Abstract
The peroxisome proliferator-activated receptor γ (PPARγ) is expressed in macrophages and plays an important role in suppressing the inflammatory response. Lipopolysaccharides (LPS), which activate Toll-like receptor 4 (TLR4), reduced PPARγ expression and function in peritoneal macrophages and macrophage cell lines. Moreover, pretreatment with the synthetic PPARγ ligand, rosiglitazone did not prevent LPS-mediated downregulation of PPARγ. Inhibition of PPARγ expression was not blocked by cycloheximide, indicating that de novo protein synthesis is not required for LPS-mediated suppression of PPARγ. Destabilization of PPARγ messenger RNA (mRNA) was not observed in LPS-stimulated macrophages, suggesting that LPS regulates the synthesis of PPARγ mRNA. LPS had no effect on PPARγ expression in macrophages from TLR4 knockout mice, whereas LPS inhibited PPARγ expression in cells that had been reconstituted to express functional TLR4. Targeting the TLR4 pathway with inhibitors of MEK1/2, p38, JNK and AP-1 had no effect on PPARγ downregulation by LPS. However, inhibitors that target NEMO, IκB and NF-κB abolished LPS-mediated downregulation of PPARγ in LPS-stimulated macrophages. Our data indicate that activation of TLR4 inhibits PPARγ mRNA synthesis by an NF-κB-dependent mechanism. Low-density genomic profiling of macrophage-specific PPARγ knockout cells indicated that PPARγ suppresses inflammation under basal conditions, and that loss of PPARγ expression is sufficient to induce a proinflammatory state. Our data reveal a regulatory feedback loop in which PPARγ represses NF-κB-mediated inflammatory signalling in unstimulated macrophages; however, upon activation of TLR4, NF-κB drives down PPARγ expression and thereby obviates any potential anti-inflammatory effects of PPARγ in LPS-stimulated macrophages.
Keywords: inflammatory, macrophage, nuclear factor-κB, Peroxisome proliferator-activated receptor γ, Toll-like receptor
Introduction
Peroxisome proliferator-activated receptor γ (PPARγ) is a nuclear receptor implicated in the control of diverse diseases such as type II diabetes, atherosclerosis and inflammatory bowel disease. PPARγ has been studied in the greatest detail in adipocytes, where it plays a key role in glucose homeostasis and adipocyte differentiation.1,2 It is also expressed in macrophages, where its activation regulates lipid metabolism.3–6 Macrophage PPARγ has recently been implicated in muscle and hepatic insulin sensitivity7,8 and in inflammation.9
PPARγ has been clinically exploited as the target of the thiazolidinedione (TZD) class of drugs for the treatment of type II diabetes. Recently, PPARγ has emerged as a potential target for treatment of inflammatory diseases such as ulcerative colitis, atherosclerosis, asthma and rheumatoid arthritis. In particular, TZDs have been recognized for their ability to reduce inflammatory gene expression in macrophages and, when administered before the onset of inflammation, TZDs exhibit beneficial effects on experimental models of inflammation such as colitis,10–15 atherosclerosis,16–19 asthma,20–22 psoriasis,23 myocarditis24,25 and allergic encephalomyelitis.26,27 The extent of regulation of inflammation by TZDs has differed among reports because of differences in the concentration, duration and identity of the TZDs (rosiglitazone, pioglitazone) employed, as well as the nature and dosage of the agents that were employed to induce inflammation.9 Collectively, it is clear that TZDs can reduce inflammation in both a PPARγ-dependent and PPARγ-independent manner, with the latter resulting from the use of high concentrations.5,9,28 Moreover, myeloid-specific PPARγ knockout animals have clearly demonstrated an anti-inflammatory role of PPARγ in macrophages. Deletion of macrophage PPARγ increases proinflammatory gene expression and leads to increased susceptibility to experimental models of ulcerative colitis29 and atherosclerosis.4 Overall, studies in vitro and in vivo demonstrate the importance of macrophage PPARγ in regulating inflammation.
While targeting PPARγ shows promise in suppressing inflammation in cell lines and experimental models, the therapeutic efficacy of TZDs in the treatment of established inflammation is less clear. For example, preventive administration of TZDs provides beneficial effects in experimental models of ulcerative colitis, but provides little or no value when given after the onset of the disease.10,12–15 Similar results were observed in a small open-ended clinical trial in which patients with moderate colitis receiving rosiglitazone experienced only modest improvement.30 Adding to the confusion concerning the potential therapeutic efficacy of PPARγ agonists in the management of acute and/or chronic ulcerative colitis, a recent report indicates that long-term treatment of mice with rosiglitazone exacerbated dextran sodium sulphate (DSS)-induced colitis.31
A rational approach to evaluate the preventive and/or therapeutic potential of PPARγ ligands in ulcerative colitis and other inflammatory conditions requires a better understanding of the cellular, molecular and genomic mechanisms by which PPARγ confers anti-inflammatory properties in different cell types. A central question is framed by the paradoxical observation that PPARγ agonists can inhibit the onset of inflammation but have reduced efficacy after the onset of colitis. A potential clue comes from the work of Katayama et al., who determined that adenovirus-mediated PPARγ gene delivery into the colon restored responsiveness to PPARγ ligands after the induction of colitis.13 This observation suggests that loss of PPARγ function may attend inflammation, and that this loss of function may be the result of a decrease in PPARγ expression. This hypothesis is consistent with observations that PPARγ expression appears to be downregulated during ulcerative colitis,13 alveolar proteinosis,32 pulmonary sarcoidosis33 and endotoxin-induced acute lung injury34 and that inflammatory mediators such as interferon-γ, interleukin-6 (IL-6), IL-1 and nitric oxide inhibit PPARγ expression in adipocytes, and mesangial and biliary cells.35–39
Activation of Toll-like receptor (TLR) signalling induces nuclear factor-κB (NF-κB) and activating protein 1 (AP-1) activity and the release of proinflammatory molecules such as nitric oxide, cytokines, chemokines and cell surface adhesion molecules. These molecules not only drive the immune response but interfere with the expression of key regulatory factors involved in regulating physiological inflammation.40,41 The studies described below were undertaken to test the hypothesis that activation of macrophage TLRs inhibits PPARγ function in these cells and thereby obviates the homeostatic regulatory and anti-inflammatory properties of PPARγ agonists.
We report here that activation of TLR1/2, 4 and 5 downregulates PPARγ expression in both peritoneal macrophages and macrophage cell lines. We demonstrate that activation of the TLR4 pathway suppresses PPARγ messenger RNA (mRNA) synthesis through an NF-κB-dependent mechanism. Using macrophage-specific PPARγ knockout mice, we illustrate that loss of PPARγ expression leads to an increase in proinflammatory gene expression, thereby demonstrating that inhibition of PPARγ expression is, in and of itself, sufficient to initiate an inflammatory state. Together, our data indicate that cross-talk between PPARγ and TLR4/NF-κB signalling regulates the inflammatory response in macrophages. We propose a model in which PPARγ represses aspects of NF-κB-mediated signalling at homeostasis and/or states of low inflammation; however, during increased inflammatory signalling NF-κB drives down PPARγ expression to cancel its actions. These observations account for the finding that activation of PPARγ before inflammation suppresses the response, whereas PPARγ agonists have little or no effect when administered after the initiation of inflammation, when macrophage PPARγ is no longer expressed.
Experimental procedures
Reagents
Ultra-pure lipopolysaccharide (LPS, Escherichia coli: 0111:B4), peptidoglycan (PGN, Bacillus subtilis), flagellin (Bacillus subtilis) and muramyldipeptide (l isoform) were purchased from Invivogen (San Diego, CA) and dissolved in endotoxin-free water. U0126, SB20380, actinomycin D, cycloheximide, curcumin and SP600125 were purchased from Calbiochem, (San Diego, CA). Bay 11-7085 and ammonium pyrrolidinedithiocarbamate (PDTC) were purchased from Sigma (St Louis, MO). Rosiglitazone maleate was purchased from ChemPacific (Baltimore MD). The I-κ B kinase γ (IKKγ) NEMO binding domain (NBD) inhibitory peptide set was purchased from Imgenex (San Diego, CA).
Cell culture
The transformed macrophage cell line, RAW 264.7, [American Type Culture Collection (ATCC), Manassas, VA], 293 cells (ATCC) and 293/TLR4/MD2-CD14 cells (Invivogen), were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS). The transformed peritoneal macrophage cell line, IC-21 (ATCC) was cultured in RPMI-1640 medium with 10% FBS.
Animals
PPARγfl/fl (PPARgtm2Rev), LysMCre, TLR4lps-del (C57BL/10ScNJ) and C57BL/10J mice were purchased from Jackson Laboratories (Bar Harbor, ME). Homozygous PPARγfl/fl mice contain loxp sites on either side of exons 1 and 2 of the PPARγ gene.3 LysMCre mice contain Cre recombinase expression driven by the lysozyme M promoter specifically in the myelomonocytic lineage.42 TLR4lps-del mice have a deletion of the Tlr4 gene and so have a defective response to LPS. C57BL/10J mice are the controls for TLR4lps-del mice. All mice were maintained as part of an American Association for Accreditation of Laboratory Animal Care facility. Animal experimentation was conducted in accordance with accepted standards of humane animal care according to protocols approved by the Mayo Clinic College of Medicine Institutional Animal Care and Use Committee.
Generation of myeloid-specific PPARγ knockout mice
Our laboratory generated mice with PPARγ expression deleted in myeloid cells by crossing PPARγfl/fl and LysMCre+/+ mice (C57BL/6J background). Mice were interbred to achieve LysMCre−/−/PPARγfl/fl and LysMCre+/−/PPARγfl/fl mice, which were subsequently used in all experiments at the age of 6 weeks. All animals were genotyped by polymerase chain reaction (PCR) using genomic DNA extracted from tail clips using the Extract-n-AMP PCR system (Sigma). The presence of the floxed PPARγ allele was evaluated with a primer set (oIMR1934, 5′-TGTAATGGAAGGGCAAAAGG-3′; 5′-TGGCTTCCAGTGCATAAGTT-3′) that amplifies across the downstream loxp site and yields a 214-base-pair (bp) fragment from wild-type mice and a 250-bp fragment from the floxed PPARγ allele. Mice were characterized for the presence of the LysMCre gene with primers (oIMR3066, 5′-CCCAGAAATGCCAGATTACG-3′; 5′-CTTGGGCTGCCAGAATTTCTC-3′) that amplify the Cre transgene.
Isolation of macrophages
Mice were injected intraperitoneally with 2 ml of 3·5% thioglycollate solution (BD, Spork, MD). Five days following injection, peritoneal macrophages were harvested by injecting 10 ml warm RPMI-1640 containing 10% FBS into the peritoneal cavity and withdrawing with a sterile pipette. The peritoneal exudate was centrifuged at 92·4 g and washed three times with complete media before plating onto 60-mm tissue culture plates. Macrophages were allowed to adhere to culture plates at 37° in an atmosphere of 5% CO2 for 1 hr. Cells were washed vigorously three times with phosphate-buffered saline to remove non-adherent cells. Cultures were maintained overnight in complete RMPI-1640 media to form macrophage monolayers before treatments were added.
Isolation of colon crypts
The colon was dissected from caecum to rectum, flushed with phosphate-buffered saline (4°), and soaked in Hanks’ balanced salt solution (HBSS) without Ca2+ or Mg2+. The caecum and rectum were removed and the colon was longitudinally opened to expose the lumenal surface. Each colon was washed three times for 5 min at room temperature with HBSS containing 25 mm HEPES and 1% FBS. The colon was then shaken vigorously in HBBS–20 mm ethylenedimainetetraacetic acid for 15 min at 37°. Supernatants containing individual crypts were centrifuged at 1000 g for 5 min at 4°. The pellets (pure crypt fraction) were immediately processed for RNA and/or protein.
Dextran sodium sulphate model of colitis
Dextran sodium sulfate (molecular weight 36 000–50 000; MP Biomedicals, Solon, OH) was given ad libitum in filter-purified drinking water to 6-week-old LysMCre−/−/PPARγfl/fl and LysMCre+/−/PPARγfl/fl mice for 7 days according to the well-established procedure of Okayasu et al.43 Water consumption was monitored daily to ensure that all groups consumed an equivalent volume. Body weight of each individual mouse was recorded daily. Weight loss was calculated as the per cent weight loss from days 0 and 7. Statistical analysis was carried out with Mann–Whitney rank sum analysis.
Western blot analysis
Total cell lysates were prepared by briefly sonicating cell lysates prepared in 1 × lysis buffer (Roche, Indianapolis, IN). Protein concentrations were determined using a BioRad protein assay kit (BioRad Laboratories, Hercules, CA). Protein samples were resolved on 10% precast Tris–glycine gels (Invitrogen, Carlsbad, CA) and were electrophoretically transferred to polyvinylidene fluoride membrane. Blots were stained essentially as previously described44,45 with primary antibodies against PPARγ (Santa Cruz Biotechnology, Santa Cruz, CA), and actin (Santa Cruz). Secondary antibodies (depending on the primary antibody used) were horseradish peroxidase-conjugated anti-mouse (Santa Cruz) and anti-rabbit (Jackson Immunoresearch, West Grove, PA). Antigen–antibody complexes were detected with the ECL Plus chemiluminescent system (Amersham Bioscience, Piscataway, NJ) and visualized with film.
Immunohistochemistry
Slides containing 5-μm sections of formalin-fixed and paraffin-embedded mouse colons were processed using routine histochemistry procedures. Following deparaffinization and hydration, slides were unmasked by heat-mediated antigen retrieval using DAKO® Target Retrieval Solution according to the manufacturer’s instructions. Slides were then stained with anti-F4/80 (AbD Serotec, C1:A3-1) using the VectaStain ABC Kit (Vector Laboratories, Burlingame, CA; pK-4004) as detailed by the manufacturer. Immunostaining was detected with the DAKO® Liquid DAB+ Substrate–Chromogen System. Cover glasses were mounted on glass slides using Clarion™ mounting medium (Sigma).
NF-κB DNA binding enzyme-linked immunosorbent assay
Peritoneal macrophage cultures were pretreated for 1 hr with or without NBD, PDTC, or Bay 11-7085 and then incubated with or without LPS for an additional 6 hr. Nuclear extracts were prepared with the Nuclear Extraction Kit (Panomics, Fremont, CA) and protein concentration was quantified using the BioRad Protein Assay. All nuclear extracts were stored at −80° in aliquots until use. Binding of the NF-κB p65 (RelA) subunit to a NF-κB consensus binding site was measured in nuclear extracts (10 μg) using the NF-κB Transcription Factor ELISA Kit (Panomics) as detailed by the manufacturer.
Real-time polymerase chain reaction analysis
The abundance of individual mRNAs was determined with two-step quantitative reverse transcriptase-mediated real-time PCR (qPCR). Equal aliquots of total RNA from samples were converted to cDNA with the High-Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA), and qPCR were performed in triplicate with 10 ng cDNA and the TaqMan® Universal PCR master mix (Applied Biosystems). The primers and probe detecting mPPARγ, mNOS2A, murine tumour necrosis factor-α (mTNF-α), mCD36, mGAPDH, hIL-8 and hGAPDH were purchased from Applied Biosystems. For simultaneous measurement of inflammatory mRNAs, custom TaqMan® Low Density Arrays were purchased through Applied Biosystems. Ninety nanograms of cDNA per sample were loaded into each port. All amplification data were collected with an Applied Biosystems Prism 7900 sequence detector and analysed with Sequence Detection System software (Applied Biosystems). Data were normalized to GAPDH and mRNA abundance was calculated using the ΔΔCT method.46
Results
PPARγ regulation by TLR agonists
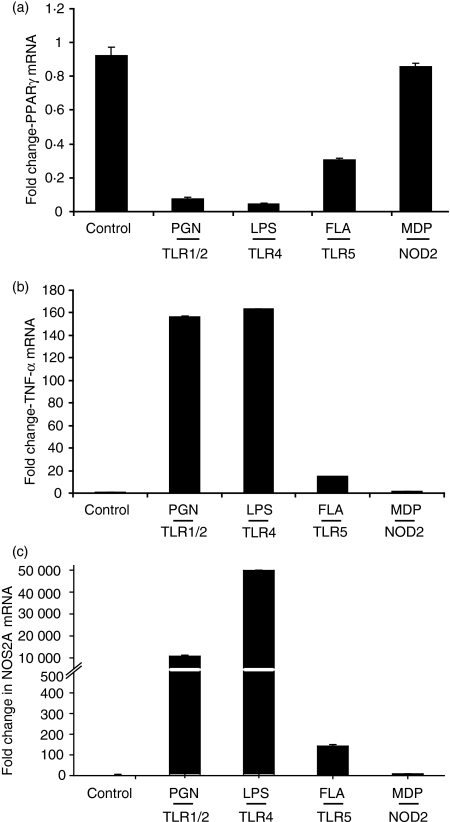
The consequence of TLR signalling activation on PPARγ expression in peritoneal macrophages was evaluated using agonists for TLR1/2, -4, -5 and -9. Figure 1(a) shows that treatment of peritoneal macrophages with agonists for TLR1/2 (PGN) and TLR4 (LPS) repressed PPARγ mRNA, with a > 90% reduction within 6 hr of treatment. Similar findings were observed with other agonists of TLR2 including the synthetic lipoprotein FSL-1 and lipoteichoic acid (LTA) (data not shown). The TLR5 agonist flagellin reduced PPARγ mRNA abundance to a lesser extent (65%), whereas, muramyldipeptide the ligand for nucleotide-binding oligomerization domain containing 2 protein (NOD2) had no significant effect on PPARγ expression. The extent of PPARγ downregulation directly correlated with the level of agonist-induced proinflammatory gene expression, as assessed by the induction of TNF-α (Fig. 1b) and NOS2A (Fig. 1c). These data indicate that TLRs play an important role in controlling the expression of PPARγ in macrophages.
Figure 1.
Toll-like receptor (TLR) ligands and cytokines differentially downregulate peroxisome proliferator-activated receptor γ (PPARγ) expression in peritoneal macrophages. Peritoneal macrophages were stimulated for 6 hr with the TLR ligands: peptidoglycan (PGN; 5 μg/ml), lipopolysaccharide (LPS; 0·1 μm), flagellin (FLA; 5 μg/ml), and muramyldipeptide (MDP; 1 μg/ml). Quantitative polymerase chain reaction was used to measure PPARγ (a), tumour necrosis factor-α (TNF-α) (b) and NOS2A (c) messenger RNA abundance from each sample. PPARγ messenger RNA was normalized to GAPDH and expressed as fold change relative to the control. Each bar represents mean ± SD, n = 3.
LPS-mediated downregulation of macrophage PPARγ
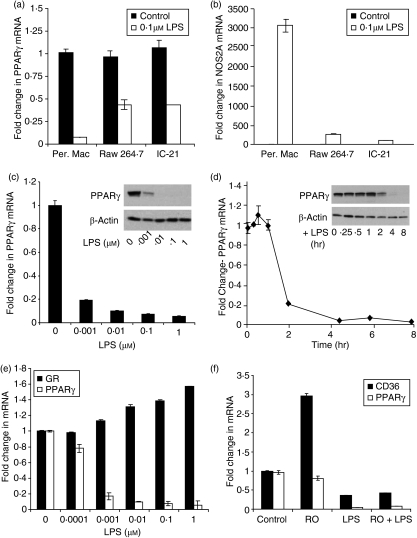
Our initial studies focused on characterizing TLR4-mediated regulation of PPARγ in macrophages. This focus on TLR4 was motivated by the unequivocal importance of TLR4 as a mediator of innate immunity in macrophages and the number of studies that link TLR4 to susceptibility to inflammatory bowel disease.47–50Figure 2(a) illustrates that LPS downregulated PPARγ mRNA expression in thioglycollate-elicited peritoneal macrophages as well as the macrophages cell lines, RAW 264.7 and IC-21. Downregulation was greatest in the primary macrophages with > 90% reduction within 6 hr, whereas 50% inhibition of PPARγ mRNA expression was observed in the established cell lines. LPS also reduced PPARγ expression in non-thioglycollate-elicited (resting) macrophages by > 90% within 6 hr (data not shown). The extent of PPARγ regulation correlated with induction of the classical LPS target gene, NOS2A, which was higher in primary macrophages than in the cell lines (Fig. 2b). The reduction in LPS activity in the macrophage cell lines likely reflects their transformed nature. It is important to note that the origins of the macrophage cell lines are distinct. Raw 264.7 are tumour-derived macrophages from BALB/c mice, whereas IC-21 cells are peritoneal macrophages from C57BL/6J mice. These results indicate that LPS can downregulate PPARγ expression in both normal and tumour macrophages.
Figure 2.
Lipopolysaccharide (LPS) regulates peroxisome proliferator-activated receptor γ (PPARγ) expression and activity in macrophages. (a,b) Quantitative polymerase chain reaction (qPCR) was used to measure PPARγ (a) and NOS2A (b) messenger RNA (mRNA) in peritoneal macrophages and the macrophage cell lines RAW 264.7 and IC-21 treated with or without 0·1 μm LPS for 6 hr. (c) PPARγ mRNA abundance was measured by qPCR in peritoneal macrophages treated with varying concentrations of LPS for 6 hr. Inset, 20 μg of total lysate was resolved, and Western blotting used to stain for PPARγ and β-actin. (d) PPARγ mRNA expression was measured in macrophages exposed to increasing time of LPS. Inset, 20 μg of total lysate was analysed by Western blot for PPARγ and β-actin. (e) PPARγ and glucocorticoid receptor (GR) mRNA abundance was quantified by qPCR in macrophages treated with increasing concentrations of LPS. Data are expressed as fold change from control for each individual probe. (f) Cultures of macrophages pretreated with vehicle or 1 μm rosiglitazone for 2 hr were incubated ± LPS for 24 hr, and PPARγ and CD36 mRNA abundance was measured by qPCR. In all experiments, mRNA abundance was normalized to GAPDH and represents the mean ± SD, n = 3.
The LPS reduced both PPARγ mRNA and protein in a concentration-dependent and time-dependent manner. Maximum inhibition was observed when cells were treated for 6 hr with 0·01–0·1 μm LPS (Fig. 2c). Maximum inhibition of > 90% occurred within 5 hr of treatment (Fig. 2d). LPS specifically downregulated PPARγ mRNA, but had no significant effect on expression of the glucocorticoid receptor, a well-known anti-inflammatory mediator (Fig. 2e). Reduction in PPARγ expression by LPS treatment should translate to the loss of transcriptional regulation of bona fide PPARγ target genes in response to PPARγ ligands. The TZD PPARγ agonist rosiglitazone induced the well-characterized PPARγ target gene, CD36,5 in untreated peritoneal macrophages (Fig. 2f). However, induction of CD36 was inhibited by LPS, which was consistent with the LPS-mediated decrease in PPARγ mRNA and protein expression. LPS also inhibited basal expression of CD36, which is known to be affected by PPARγ. Moreover, pretreatment with rosiglitazone failed to block LPS-mediated downregulation of PPARγ (Fig. 2f). Collectively, our data provide compelling evidence that LPS negatively regulates the expression and function of PPARγ in macrophages.
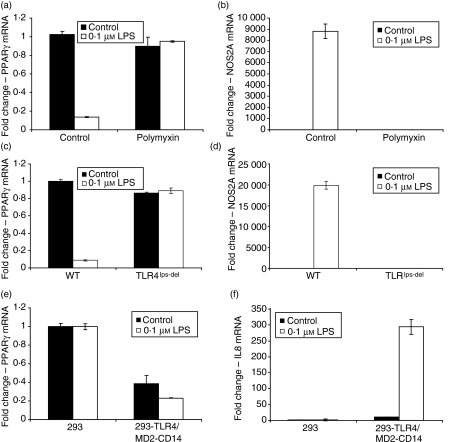
Activation of the TLR4 pathway by LPS has been studied in detail. Although highly purified LPS was employed in our studies, we confirmed that the observed effect of LPS on PPARγ expression was mediated specifically through TLR4 and was not the result of a contaminant that might activate some other pathway. First, cotreatment of primary macrophages with the LPS antagonist polymyxin blocked both downregulation of PPARγ (Fig. 3a) and induction of NOS2A (Fig. 3b). Furthermore, LPS inhibition of PPARγ (Fig. 3c) and induction of NOS2A (Fig. 3d) was not observed in macrophages from TLR4 knockout mice. LPS also inhibited PPARγ expression in human embryonic kidney cells that have been engineered to stably overexpress a functional TLR4 pathway (HEK293/TLR4/MD2-CD14), whereas LPS had no effect on PPARγ expression in wild-type HEK293 cells, which do not express TLR4 (Fig. 3e). The overall level of LPS-mediated reduction in PPARγ in the TLR4-responsive HEK293 cells was similar to that observed in the transformed macrophage cell lines, but was less robust than the response observed in primary peritoneal macrophages. The weaker response in the HEK293/TLR4/MD2-CD14 cells may result from intrinsic differences in TLR4 signalling among epithelial and macrophage cell types. For example, LPS treatment of TLR4-responsive HEK293 cells does not induce NOS2A (data not shown), and TLR4 activity in such cells in response to LPS was measured by IL-8 induction (Fig. 3f). Nevertheless, these data indicate that a reconstituted TLR4 signalling pathway inhibits PPARγ expression, irrespective of the cellular background. Collectively, these data demonstrate that the effect of LPS on PPARγ expression is specifically mediated through TLR4.
Figure 3.
Downregulation of peroxisome proliferator-activated receptor γ (PPARγ) by lipopolysaccharide (LPS) is Toll-like receptor 4 (TLR4) dependent. (a,b) Quantitative polymerase chain reaction (qPCR) was used to measure PPARγ (a) and NOS2A (b) abundance in primary macrophage cultures treated with vehicle and the LPS antagonist polymyxin (50 μg/ml) ± LPS for 6 hr. (c,d) Macrophages from wild-type mice expressing TLR4 (C57BL/10J) and TLR4 knockout mice (TLR4lps-del, C57BL/10ScNJ) were stimulated with vehicle or LPS for 6 hr. The qPCR was used to measure PPARγ (c) and NOS2A (d) messenger RNA (mRNA) abundance. (e,f) Wild-type 293 cells lacking TRL4 expression and 293 cells engineered to express TLR4, MD2, and CD14 (293/TLR4/MD2-CD14) were exposed to vehicle or LPS for 6 hr and expression of PPARγ (e) and interleukin-8 (IL-8) (f) mRNA was determined by qPCR. In all experiments, mRNA was normalized to GAPDH and expressed as fold change relative to the control. Each bar represents mean ± SD, n = 3.
LPS inhibits macrophage PPARγ mRNA synthesis
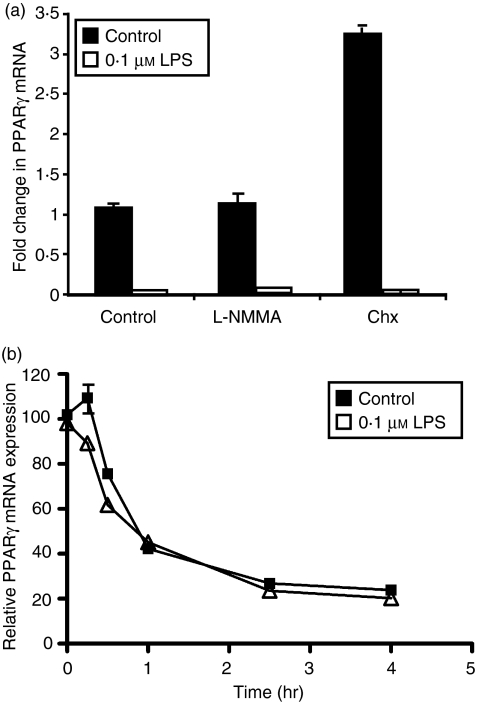
Activation of macrophages through TLR4 signalling leads to rapid induction of proinflammatory cytokines such as IL-6, IL-1β and interferon-γ, in addition to NOS2A, resulting in synthesis of proinflammatory nitric oxide. These inflammatory agents have been reported to affect PPARγ expression in other cell types,35–39 but not in macrophages. The role of nitric oxide was evaluated to determine if downregulation of PPARγ occurs secondary to the massive LPS-mediated induction of NOS2A and the resultant increase in nitric oxide, which is well-known to inactivate transcription factors. However, the nitric oxide inhibitor l-NG-monomethyl arginine citrate (l-NNMA) had no effect on LPS regulation of PPARγ mRNA in LPS-treated macrophages (Fig. 4a).
Figure 4.
Lipopolysaccharide (LPS)-mediated downregulation of peroxisome proliferator-activated receptor γ (PPARγ) occurs at the promoter level. (a) Macrophages were exposed to vehicle, the nitric oxide inhibitor l-NNMA (500 μm), and the protein synthesis inhibitor cycloheximide (5 μg/ml) ± LPS for 6 hr. Quantitative polymerase chain reaction (qPCR) was used to measure PPARγ messenger RNA (mRNA) abundance. (b) Macrophage cultures were pretreated with LPS for 1 hr and then subsequently exposed to actinomycin D (5 μg/ml) for the indicated times. At each time-point, RNA was harvested and PPARγ expression was determined by qPCR. In all experiments, the abundance of PPARγ mRNA was normalized to GAPDH and expressed as fold change relative to the control. Each bar represents mean ± SD, n = 3.
Furthermore, treatment of primary macrophages with the protein synthesis inhibitor cycloheximide had no effect on LPS-mediated inhibition of PPARγ expression (Fig. 4a). These data illustrate that de novo protein synthesis is not required for LPS-mediated downregulation of PPARγ, confirming our conclusion that this process is not a secondary consequence of induction of NOS2A. Furthermore, the observation that protein synthesis is not required for downregulation of PPARγ also eliminates the possibility that LPS-mediated induction of cytokines is required for suppression of PPARγ expression.
The half-life of PPARγ mRNA was determined in primary macrophages after the addition of the RNA synthesis inhibitor actinomycin D (Fig. 4b). The apparent rates of PPARγ mRNA degradation were similar in untreated and LPS-treated macrophages, indicating that LPS has no significant effect on PPARγ mRNA stability. These data indicate that regulation of PPARγ mRNA expression by LPS is the result of a direct effect of LPS signalling (i.e. does not require de novo protein synthesis) and that LPS has no effect on the rate of mRNA degradation. The simplest explanation of these data is that LPS activates a transcription factor that represses PPARγ transcription.
Activation of NF-κB signalling downregulates PPARγ expression
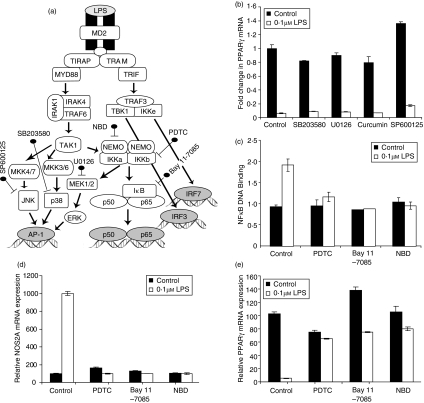
Our studies next focused on the molecular mechanism that links TLR4 activation by LPS to inhibition of PPARγ mRNA synthesis. TLR4 signalling can lead to activation of the proinflammatory transcription factor AP-1 through MEK1/2-, p38 MAPK- and JNK-dependent phosphorylation; whereas IKK-dependent processes lead to NF-κB activation,40,41 as illustrated in Fig. 5(a). Inhibitors of p38 MAPK (SB203580), MEK1/2 (U0126), JNK (SP600125) and AP-1 (curcumin) were used to determine if any of these pathways is required for LPS-mediated regulation of PPARγ. Exposure of peritoneal macrophages to these inhibitors for 6 hr had no effect on PPARγ regulation by LPS (Fig. 5b), indicating that TLR4-dependent activation of AP-1 is not involved in downregulation of PPARγ expression by LPS.
Figure 5.
Nuclear factor-κB (NF-κB) signalling mediates downregulation of peroxisome proliferator-activated receptor γ (PPARγ) expression by lipopolysaccharide (LPS). (a) The Toll-like receptor 4 (TLR4) signalling pathway activates AP-1 and NF-κB through MEK1/2-, p38-, JNK- and IKK-dependent processes. Chemical inhibitors targeting these proteins within each axis are shown in bold. (b) Peritoneal macrophage cultures were pretreated for 1 hr with inhibitors of the MEK/JNK/AP-1 signalling axis that include p38 (SB203580, 20 μm), MEK1/2 (U0126, 10 μm), AP-1 (curcumin, 20 μm), JNK (SP600125, 10 μm) and subsequently exposed to 6 hr ± LPS. Quantitative polymerase chain reaction (qPCR) was used to measure PPARγ mRNA abundance from each sample. (c–e) Cultures of peritoneal macrophages were pretreated for 1 hr with inhibitors upstream of NF-κB that include IKK (Bay 11-7085, 5 μm), IκB (NBD, 100 μm), and NF-κB (PDTC, 100 μm), and then exposed to an additional 6 hr ± LPS. NF-κB activity was determined by measurement of NF-κB DNA binding activity (c) and induction of NOS2A mRNA. Expression of NOS2A (d) and PPARγ mRNA (e) was determined by qPCR. All data are expressed as fold change relative to the control. Each bar represents mean ± SD, n = 3.
We next evaluated the role of NF-κB in LPS-regulated PPARγ expression. To this end, we determined the effect of inhibitors that target NEMO (NBD, NEMO binding domain), the IKK complex (PDTC) and IκB (Bay 11-7085), upstream components required for the activation of NF-κB (Fig. 5a). The efficacy of the inhibitors is shown through inhibition of LPS-induced NF-κB DNA binding activity (Fig. 5c) and upregulation of NOS2A levels (Fig. 5d). Each of these NF-κB inhibitors effectively blocked LPS-mediated downregulation of PPARγ (Fig. 5e). Collectively, these results indicate that activation of NF-κB by TLR4 signalling mediates the downregulation of macrophage PPARγ expression.
Genomic consequence of PPARγ loss in macrophages
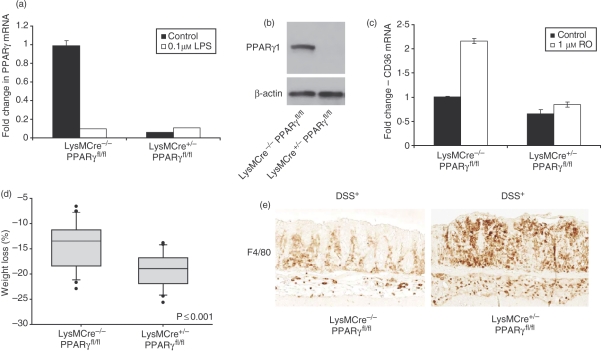
To understand the genomic impact of loss of PPARγ on inflammatory signalling, macrophage-specific PPARγ knockout mice were generated by breeding mice that are homozygous for a ‘floxed’ PPARγ allele (PPARγfl/fl) to transgenic mice that express Cre recombinase under the control of the macrophage-specific LysM promoter (LysMCre+/−). Macrophages from homozygous knockout mice (LysMCre+/−/PPARγfl/fl) exhibited > 90% reduction in PPARγ mRNA (Fig. 6a) and protein (Fig. 6b). The loss of PPARγ expression in macrophages from knockout mice resulted in loss of PPARγ transcriptional activity, as evidenced by a reduction in basal levels of well-known PPARγ target gene CD36 in LysMCre+/−/PPARγfl/fl mice. Moreover, rosiglitazone induced CD36 in peritoneal macrophages from LysMCre−/−/PPARγfl/fl but had no effect on CD36 expression in LysMCre+/−/PPARγfl/fl (knockout) mice (Fig. 6c).
Figure 6.
Characterization of myeloid-specific peroxisome proliferator-activated receptor γ (PPARγ) knockout (KO) mice. (a) Peritoneal macrophages from wild-type (WT; LysMCre−/−/PPARfl/fl) and myeloid-specific PPARγ KO mice (LysMCre+/−/PPARfl/fl) were analysed for PPARγ messenger RNA (mRNA) abundance by quantitative polymerase chain reaction (qPCR). (b) Western blotting was used to stain 20 μg of total lysate prepared from the macrophage cultures for PPARγ and β-actin expression. (c) Macrophage cultures from WT and KO mice were treated with rosiglitazone (1 μm) or vehicle for 24 hr. RNA was extracted and assayed for the mRNA abundance of PPARγ and the target gene, CD36. All mRNA was normalized to GAPDH and expressed as fold change relative to the control. Each bar represents mean ± SD, n = 3. (d,e) LysMCre−/−/PPARfl/fl and LysMCre+/−/PPARfl/fl mice were given acute colitis by administration of 3·5% dextran sodium sulphate (DSS) in drinking water for 7 days. Weight loss (d) was calculated as the per cent weight loss from days 0 and 7. Statistical analysis was carried out with Mann–Whitney rank sum analysis. (e) Representative sections of colons of DSS-treated LysMCre−/−/PPARfl/fl and LysMCre+/−/PPARfl/fl mice stained for macrophage-specific antibody F4/80 antibody.
To further characterize the mice for loss of functional activity, LysMCre+/−/PPARγfl/fl mice were subjected to acute colitis by exposure to 3·5% DSS, and disease activity was evaluated as a function of weight loss and macrophage infiltration after 7 days of DSS. The macrophage-specific PPARγ knockout mice (LysMCre+/−/PPARγfl/fl) exhibited a statistically significant weight loss (Fig. 6d) compared to control littermates (LysMCre−/−/PPARγfl/fl). Colons of DSS-treated LysMCre+/−/PPARγfl/fl knockout mice displayed increased epithelial damage and macrophage infiltration over their control littermates (Fig. 6e). These data are consistent with published data that indicate that loss of PPARγ from macrophages predisposes to experimental colitis.29
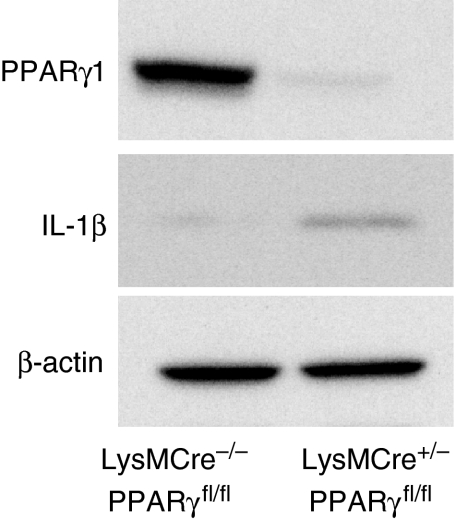
In an effort to understand how loss of PPARγ expression affects inflammatory signalling in macrophages, we isolated peritoneal macrophages from LysMCre+/−/PPARγfl/fl knockout mice and from LysMCre−/−/PPARγfl/fl littermates. The cells were maintained in culture with or without LPS for 6 hr, at which time RNA was extracted. Quantitative real-time PCR was employed to measure the expression of 80 genes through the use of Custom Taqman® low density arrays (supplemental Table S1). Each gene is classified by its biological function according to the Panther Classification System.51 The majority of genes represent chemokines, cytokines and other inflammatory mediators. Target genes regulated basally by PPARγ expression greater than twofold are listed in Table 1. Data are represented as average fold change in basal gene expression of knockout animals over wild-type. The basal levels of PPARγ and its target genes CD365,52 and ABCG13 were dramatically reduced in macrophage-specific PPARγ knockout mice. In detail, our results indicate that PPARγ negatively regulates the expression of several members of the CC (CCL17, CCL22) and CXC (CXCl2, CXCl4, CXCl5, CXCl14) subfamilies of chemokines, which are important for chemoattraction of neutrophils, monocytes and T lymphocytes. CCL17 and CCL22 are ligands for CCR4 that act as chemoattractants to draw T lymphocytes to the site of inflammation. CXCL2 and CXCL5 bind IL-8 receptor β and are involved in neutrophil attraction and activation. Our results also indicate that the basal expression of cytokines involved in the immune response such as IL-1β, IL-6, IL-12a and Csf3 and well-established mediators of inflammation such as PTGS2 (cyclooxygenase-2), NOS2A and C3 were elevated in macrophages deficient in PPARγ. Lastly, PPARγ knockout decreased the basal levels of MMP9, a regulator of matrix remodelling that is a key player in chronic inflammation and whose expression is elevated in inflammatory conditions such as inflammatory bowel disease.53–55 The increase in the basal level of IL-1β in PPARγ knockout macrophages was confirmed at the protein level by Western blot analysis (Fig. 7).
Table 1.
Genes regulated by peroxisome proliferator-activated receptor γ expression in peritoneal macrophages
| Gene title | Gene symbol | KO fold Δ over WT – LPS | KO fold Δ over WT + LPS | GO function-biological process |
|---|---|---|---|---|
| Peroxisome proliferator activated receptor γ | Pparg | −212·95 | −1·6 | Transcription/lipid metabolism |
| CD36 antigen | Cd36 | −11·5 | −1·49 | Lipid and fatty acid metabolism |
| ATP-binding cassette, subfamily G, member 1 | Abcg1 | −6·87 | −1·4 | Lipid and cholesterol transport |
| Chemokine (C-C motif) ligand 17 | Ccl17 | 129·54 | 3·67 | Chemotaxis, inflammatory response |
| Chemokine (C-C motif) ligand 22 | Ccl22 | 18·16 | 3·21 | Chemotaxis, inflammatory response |
| Chemokine (C-X-C motif) ligand 2 | Cxcl2 | 84·88 | 9·9 | Chemotaxis, inflammatory response |
| Chemokine (C-X-C motif) ligand 4 | Cxcl4 | 14·42 | 5 | Chemotaxis, inflammatory response |
| Chemokine (C-X-C motif) ligand 5 | Cxcl5 | 87·29 | 23·50 | Chemotaxis, inflammatory response |
| Chemokine (C-X-C motif) ligand 14 | Cxcl14 | 3·93 | 4·29 | Chemotaxis, inflammatory response |
| CC chemokine receptor 7 | Ccr7 | 5·14 | 3·79 | Chemotaxis, inflammatory response |
| Interleukin-6 | Il6 | 16 372 | 65·4 | Cytokine activity |
| Interleukin-1 beta | Il1b | 789·67 | 8·37 | Cytokine activity, signal transduction |
| Interleukin-12, p35 | Il12a | 92·9 | 1·53 | Cytokine activity |
| Colony stimulating factor 3 | Csf3 | 5790 | 11·14 | Immune and defence response, positive regulator of cell proliferation |
| IL-2 receptor alpha subunit | Il2ra | 15·7 | 7·62 | Immune response, cell cycle progression |
| Prostaglandin G/H synthase and cyclooxygenase | Ptgs2 | 1738 | 43·69 | Prostaglandin and fatty acid biosynthesis, cyclooxygenase |
| Nitric oxide synthase 2a | Nos2a | 2·56 | 0·99 | Inflammatory and defence response, nitric oxide synthesis, electron transport |
| Complement component C3 | C3 | 4·96 | 2·56 | Inflammatory response, complement activation |
| Matrix metalloproteinase 9 | Mmp9 | 14·89 | 6·89 | Macrophage differentiation, proteolysis, extracellular matrix organization and biogenesis |
GO, Gene Ontology; KO, knockout; LPS, lipopolysaccharide; WT, wild-type.
Figure 7.
Confirmation of macrophage peroxisome proliferator-activated receptor γ (PPARγ) genetic targets at the protein level. Western blot of total lysate (20 μg) of peritoneal macrophage cultures from wild-type (LysMCre−/−/PPARfl/fl) and myeloid-specific PPARγ KO mice (LysMCre+/−/PPARfl/fl) stained for interleukin-1β (IL-1β), PPARγ and β-actin expression.
Importantly, while these data indicate a significant repressive effect of PPARγ expression on the basal level of certain proinflammatory genes, a less pronounced effect was observed in the presence of LPS. The significant increase in proinflammatory gene expression observed in the PPARγ knockout macrophages at the basal level was severely reduced or lost in the presence of LPS. For example, PPARγ knockout macrophages exhibited a 14·42-fold increase in Cxcl14 expression over wild-type macrophages at the basal level (absence of LPS), but only a fivefold increase in the presence of LPS. Table 1 illustrates that this trend applies to each inflammatory gene, except for Cxcl4, whose levels remained consistently higher in the knockout animals in the presence and absence of LPS. Collectively, these data indicate that activation of PPARγ by endogenous ligands is important for suppressing inflammatory signalling at a physiological state but may have little impact in the context of inflammatory stimuli.
Discussion
Toll-like receptors are important mediators of innate immunity. Such receptors drive the inflammatory response by activation of proinflammatory transcription factors such as NF-κB and AP-1. In addition, our data indicate that TLR1/2, -4 and -5 inhibit the expression of an anti-inflammatory transcription factor, PPARγ. Our mechanistic studies have focused on TLR4, which has a well-known role in the development of inflammatory conditions such as sepsis, atherosclerosis and ulcerative colitis.56–60 Our data indicate that activation of TLR4 by LPS downregulates PPARγ through IκB-mediated activation of NF-κB. Since activation of NF-κB is characteristic of all TLRs, we speculate that inhibition of PPARγ expression by TLR1/2 and TLR5 agonists proceeds by the same mechanism that we have described for TLR4. We further speculate that a similar mechanism accounts for the observation that LPS reduces PPARγ levels in brown adipocytes and lung cells.34,39
In contrast to the effects of LPS on PPARγ expression in macrophages, brown adipocytes and lung cells, it has been reported that PPARγ immunostaining in colonic epithelial cells was decreased in TLR4 knockout mice.61 This observation implies that TLR4 may induce PPARγ in some cell types. To test this hypothesis, we quantified PPARγ mRNA and protein in extracts from purified colonic epithelial cells from wild-type and TLR4 knockout mice. We observed no difference in either PPARγ mRNA or protein in colonic epithelial cells from TLR4-deficient and control mice (supplemental Fig. S1a, b). We conclude that TLR4 does not regulate basal PPARγ expression in the colonic epithelium.
Cross-talk between NF-κB and PPARγ has been reported at the signal transduction level. For example, in adipose tissue the RelA subunit of NF-κB binds to PPARγ and prevents PPARγ/RXRα binding to target genes.38 Conversely, binding of PPARγ to RelA results in nuclear export of NF-κB and consequent attenuation of NF-κB-mediated inflammatory gene expression.62 Furthermore, SUMOylated PPARγ is recruited to promoters of inflammatory NF-κB target genes and inhibits the expression of these genes by a mechanism that appears to involve corepressor recruitment to SUMOylated PPARγ.63 Basal inflammatory signals are controlled by a yin/yang mechanism in which the activities of NF-κB and PPARγ depend on the relative abundance and the activity of the other.
Our studies add a new dimension to this model. Upon activation of TLR4 in macrophages, cross-talk between PPARγ and NF-κB is determined not by protein–protein interaction between the two, but by an NF-κB-mediated effect on PPARγ mRNA synthesis. This NF-κB-dependent loss of PPARγ expression secondary to TLR4 activation essentially resets the balance between the proinflammatory effects of NF-κB and the anti-inflammatory effects of PPARγ, establishing a synergistic proinflammatory signalling state, which affects macrophage function. First, downregulation of PPARγ by either activation of TLRs or genetic knockout alters the expression of PPARγ target genes that are involved in low-density lipoprotein/cholesterol metabolism, such as CD36 and ABCG1. These observations implicate TLR4 in the development of atherosclerotic foam cells through an NF-κB/PPARγ-dependent mechanism. Second, reduction in macrophage PPARγ leads to dramatic increases in basal levels of proinflammatory molecules, including chemokines, interleukins and other inflammatory molecules. These findings suggest that the anti-inflammatory actions of endogenous PPARγ are critical for maintaining basal or ‘physiological’ inflammation, but are not sufficient for suppressing stimulus (LPS)-induced inflammation as a result of inhibition of PPARγ expression and function by TLR signalling. Based on these data, we propose a model in which PPARγ normally suppresses NF-κB-mediated physiological inflammation and control of physiological processes such as lipid metabolism and atherosclerosis (Fig. 8). However, upon injury or a deregulated response such as in inflammatory bowel disease, activation of NF-κB suppresses PPARγ expression, neutralizing the anti-inflammatory and physiological actions of PPARγ and enhancing the inflammatory response. In the context of inflammatory diseases mediated by macrophages, such as ulcerative colitis, atherosclerosis and pulmonary sarcoidosis, PPARγ downregulation should be considered an important component by which NF-κB drives inflammation. Moreover, targeting macrophage PPARγ in these diseases with therapeutic doses of TZDs may have little efficacy after the onset of inflammation because of the downregulation of PPARγ. This model explains, at least in part, the observation that TZDs may block inflammation in experimental models of colitis but have little or no effect on established inflammation. A similar model has been proposed in multiple sclerosis, where an inflammation-mediated reduction in PPARγ levels reduces the efficacy of PPARγ agonists.64
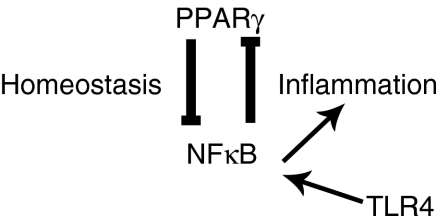
Figure 8.
Negative feedback loop of nuclear factor-κB (NF-κB) on peroxisome proliferator-activated receptor γ (PPARγ) expression. The working hypothesis predicts that PPARγ suppresses NF-κB-mediated physiological inflammation; however, upon activation of TLR4 signalling by LPS, increased activity of NF-κB downregulates PPARγ expression and anti-inflammatory actions, thus allowing a full inflammatory response.
Downregulation of PPARγ by TLR4 may also be relevant to other diseases such as obesity-related insulin resistance. Obesity is associated with macrophage accumulation and proinflammatory molecule release in adipose tissue, and these events are thought to drive insulin resistance.65–68 Inflammation-associated insulin resistance has been proposed to be mediated, at least in part, by an obesity-related increase in free fatty acids.69–71 Free fatty acids can activate TLR4 on macrophages to induce an inflammatory response, and so may be responsible for increased macrophage accumulation and proinflammatory signalling in adipose tissue during obesity.72–75 This hypothesis is consistent with recent studies that report that macrophage PPARγ is required to maintain glucose homeostasis and insulin sensitivity, and that the insulin-sensitizing effects of TZD PPARγ agonists are mediated, at least in part, through anti-inflammatory effects on macrophages.7,8 We speculate that obesity-related insulin resistance may arise by a free fatty acid-mediated TLR4-dependent downregulation of macrophage PPARγ.
In conclusion, our data describe a novel regulatory feedback loop between PPARγ and TLR4/NF-κB signalling in macrophages. In this loop, PPARγ represses NF-κB-mediated inflammatory signalling to maintain proper control of physiological inflammation and allow for the execution of physiological functions of PPARγ in macrophages such as lipid metabolism. However, in an increased inflammatory state, NF-κB drives down PPARγ expression to nullify its actions and accelerate the inflammatory process. The downregulation of PPARγ by TLR4 signalling may significantly impact the therapeutic efficacy of TZDs in inflammatory diseases and may play an important role in metabolic diseases such as obesity-related insulin resistance.
Acknowledgments
We thank Jennifer H. Havens for valuable technical assistance in the execution of these experiments. We acknowledge Laura Lewis-Tuffin for her critical review of the manuscript. The support of the Crohn’s and Colitis Foundation is gratefully acknowledged, as well as the support of the Mayo Foundation, which maintains most of the infrastructure without which these experiments could not have been carried out.
Supplementary material
The following supplementary material is available for this article online:
Basal levels of PPARγ in colonic epithelial cells are unaffected by TLR4 expression.
Genes analyzed by real-time PCR in macrophages from wild-type and myeloid-specific PPARγ knockout mice
This material is available as part of the online article from http://www.blackwell-synergy.com.
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–56. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 2.Hu E, Tontonoz P, Spiegelman BM. Transdifferentiation of myoblasts by the adipogenic transcription factors PPAR gamma and C/EBP alpha. Proc Natl Acad Sci USA. 1995;92:9856–60. doi: 10.1073/pnas.92.21.9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akiyama TE, Sakai S, Lambert G, et al. Conditional disruption of the peroxisome proliferator-activated receptor gamma gene in mice results in lowered expression of ABCA1, ABCG1, and apoE in macrophages and reduced cholesterol efflux. Mol Cell Biol. 2002;22:2607–19. doi: 10.1128/MCB.22.8.2607-2619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babaev VR, Yancey PG, Ryzhov SV, Kon V, Breyer MD, Magnuson MA, Fazio S, Linton MF. Conditional knockout of macrophage PPARgamma increases atherosclerosis in C57BL/6 and low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2005;25:1647–53. doi: 10.1161/01.ATV.0000173413.31789.1a. [DOI] [PubMed] [Google Scholar]
- 5.Chawla A, Barak Y, Nagy L, Liao D, Tontonoz P, Evans RM. PPAR-gamma dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nat Med. 2001;7:48–52. doi: 10.1038/83336. [DOI] [PubMed] [Google Scholar]
- 6.Lee CH, Evans RM. Peroxisome proliferator-activated receptor-gamma in macrophage lipid homeostasis. Trends Endocrinol Metab. 2002;13:331–5. doi: 10.1016/s1043-2760(02)00668-9. [DOI] [PubMed] [Google Scholar]
- 7.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–20. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hevener AL, Olefsky JM, Reichart D, et al. Macrophage PPAR gamma is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J Clin Invest. 2007;117:1658–69. doi: 10.1172/JCI31561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore KJ, Fitzgerald ML, Freeman MW. Peroxisome proliferator-activated receptors in macrophage biology: friend or foe? Curr Opin Lipidol. 2001;12:519–27. doi: 10.1097/00041433-200110000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Su CG, Wen X, Bailey ST, et al. A novel therapy for colitis utilizing PPAR-gamma ligands to inhibit the epithelial inflammatory response. J Clin Invest. 1999;104:383–9. doi: 10.1172/JCI7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desreumaux P, Dubuquoy L, Nutten S, et al. Attenuation of colon inflammation through activators of the retinoid X receptor (RXR)/peroxisome proliferator-activated receptor gamma (PPARgamma) heterodimer. A basis for new therapeutic strategies. J Exp Med. 2001;193:827–38. doi: 10.1084/jem.193.7.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saubermann LJ, Nakajima A, Wada K, et al. Peroxisome proliferator-activated receptor gamma agonist ligands stimulate a Th2 cytokine response and prevent acute colitis. Inflamm Bowel Dis. 2002;8:330–9. doi: 10.1097/00054725-200209000-00004. [DOI] [PubMed] [Google Scholar]
- 13.Katayama K, Wada K, Nakajima A, et al. A novel PPAR gamma gene therapy to control inflammation associated with inflammatory bowel disease in a murine model. Gastroenterology. 2003;124:1315–24. doi: 10.1016/s0016-5085(03)00262-2. [DOI] [PubMed] [Google Scholar]
- 14.Lytle C, Tod TJ, Vo KT, Lee JW, Atkinson RD, Straus DS. The peroxisome proliferator-activated receptor gamma ligand rosiglitazone delays the onset of inflammatory bowel disease in mice with interleukin 10 deficiency. Inflamm Bowel Dis. 2005;11:231–43. doi: 10.1097/01.mib.0000160805.46235.eb. [DOI] [PubMed] [Google Scholar]
- 15.Schaefer KL, Denevich S, Ma C, et al. Intestinal antiinflammatory effects of thiazolidenedione peroxisome proliferator-activated receptor-gamma ligands on T helper type 1 chemokine regulation include nontranscriptional control mechanisms. Inflamm Bowel Dis. 2005;11:244–52. doi: 10.1097/01.mib.0000160770.94199.9b. [DOI] [PubMed] [Google Scholar]
- 16.Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell. 1998;93:229–40. doi: 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
- 17.Tontonoz P, Nagy L, Alvarez JG, Thomazy VA, Evans RM. PPARgamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell. 1998;93:241–52. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- 18.Li AC, Brown KK, Silvestre MJ, Willson TM, Palinski W, Glass CK. Peroxisome proliferator-activated receptor gamma ligands inhibit development of atherosclerosis in LDL receptor-deficient mice. J Clin Invest. 2000;106:523–31. doi: 10.1172/JCI10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Chawla A. Role of PPARgamma in macrophage biology and atherosclerosis. Trends Endocrinol Metab. 2004;15:500–5. doi: 10.1016/j.tem.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Hammad H, de Heer HJ, Soullie T, Angeli V, Trottein F, Hoogsteden HC, Lambrecht BN. Activation of peroxisome proliferator-activated receptor-gamma in dendritic cells inhibits the development of eosinophilic airway inflammation in a mouse model of asthma. Am J Pathol. 2004;164:263–71. doi: 10.1016/s0002-9440(10)63116-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woerly G, Honda K, Loyens M, Papin JP, Auwerx J, Staels B, Capron M, Dombrowicz D. Peroxisome proliferator-activated receptors alpha and gamma down-regulate allergic inflammation and eosinophil activation. J Exp Med. 2003;198:411–21. doi: 10.1084/jem.20021384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trifilieff A, Bench A, Hanley M, Bayley D, Campbell E, Whittaker P. PPAR-alpha and -gamma but not -delta agonists inhibit airway inflammation in a murine model of asthma: in vitro evidence for an NF-kappaB-independent effect. Br J Pharmacol. 2003;139:163–71. doi: 10.1038/sj.bjp.0705232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demerjian M, Man MQ, Choi EH, et al. Topical treatment with thiazolidinediones, activators of peroxisome proliferator-activated receptor-gamma, normalizes epidermal homeostasis in a murine hyperproliferative disease model. Exp Dermatol. 2006;15:154–60. doi: 10.1111/j.1600-0625.2006.00402.x. [DOI] [PubMed] [Google Scholar]
- 24.Hasegawa H, Takano H, Zou Y, Qin Y, Hizukuri K, Odaka K, Toyozaki T, Komuro I. Pioglitazone, a peroxisome proliferator-activated receptor gamma activator, ameliorates experimental autoimmune myocarditis by modulating Th1/Th2 balance. J Mol Cell Cardiol. 2005;38:257–65. doi: 10.1016/j.yjmcc.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Yuan ZY, Liu Y, Liu Y, Zhang JJ, Kishimoto C, Wang YN, Ma AQ, Liu ZQ. PPAR-gamma ligands inhibit the expression of inflammatory cytokines and attenuate autoimmune myocarditis. Chin Med J (Engl) 2004;117:1253–5. [PubMed] [Google Scholar]
- 26.Natarajan C, Bright JJ. Peroxisome proliferator-activated receptor-gamma agonists inhibit experimental allergic encephalomyelitis by blocking IL-12 production, IL-12 signaling and Th1 differentiation. Genes Immun. 2002;3:59–70. doi: 10.1038/sj.gene.6363832. [DOI] [PubMed] [Google Scholar]
- 27.Diab A, Deng C, Smith JD, Hussain RZ, Phanavanh B, Lovett-Racke AE, Drew PD, Racke MK. Peroxisome proliferator-activated receptor-gamma agonist 15-deoxy-Delta(12,14)-prostaglandin J(2) ameliorates experimental autoimmune encephalomyelitis. J Immunol. 2002;168:2508–15. doi: 10.4049/jimmunol.168.5.2508. [DOI] [PubMed] [Google Scholar]
- 28.Crosby MB, Svenson JL, Zhang J, Nicol CJ, Gonzalez FJ, Gilkeson GS. Peroxisome proliferation-activated receptor (PPAR)gamma is not necessary for synthetic PPARgamma agonist inhibition of inducible nitric-oxide synthase and nitric oxide. J Pharmacol Exp Ther. 2005;312:69–76. doi: 10.1124/jpet.104.074005. [DOI] [PubMed] [Google Scholar]
- 29.Shah YM, Morimura K, Gonzalez FJ. Expression of peroxisome proliferator-activated receptor-gamma in macrophage suppresses experimentally induced colitis. Am J Physiol Gastrointest Liver Physiol. 2007;292:G657–66. doi: 10.1152/ajpgi.00381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis JD, Lichtenstein GR, Stein RB, et al. An open-label trial of the PPAR-gamma ligand rosiglitazone for active ulcerative colitis. Am J Gastroenterol. 2001;96:3323–8. doi: 10.1111/j.1572-0241.2001.05333.x. [DOI] [PubMed] [Google Scholar]
- 31.Ramakers JD, Verstege MI, Thuijls G, Te Velde AA, Mensink RP, Plat J. The PPARgamma agonist rosiglitazone impairs colonic inflammation in mice with experimental colitis. J Clin Immunol. 2007;27:275–83. doi: 10.1007/s10875-007-9074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonfield TL, Farver CF, Barna BP, Malur A, Abraham S, Raychaudhuri B, Kavuru MS, Thomassen MJ. Peroxisome proliferator-activated receptor-gamma is deficient in alveolar macrophages from patients with alveolar proteinosis. Am J Respir Cell Mol Biol. 2003;29:677–82. doi: 10.1165/rcmb.2003-0148OC. [DOI] [PubMed] [Google Scholar]
- 33.Culver DA, Barna BP, Raychaudhuri B, et al. Peroxisome proliferator-activated receptor gamma activity is deficient in alveolar macrophages in pulmonary sarcoidosis. Am J Respir Cell Mol Biol. 2004;30:1–5. doi: 10.1165/rcmb.2003-0304RC. [DOI] [PubMed] [Google Scholar]
- 34.Liu D, Zeng BX, Shang Y. Decreased expression of peroxisome proliferator-activated receptor gamma in endotoxin-induced acute lung injury. Physiol Res. 2006;55:291–9. doi: 10.33549/physiolres.930822. [DOI] [PubMed] [Google Scholar]
- 35.Waite KJ, Floyd ZE, Arbour-Reily P, Stephens JM. Interferon-gamma-induced regulation of peroxisome proliferator-activated receptor gamma and STATs in adipocytes. J Biol Chem. 2001;276:7062–8. doi: 10.1074/jbc.M007894200. [DOI] [PubMed] [Google Scholar]
- 36.Harada K, Isse K, Kamihira T, Shimoda S, Nakanuma Y. Th1 cytokine-induced downregulation of PPARgamma in human biliary cells relates to cholangitis in primary biliary cirrhosis. Hepatology. 2005;41:1329–38. doi: 10.1002/hep.20705. [DOI] [PubMed] [Google Scholar]
- 37.Crosby MB, Zhang J, Nowling TM, Svenson JL, Nicol CJ, Gonzalez FJ, Gilkeson GS. Inflammatory modulation of PPAR gamma expression and activity. Clin Immunol. 2006;118:276–83. doi: 10.1016/j.clim.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 38.Suzawa M, Takada I, Yanagisawa J, et al. Cytokines suppress adipogenesis and PPAR-gamma function through the TAK1/TAB1/NIK cascade. Nat Cell Biol. 2003;5:224–30. doi: 10.1038/ncb942. [DOI] [PubMed] [Google Scholar]
- 39.Mracek T, Cannon B, Houstek J. IL-1 and LPS but not IL-6 inhibit differentiation and downregulate PPAR gamma in brown adipocytes. Cytokine. 2004;26:9–15. doi: 10.1016/j.cyto.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 41.Trinchieri G, Sher A. Cooperation of Toll-like receptor signals in innate immune defence. Nat Rev Immunol. 2007;7:179–90. doi: 10.1038/nri2038. [DOI] [PubMed] [Google Scholar]
- 42.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–77. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 43.Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- 44.Chen L, Necela BM, Su W, Yanagisawa M, Anastasiadis PZ, Fields AP, Thompson EA. Peroxisome proliferator-activated receptor gamma promotes epithelial to mesenchymal transformation by Rho GTPase-dependent activation of ERK1/2. J Biol Chem. 2006;281:24575–87. doi: 10.1074/jbc.M604147200. [DOI] [PubMed] [Google Scholar]
- 45.Chen L, Bush CR, Necela BM, Su W, Yanagisawa M, Anastasiadis PZ, Fields AP, Thompson EA. RS5444, a novel PPARgamma agonist, regulates aspects of the differentiated phenotype in nontransformed intestinal epithelial cells. Mol Cell Endocrinol. 2006;251:17–32. doi: 10.1016/j.mce.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 47.De Jager PL, Franchimont D, Waliszewska A, et al. The role of the Toll receptor pathway in susceptibility to inflammatory bowel diseases. Genes Immun. 2007;8:387–97. doi: 10.1038/sj.gene.6364398. [DOI] [PubMed] [Google Scholar]
- 48.Gazouli M, Mantzaris G, Kotsinas A, Zacharatos P, Papalambros E, Archimandritis A, Ikonomopoulos J, Gorgoulis VG. Association between polymorphisms in the Toll-like receptor 4, CD14, and CARD15/NOD2 and inflammatory bowel disease in the Greek population. World J Gastroenterol. 2005;11:681–5. doi: 10.3748/wjg.v11.i5.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oostenbrug LE, Drenth JP, de Jong DJ, et al. Association between Toll-like receptor 4 and inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:567–75. doi: 10.1097/01.mib.0000161305.81198.0f. [DOI] [PubMed] [Google Scholar]
- 50.Browning BL, Huebner C, Petermann I, Gearry RB, Barclay ML, Shelling AN, Ferguson LR. Has toll-like receptor 4 been prematurely dismissed as an inflammatory bowel disease gene? Association study combined with meta-analysis shows strong evidence for association. Am J Gastroenterol. 2007;102:2504–12. doi: 10.1111/j.1572-0241.2007.01463.x. [DOI] [PubMed] [Google Scholar]
- 51.Thomas PD, Kejariwal A, Campbell MJ, et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–41. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welch JS, Ricote M, Akiyama TE, Gonzalez FJ, Glass CK. PPARgamma and PPARdelta negatively regulate specific subsets of lipopolysaccharide and IFN-gamma target genes in macrophages. Proc Natl Acad Sci USA. 2003;100:6712–7. doi: 10.1073/pnas.1031789100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Medina C, Videla S, Radomski A, et al. Increased activity and expression of matrix metalloproteinase-9 in a rat model of distal colitis. Am J Physiol Gastrointest Liver Physiol. 2003;284:G116–22. doi: 10.1152/ajpheart.00036.2002. [DOI] [PubMed] [Google Scholar]
- 54.Medina C, Santana A, Paz MC, Diaz-Gonzalez F, Farre E, Salas A, Radomski MW, Quintero E. Matrix metalloproteinase-9 modulates intestinal injury in rats with transmural colitis. J Leukoc Biol. 2006;79:954–62. doi: 10.1189/jlb.1005544. [DOI] [PubMed] [Google Scholar]
- 55.Gao Q, Meijer MJ, Kubben FJ, et al. Expression of matrix metalloproteinases-2 and -9 in intestinal tissue of patients with inflammatory bowel diseases. Dig Liver Dis. 2005;37:584–92. doi: 10.1016/j.dld.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 56.Bosshart H, Heinzelmann M. Targeting bacterial endotoxin: two sides of a coin. Ann N Y Acad Sci. 2007;1096:1–17. doi: 10.1196/annals.1397.064. [DOI] [PubMed] [Google Scholar]
- 57.Li H, He Y, Zhang J, Sun S, Sun B. Lipopolysaccharide regulates toll-like receptor 4 expression in human aortic smooth muscle cells. Cell Biol Int. 2007;31:831–5. doi: 10.1016/j.cellbi.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 58.Shi D, Das J, Das G. Inflammatory bowel disease requires the interplay between innate and adaptive immune signals. Cell Res. 2006;16:70–4. doi: 10.1038/sj.cr.7310009. [DOI] [PubMed] [Google Scholar]
- 59.Fukata M, Michelsen KS, Eri R, et al. Toll-like receptor-4 is required for intestinal response to epithelial injury and limiting bacterial translocation in a murine model of acute colitis. Am J Physiol Gastrointest Liver Physiol. 2005;288:G1055–65. doi: 10.1152/ajpgi.00328.2004. [DOI] [PubMed] [Google Scholar]
- 60.Fort MM, Mozaffarian A, Stover AG, et al. A synthetic TLR4 antagonist has anti-inflammatory effects in two murine models of inflammatory bowel disease. J Immunol. 2005;174:6416–23. doi: 10.4049/jimmunol.174.10.6416. [DOI] [PubMed] [Google Scholar]
- 61.Dubuquoy L, Jansson EA, Deeb S, et al. Impaired expression of peroxisome proliferator-activated receptor gamma in ulcerative colitis. Gastroenterology. 2003;124:1265–76. doi: 10.1016/s0016-5085(03)00271-3. [DOI] [PubMed] [Google Scholar]
- 62.Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AG, Pettersson S, Conway S. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–12. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 63.Pascual G, Fong AL, Ogawa S, et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–63. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klotz L, Schmidt M, Giese T, Sastre M, Knolle P, Klockgether T, Heneka MT. Proinflammatory stimulation and pioglitazone treatment regulate peroxisome proliferator-activated receptor gamma levels in peripheral blood mononuclear cells from healthy controls and multiple sclerosis patients. J Immunol. 2005;175:4948–55. doi: 10.4049/jimmunol.175.8.4948. [DOI] [PubMed] [Google Scholar]
- 65.Wellen KE, Hotamisligil GS. Obesity-induced inflammatory changes in adipose tissue. J Clin Invest. 2003;112:1785–8. doi: 10.1172/JCI20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu H, Barnes GT, Yang Q, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–30. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arkan MC, Hevener AL, Greten FR, et al. IKK-beta links inflammation to obesity-induced insulin resistance. Nat Med. 2005;11:191–8. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- 69.Boden G, She P, Mozzoli M, et al. Free fatty acids produce insulin resistance and activate the proinflammatory nuclear factor-kappaB pathway in rat liver. Diabetes. 2005;54:3458–65. doi: 10.2337/diabetes.54.12.3458. [DOI] [PubMed] [Google Scholar]
- 70.Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115:1111–9. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shah PK. Innate immune pathway links obesity to insulin resistance. Circ Res. 2007;100:1531–3. doi: 10.1161/CIRCRESAHA.107.101104. [DOI] [PubMed] [Google Scholar]
- 72.Lee JY, Sohn KH, Rhee SH, Hwang D. Saturated fatty acids, but not unsaturated fatty acids, induce the expression of cyclooxygenase-2 mediated through Toll-like receptor 4. J Biol Chem. 2001;276:16683–9. doi: 10.1074/jbc.M011695200. [DOI] [PubMed] [Google Scholar]
- 73.Hwang D. Modulation of the expression of cyclooxygenase-2 by fatty acids mediated through toll-like receptor 4-derived signaling pathways. FASEB J. 2001;15:2556–64. doi: 10.1096/fj.01-0432com. [DOI] [PubMed] [Google Scholar]
- 74.Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest. 2006;116:3015–25. doi: 10.1172/JCI28898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suganami T, Tanimoto-Koyama K, Nishida J, et al. Role of the Toll-like receptor 4/NF-kappaB pathway in saturated fatty acid-induced inflammatory changes in the interaction between adipocytes and macrophages. Arterioscler Thromb Vasc Biol. 2007;27:84–91. doi: 10.1161/01.ATV.0000251608.09329.9a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Basal levels of PPARγ in colonic epithelial cells are unaffected by TLR4 expression.
Genes analyzed by real-time PCR in macrophages from wild-type and myeloid-specific PPARγ knockout mice