30.Roura-Mir C, Wang L, Cheng TY, et al. Mycobacterium tuberculosis regulates CD1 antigen presentation pathways through TLR-2. J Immunol. 2005;175:1758–66. doi: 10.4049/jimmunol.175.3.1758. [DOI] [PubMed] [Google Scholar]
- 31.Mueller E, Sarraf P, Tontonoz P, et al. Terminal differentiation of human breast cancer through PPAR gamma. Mol Cell. 1998;1:465–70. doi: 10.1016/s1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- 32.Calabi F, Jarvis JM, Martin L, Milstein C. Two classes of CD1 genes. Eur J Immunol. 1989;19:285–92. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- 33.Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–8. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 34.Szatmari I, Torocsik D, Agostini M, Nagy T, Gurnell M, Barta E, Chatterjee K, Nagy L. PPARgamma regulates the function of human dendritic cells primarily by altering lipid metabolism. Blood. 2007;110:3271–80. doi: 10.1182/blood-2007-06-096222. [DOI] [PubMed] [Google Scholar]
- 35.Kliewer SA, Sundseth SS, Jones SA, et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc Natl Acad Sci USA. 1997;94:4318–23. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–7. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 37.Deguchi H, Fernandez JA, Hackeng TM, Banka CL, Griffin JH. Cardiolipin is a normal component of human plasma lipoproteins. Proc Natl Acad Sci USA. 2000;97:1743–8. doi: 10.1073/pnas.97.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anliker B, Chun J. Cell surface receptors in lysophospholipid signaling. Semin Cell Dev Biol. 2004;15:457–65. doi: 10.1016/j.semcdb.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Faveeuw C, Fougeray S, Angeli V, et al. Peroxisome proliferator-activated receptor gamma activators inhibit interleukin-12 production in murine dendritic cells. FEBS Lett. 2000;486:261–6. doi: 10.1016/s0014-5793(00)02319-x. [DOI] [PubMed] [Google Scholar]
- 40.Nencioni A, Grunebach F, Zobywlaski A, Denzlinger C, Brugger W, Brossart P. Dendritic cell immunogenicity is regulated by peroxisome proliferator-activated receptor gamma. J Immunol. 2002;169:1228–35. doi: 10.4049/jimmunol.169.3.1228. [DOI] [PubMed] [Google Scholar]
- 41.Tigyi G, Parrill AL. Molecular mechanisms of lysophosphatidic acid action. Prog Lipid Res. 2003;42:498–526. doi: 10.1016/s0163-7827(03)00035-3. [DOI] [PubMed] [Google Scholar]