Abstract
GATA-3 is the master transcription factor for T helper 2 (Th2) cell differentiation and is critical for the expression of Th2 cytokines. Little is known, however, about the nature of the functional molecular complexes of GATA-3. We identified a high-mobility group (HMG)-box type transcription factor, lymphoid enhancer factor 1 (LEF-1), in the GATA-3 complex present in Th2 cells using a Flag-calmodulin-binding peptide (CBP)-tag based proteomics method. The interaction between GATA-3 and LEF-1 was confirmed by co-immunoprecipitation experiments using LEF-1-introduced T-cell lineage TG40 cells. The HMG-box domain of LEF-1 and two zinc finger domains of GATA-3 were found to be important for the physical association. The introduction of LEF-1 into developing Th2 cells resulted in the suppression of Th2 cytokine production. The suppression was significantly lower in the cells into which a HMG-box-deleted LEF-1 mutant was introduced. Moreover, LEF-1 inhibited the binding activity of GATA-3 to the interleukin (IL)-5 promoter. These results suggest that LEF-1 is involved in the GATA-3 complex, while also regulating the GATA-3 function, such as the induction of Th2 cytokine expression via the inhibition of the DNA-binding activity of GATA-3.
Keywords: GATA-3, HMG box, LEF-1, Th2, Wnt/β-catenin
Introduction
CD4 T cell-dependent immune responses are controlled by at least three distinct helper T-cell subsets, T helper 1 (Th1), Th2 and Th17.1–6 Th1 cells produce a large amount of interferon (IFN)-γ to control cell-mediated immunity against intracellular pathogens, whereas Th2 cells produce IL-4, IL-5 and IL-13 and are involved in humoral immunity.2–4 Th17 is a recently identified helper T-cell subpopulation that is required for the exclusion of extracellular pathogens and is associated with inflammatory autoimmune responses.5,6
Several transcription factors that control Th1/Th2/Th17 cell differentiation have been identified. Of these, GATA-3 appears to be a key factor for Th2 cell differentiation,7–9 as is T-cell-specific T-box transcription factor (T-bet) for Th1,10 and retinoid-related orphan receptor gamma t (RORγt) for Th17.11 Recently, chromatin modification of the IL-4/IL-5/IL-13 loci in developing Th2 cells,12–14 the IFN-γ locus in Th1 cells,15 and the IL-17 locus in Th17 cells has been demonstrated.16 The histone modifications at the Th2 cytokine gene loci are primarily mediated by GATA-3 in CD4 and CD8 T cells.14,17,18 The binding of GATA-3 at the conserved GATA-3 response element (CGRE) region appears to initiate the long-range histone acetylation accompanied by non-coding transcription.14
GATA-3 is abundantly expressed in T lymphocytes and the embryonic brain.19,20 The expression of GATA-3 is required for the development of T cells in the thymus21,22 and for β-selection and CD4 single positive thymocyte development.23 In peripheral T cells, the activation of signal transducer and activator of transcription 6 (STAT6) induces GATA-3 mRNA expression, and the ectopic expression of GATA-3 results in Th2 cell differentiation even in the absence of STAT6.9,24 In addition to transcriptional regulation, the expression of GATA-3 is also regulated by a post-transcriptional mechanism. We recently reported that the rat sarcoma virus oncogene 1 (Ras)-extracellular regulatory kinase (ERK) mitogen-activated protein kinase (MAPK) cascade25 and mammalian polycomb group protein B lymphoma Mo-MLV insertion region 1 (Bmi1) controls GATA-3 stability through the ubiquitin/proteasome-dependent pathway.26
GATA-3 belongs to a family of zinc finger transcription factors. The mammalian GATA- family consists of six proteins (GATA-1 to GATA-6) that share a zinc finger domain (Cys-X2-Cys-X17-Cys-X2-Cys, where X represents any amino acid residue) and bind to the consensus sequence 5′-(A/T)GATA(A/G)-3′.27 The zinc finger domain of the GATA protein contains two finger domains, a C-terminal finger (C-finger) and an N-terminal finger (N-finger). The C-finger is responsible for DNA binding, whereas the stabilization of DNA binding is regulated by the N-finger, which also physically interacts with the other zinc finger proteins. Recently, we identified a novel conserved sequence YxKxHxxxPP located immediately downstream of the C-finger, which is essential for DNA binding, supporting Th2 cell differentiation and chromatin remodelling of the Th2 cytokine gene loci.28
Lymphoid enhancer factor 1 (LEF-1) is a member of the LEF-1/transcription factor (TCF) subfamily of high-mobility group (HMG) domain proteins and recognizes a 5′CCTTTGAACT sequence.29,30 There are four genes, Lef1, Tcf3, Tcf4, and Tcf7 (TCF-1) in the LEF-1/TCF family, which are the products of various different splice variants.31 These family members show homology in the N-terminal and C-terminal regions. The N-terminal region contains a β-catenin-binding domain whereas the HMG DNA-binding domain is located in the C-terminal region of the protein. The other amino acid sequences of the protein are very diverse, and have different functional motifs, such as the C-terminal binding protein (CtBP)-binding motif,32 the p300 interacting domain,33 and the E-tail motif (CRARF motif).34 These functional motifs are present only in some isoforms of the LEF-1/TCF molecules. These LEF-1/TCF proteins show various activities when they are expressed ectopically35 and they also have a regulatory function on some promoters of the target genes.33
LEF-1 is expressed in lymphoid cells and also in many cells undergoing the processes of organogenesis in the developing mouse.36,37 The HMG-box domain of LEF-1 facilitates the assembly of the DNA-bound enhanceosome by inducing DNA bending,38 whereas the activation domain of LEF-1 contributes to enhanceosome-mediated enhancement of downstream T-cell receptor (TCR)-α promoter activity.39,40 LEF-1 acts as an architectural transcriptional factor, which mediates wingless/int-1 (Wnt)/β-catenin signalling.41–43 In response to Wnt signals, LEF-1 interacts with and stabilizes β-catenin at its N-terminal region, and hence activates Wnt-response gene transcription.44 In association with the co-repressor Groucho, LEF-1 represses the Wnt-responsive genes.45 In addition, LEF-1 can interact with the cofactor ALY and other transcription factors to activate enhancers of specific target genes, in the absence of Wnt signalling.
In the present study, we demonstrate that LEF-1 associates with GATA-3. We found the zinc finger region of GATA-3 and the HMG box of LEF-1 to play important roles in the association of these two molecules. LEF-1 suppresses the transcription of the Th2 cytokine genes in developing Th2 cells. Our results may contribute to a better understanding of the regulation of GATA-3 activity in Th2 cells.
Materials and methods
Generation of Flag-tagged GATA-3 and its expression in a Th2 cell line
Flag-calmodulin-binding peptide (CBP)-tagged cDNA for human GATA-3 was inserted into a multi-cloning site of the pMx-internal ribosome entry site (IRES)-green fluorescent protein (GFP) vector. The retrovirus vector-containing culture supernatant was prepared as described previously.46 A Th2 cell line, D10G4.1, was stimulated in vitro with immobilized anti-TCR-β monoclonal antibody (mAb) for 2 days, and then the stimulated cells were infected with a retrovirus containing a control mock (pMX-IRES-GFP) or the Flag-CBP-GATA-3 gene (pMx-Flag-CBP-GATA-3-IRES-GFP). Three days after infection, GFP-positive retrovirus-infected cells were sorted with a FACSVantage (Becton Dickinson, San Jose, CA). For infection of the primary CD4 T cells, the pMX-IRES-human nerve growth factor (NGF) receptor p75 (hNGFR) plasmid was used.17
Identification of LEF-1 in a GATA-3 complex in a Th2 cell line
Cell lysates were prepared from Flag-tagged-GATA-3 or mock-infected D10G4·1 cells as previously described.47 The nuclear fraction was solubilized with the following protease inhibitor-containing buffer: 20 mm HEPES–NaOH (pH 7·5), 200 mm NaCl, 1·5 mm MgCl2, 1·5 mm CaCl2, 1% Nonidet P-40 (NP-40), 1 mm Na2VO3, 1 mm NaF and protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN), and lysed on ice for 30 min with gently shaking. The insoluble materials were removed by centrifugation and the supernatant was filtrated and diluted 1 : 4 with the protease inhibitor-containing buffer without NP-40. The GATA-3-associated molecules were investigated using the protocol based on the tandem affinity purification method.48 The final samples were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) and the candidate was identified by peptide mass fingerprinting with matrix assisted laser desorption ionization-time of flight-mass spectrum (MALDI-TOF/MS) as previously described.49
Expression plasmid and transfection
The Flag-tagged GATA-3 mutants (pCMV2-Flag) and Myc-tagged LEF-1 mutants (pCMV-Myc) were generated by a polymerase chain reaction (PCR)-based mutation. Human embryonic kidney 293 (HEK293) T cells were transfected using a FuGENE reagent (Roche Applied Science) according to the manufacturer’s protocol.
Cell lysis, immnunoprecipitation and immunoblotting
The protein extracts from the transfected cells were prepared by lysing cells in 200 μl/well (35-mm dish) of RIPA buffer [50 mm Tris–HCl, pH7·4, 1%NP-40, 0·25% Na-deoxycholate, 150 mm NaCl, 1 mm ethylenediaminetetraacetic acid (EDTA), pH 8·0, 1mm phenylmethylsulphonyl fluoride (PMSF), Aprotinin (1μg/ml; Roche Applied Science), Leupeptin (1μg/ml; Roche Applied Science), 1 mm Na3VO4 and 1 mm NaF]. For the co-immunoprecipitation analysis, the cell lysates were subjected to the pre-clear process with protein G-sepharose at 4° for 1 hr by rotating. The pre-cleared extracts were then subjected to co-immunoprecipitation with anti-Flag mAb (F 3165; Sigma, St Louis, MO) or anti-Myc mAb (M047-3; Medical and Biological Laboratories Co. Ltd, Nagoya, Japan) at 4° for 2 hr, and then the immunocomplexes were precipitated with protein G-sepharose beads at 4° for 1 hr. For the immunoblot analysis, the immunoprecipitates were eluted from beads in SDS-gel loading buffer and run on 10% or 4–20% gradient polyacrylamide gels. After electrophoresis, the proteins were transferred onto nitrocellulose membranes and blotted with anti-Myc mAb, anti-Flag mAb, anti-GATA-3 mAb (sc-22206; Santa Cruz Biotechnology, Santa Cruz, CA), or anti-tubulin-α mAb (MS-581-P, Neo Markers, Fremont, CA) primary antibodies. Horseradish peroxidase-labelled anti-mouse immunoglobulin G (IgG) was used for the secondary antibody (GE Healthcare, Buckinghamshire, UK). The antibody–antigen complexes were visualized by chemiluminescence (GE Healthcare).
In vitro Th2 cell differentiation and the introduction of LEF-1
Splenic CD4 T cells were purified using magnetic beads and an Auto-MACS Sorter (Miltenyi Biotec, Bergisch Gladbach, Germany) yielding a purity of >98%. For Th2 cell differentiation, the purified CD4 T cells (1·5 × 106) were stimulated for 2 days with immobilized anti-TCR mAb (H57-597, 3 μg/ml) in the presence of IL-2 (25 U/ml), IL-4 (100 U/ml), and anti-IFN-γ mAb (R4.6A2, 25% culture supernatant). The pMX-IRES-human NGF receptor p75 (hNGFR) plasmid was generated from the pMX-IRES-GFP plasmid by replacing the enhanced green fluorescent protein (EGFP) with the cytoplasmic region-deleted hNGFR cDNA. The method for the generation of virus supernatant and infection has been described previously.46 Virus supernatant was added to the Th2 cell differentiation cultures on day 2. After culturing for three more days, the cells were harvested, restimulated and subjected to reverse transcriptase (RT)-PCR and enzyme-linked immunosorbent assay (ELISA). The infected cells were enriched by magnetic antibody cell sorting (MACS) with anti-human NGFR (C40-1457; BD Bioscience, San Jose, CA).
Quantitative RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA). Reverse transcription was performed using Superscript II (Invitrogen Life Technologies). For the quantitative PCR, a TaqMan universal PCR Master Mix was used for all reactions (Applied Biosystems, Foster City, CA), and the ABI Prism 7000 Sequence Detection System (Applied Biosystems) was employed. The primers and TaqMan probes for the detection of mouse LEF-1, IL-4, IL-5, IL-13 and hypoxanthine phosphoribosyltransferase (HPRT) were purchased from Applied Biosystems. The expression was normalized using the HPRT signal.
ELISA
The cytokine concentrations were assessed by ELISA as described previously.50
Chromatin immunoprecipitation (ChIP) assay
A ChIP assay was performed using the anti-acetyl histone H3 K9/14 and anti-tri-methyl histone H3 K4 as described previously.51 The primers and probes used for the quantitative PCR analysis have been described previously.14
Electrophoretic mobility shift assay (EMSA)
The electrophoretic mobility shift assay was performed using a Gel Shift Assay System (Promega, Madison, WI) as described previously.28 In brief, the nuclear extracts were incubated at 4° with 32P-labelled double-stranded oligonucleotide in DNA-binding buffer. Electrophoresis was carried out on 4% native polyacrylamide gel (0·5 × Tris-borate/EDTA (TBE) buffer, acrylamide: bisacrylamide 29 : 1), and the radioactivity was visualized by autoradiography. A supershift analysis was performed using anti-Flag mAb (M2; Sigma-Aldrich). The oligonucleotide (for the IL-5 promoter) used in this experiment was 5′-TGCTAACAATCAGATAGAGG-3′.
Results
Purification of a functional GATA-3 complex in a Th2 cell line
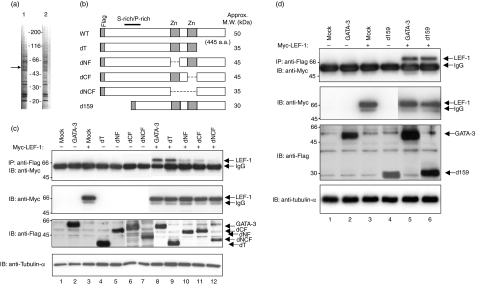
To investigate the nature of the functional GATA-3 complex, we performed a proteomics analysis. Flag/CBP-tagged wild-type GATA-3 and non-functional GATA-3 C-terminal deletion mutant (GATA-3d349)28 molecules were introduced into cells of a Th2 cell line, D10G4.1, using a retrovirus vector system. The GFP-positive retrovirus-infected cells were sorted and subjected to the process of protein purification. The GATA-3-associated molecules were purified using a protocol based on the tandem affinity purification method48 and final samples were separated by SDS-PAGE (Fig. 1a). We compared the bands shown in the wild-type (lane 1) and mutant GATA-3 (lane 2) precipitates, and a selectively darker band (indicated by an arrow) was sliced and analysed by mass spectrometry as previously described.49
Figure 1.
Both zinc finger domains of GATA-3 are important for the association with lymphoid enhancer factor 1 (LEF-1). (a) A picture of silver staining of the final sample for the identification of GATA-3-associated molecules. Lane 1: wild-type (WT) GATA-3. Lane 2: GATA-3d349. The arrow in lane 1 (50 kDa) indicates a band that was sliced and analysed by mass spectrometry. The numbers indicates the molecular mass markers. (b) A schematic representation of the wild type and GATA-3 mutants. The location of the serine (S)-proline (P)-rich transactivation domain (over the line) and zinc finger domains (Zn) are indicated. The approximate molecular weight (MW) of each mutant is also indicated. (c) The role of GATA-3 C-terminal and zinc finger regions for the association with LEF-1. 293 T cells were transfected with expression plasmids encoding Myc-tagged wild-type LEF-1 and Flag-tagged wild-type or mutant GATA-3 genes. Two days later, immunoprecipitates with anti-Flag monoclonal antibody (mAb) were subjected to immunoblotting with anti-Myc mAb (upper panel). The total lysates were also run in parallel and immunoblotted with anti-Myc mAb, anti-Flag mAb, or anti-tubulin-α mAb. (d) The N-terminal serine-rich region of GATA-3 is not essential for the binding to LEF-1. 293 T cells were transfected with expression plasmids encoding Myc-tagged wild-type LEF-1 and Flag-tagged wild-type or N-terminal deleted GATA-3. IB, immunoblotting; IgG, immunoglobulin G; IP, immunoprecipitation.
LEF-1 was identified as a GATA-3-associated molecule in a Th2 cell line
Peptide mass fingerprinting with MALDI-TOF/MS revealed that the band was an HMG DNA-binding domain containing the transcription factor LEF-1 (data not shown). To confirm the association of GATA-3 with LEF-1, we generated the wild type and various truncation mutants of GATA-3 (Fig. 1b). 293 T cells were transfected with Flag-tagged GATA-3 and Myc-tagged LEF-1, and then the lysates were immunoprecipitated with anti-Flag mAb. The precipitates were separated by SDS-PAGE, transferred to a polyvinilidene difluoride (PVDF) membrane and blotted with an anti-Myc mAb. A signal for Myc-tagged LEF-1 protein was detected in the precipitate only when GATA-3 and LEF-1 were co-transfected (Fig. 1c, lane 8), thus suggesting the interaction of GATA-3 with LEF-1 to be specific. The expression levels of GATA-3 and LEF-1 were equivalent in each lane (see anti-Myc immunoblotting and anti-Flag immunoblotting; middle panels in Fig. 1c).
The zinc finger domain of GATA-3 is important for the binding to LEF-1
To determine which domains of GATA-3 are important for the binding to LEF-1, Flag-tagged wild-type or mutant GATA-3 molecules and a Myc-tagged LEF-1 were co-transfected into 293 T cells. The interaction was assessed by immunoprecipitation with an anti-Flag mAb and immunoblotting with an anti-Myc mAb. The interactions of LEF-1 with wild-type GATA-3 and with the C-terminal region mutant dT were similar in intensity (Fig. 1c; compare lanes 8 and 9), whereas the interaction with the deletion mutants of the zinc finger domain of GATA-3 (dNF, dCF and dNCF) was apparently impaired (Fig. 1c; compare lanes 8, 10, 11 and 12). The binding with LEF-1 was almost abolished by the deletion of both zinc fingers (dNCF) (Fig. 1c, lane 12), thus suggesting that LEF-1 binds to the zinc finger regions of GATA-3. All GATA-3 mutants showed approximately equivalent expression (Fig. 1c, lower middle panel). Moreover, we assessed the possible role of the N-terminal region of GATA-3 in the binding with LEF-1. The binding of an N-terminal deletion mutant of GATA-3 (d159) was comparable to that of wild-type GATA-3 (Fig. 1d). The equivalent expression of each protein was confirmed by blotting with anti-Myc or anti-Flag mAb (Fig. 1d, middle panels). These results suggest that both zinc finger domains of GATA-3 are important for the binding to LEF-1.
The HMG box domain of LEF-1 is required for the association with GATA-3
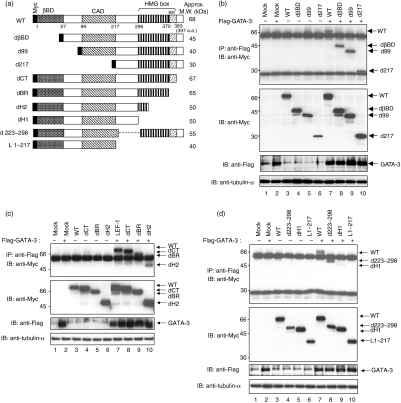
We next wanted to identify the functional domains of LEF-1 that interact with GATA-3. We generated various Myc-tagged LEF-1 mutants and determined the association with GATA-3 in a co-immunopreciptitation assay. A schematic representation of the mutants is depicted in Fig. 2a. First, we examined the association of the N-terminal region truncation mutants dβBD (where βBD is the β-catenin binding domain), d99 and d217 [deleted context-dependent activation domain (CAD)]. These three mutants were efficiently co-immunoprecipitated with GATA-3 in the transfected 293 T cells (Fig. 2b, lanes 7–10). The equivalent expression of each protein was confirmed by blotting with anti-Myc or anti-Flag mAb (Fig. 2b, lower panels). These results suggest that the N-terminal region of LEF-1 is not important for the binding to GATA-3.
Figure 2.
The high-mobility group (HMG) box of lymphoid enhancer factor 1 (LEF-1) is important for the association with GATA-3. (a) A schematic representation of the wild type (WT) and various LEF-1 mutants. The β-catenin-binding domain (βBD), context-dependent activation domain (CAD) and HMG DNA-binding domain are indicated. The numbers below the wild type indicate the amino acid positions of the respective protein domains. The approximate molecular weight (MW) of each mutant is also indicated. (b-d) The association of Flag-tagged wild-type GATA-3 with Myc-tagged wild type and various LEF-1 mutants was investigated as in Fig 1b. IB, immunoblotting; IP, immunoprecipitation.
Concurrently, we investigated the role of the C-terminal region of LEF-1, which contains the HMG box, in the association with GATA-3. The HMG box consists of four different domains, including basic region (BR), helix-1 (H1), helix-2 (H2) and helix-3 (H3). We therefore generated five mutants, dCT, dBR, dH1, dH2 and dH3, respectively. The levels of co-immunoprecipitation with GATA-3 in the deletion mutants, i.e. a mutant missing the C-terminal region (dCT) and a mutant missing the region downstream of the basic region (dBR), were similar to those of the wild-type LEF-1 (Fig. 2c). However, the interaction between GATA-3 and the dH2 deletion mutant was reproducibly weak (Fig. 2c, lane 10). Although we also generated a dH3 mutant that lacked a whole HMG box, we failed to detect the dH3 mutant band because of its molecular size; it always merged with the IgG band (data not shown). To determine the domain of interaction of LEF-1 with GATA-3 more precisely, we generated additional LEF-1 mutants, dH1, d223-298 and L1-217 (see Fig. 2a). The wild-type LEF-1 and the d223-298 mutant effectively bound to GATA-3 (Fig. 2d, lanes 7 and 8), whereas the interaction of dH1 and L1-217 was apparently reduced (Fig. 2d, lanes 9 and 10). Again, the expression levels of each mutant protein were equivalent (see middle panels). We also generated an Myc-tagged HMG-box construct, but failed to detect its protein expression in the 293 T cells (data not shown). These results suggest that the HMG-box region of LEF-1 is important for the interaction with GATA-3.
LEF-1 associates with GATA-3 in a T cell line, TG40
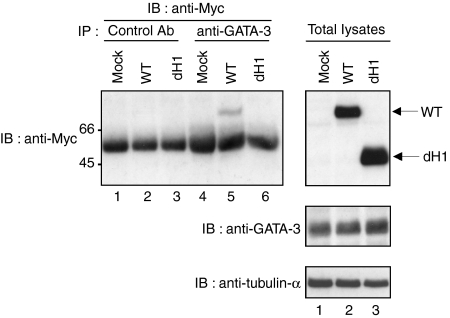
Finally, we investigated the interaction of LEF-1 with GATA-3 under more physiological conditions using a T-cell line, TG40, that expresses endogenous GATA-3. Myc-tagged wild-type LEF-1 and the dH1 mutant were introduced into TG40 cells using retroviral expression vectors. As expected, the wild-type LEF-1 proteins were co-immunoprecipitated with GATA-3 (Fig. 3, lane 5), whereas dH1 protein was not detected in the anti-GATA-3 immunoprecipitates (Fig. 3, lane 6). For controls, we used an antibody against the goat IgG fraction. No specific band was detected (Fig. 3, lanes 1–3). The protein expression levels of the wild type and the dH1 mutant of LEF-1 were comparable (Fig. 3, right panels). These results indicate that LEF-1 binds to GATA-3 via the HMG box in TG40 T cells.
Figure 3.
Association of endogenous GATA-3 with transfected lymphoid enhancer factor 1 (LEF-1) in a T-cell line. A mouse GATA-3-expressing T-cell line, TG40, was infected with retrovirus vector encoding wild-type (WT) Myc-tagged LEF-1 and the helix-1 mutant (dH1). Thereafter, the immunoprecipitates with control goat immunoglobulin G (IgG) antibody (lanes 1–3) or anti-GATA-3 antibody (lanes 4–6) were subjected to immunoblotting with anti-Myc monoclonal antibody (mAb) (left panel). The total lysates were also run in parallel as a loading control (right panel). IB, immunoblotting; IP, immunoprecipitation.
LEF-1 suppresses GATA-3 DNA-binding activity and Th2 cytokine production
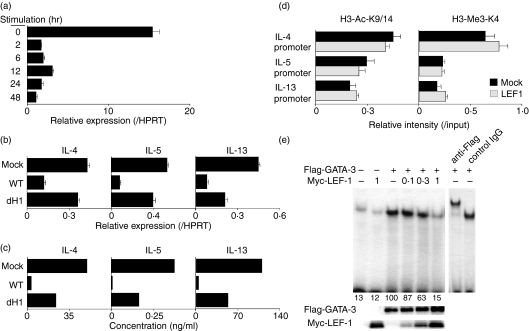
In order to assess the physiological roles of LEF-1 in the developing Th2 cells, the expression levels of LEF-1 were determined in CD4 T cells after TCR stimulation. The mRNA levels of LEF-1 were substantial in naïve CD4 T cells, and were rapidly down-regulated after the TCR stimulation (Fig. 4a). Next, the wild-type LEF-1 and dH1 mutant were introduced into CD4 T cells cultured under Th2 conditions (developing Th2 cells) using a retrovirus gene transduction system, and the mRNA expression of IL-4, IL-5 and IL-13 was examined by a quantitative RT-PCR analysis. As shown in Fig. 4(b), the mRNA expression of IL-4, IL-5 and IL-13 was suppressed dramatically by the introduction of wild-type LEF-1. The levels of suppression of IL-4, IL-5 and IL-13 expression were apparently lower in cells in which the dH1 mutant was introduced. The expression levels of wild-type LEF-1 and the dH1 mutant mRNA were similar in the infected cells (data not shown). The effects of wild-type LEF-1 and the dH1 mutant on the production of Th2 cytokines were confirmed by ELISA (Fig. 4c). These results indicate that Th2 cytokine production was negatively regulated by LEF-1 in the developing Th2 cells.
Figure 4.
Lymphoid enhancer factor 1 (LEF-1) suppresses the DNA-binding activity of GATA-3 and T helper 2 (Th2) cytokine expression in developing Th2 cells. (a) The expression levels of LEF-1 in freshly isolated splenic CD4 T cells after stimulation with immobilized anti-T-cell receptor (TCR) monoclonal antibody (mAb) in vitro were determined by a quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) analysis. (b) Splenic CD4 T cells were stimulated under Th2 culture conditions and infected with retrovirus vector encoding wild-type Myc-tagged LEF-1 and the helix-1 mutant (dH1). To examine the mRNA expression levels of Th2 cytokines in infected cells, the human nerve growth factor (NGF) receptor p75 (hNGFR)-positive cells were sorted and were stimulated with anti-TCR mAb for 4 hr and subjected to a quantitative RT-PCR analysis. (c) Th2 cytokine production in infected cells was determined by enzyme-linked immunosorbent assay (ELISA). The hNGFR+ cells were sorted and were stimulated with anti-TCR mAb for 24 hr. Thereafter, the culture supernatants were collected and subjected to ELISA. Three independent experiments were performed with similar results. (d) The levels of histone modification (H3-K9/14 acetylation and H3-K4 tri-methylation) at the Th2 cytokine gene loci were assessed in a chromatin immunoprecipitation (ChIP) analysis. (e) Flag-tagged GATA-3 and Myc-tagged LEF-1 were expressed in 293 T cells. Nuclear extracts from the transfected cells were prepared and subjected to an electrophoretic mobility shift assay (EMSA) with the GATA-binding sequence in the interleukin (IL)-5 promoter region. The relative amounts of LEF-1 in the nuclear lysates were indicated. The protein levels of GATA-3 and LEF-1 were determined by immunoblotting with anti-Flag mAb and anti-Myc mAb, respectively (lower panel). A super-shift assay was performed with control mouse immunoglobulin G (IgG) and an anti-Flag mAb (right panel). Arbitrary densitometric units are shown under each band. Three independent experiments were performed with similar results. HPRT, hypoxanthine phosphoribosyltransferase.
It is known that histone modification at the Th2 cytokine gene loci occurs during Th2 cell differentiation.14 Consequently, we wanted to know whether the introduction of LEF-1 affects histone modification of the Th2 cytokine gene loci. A ChIP assay using mAbs specific for anti-acetyl histone H3-K9/14 or anti-tri-methyl histone H3-K4 was performed. The levels of acetylation of histone H3-K9/14 or tri-methylation of histone H3-K4 at the promoter regions of IL-4, IL-5 and IL-13 were equivalent between the mock-infected Th2 cells and those into which LEF-1 had been introduced (Fig. 4d). These results suggest that the overexpression of LEF-1 did not significantly affect histone modification at the Th2 cytokine gene loci.
Finally, we examined the effect of LEF-1 on the DNA-binding activity of GATA-3 in an EMSA assay using a GATA-3-binding sequence in the IL-5 promoter. The binding of GATA-3 was inhibited by LEF-1 in a dose-dependent manner (Fig. 4e, upper left). Specific binding of the GATA-3 complex to the oligo probe was confirmed by a super-shift assay with anti-Flag mAb (Fig. 4e, upper right). The amount of GATA-3 protein and LEF-1 protein was assessed by immunoblotting with anti-Flag and anti-Myc mAb, respectively (lower panel). Thus, LEF-1 may affect the transcription of Th2 cytokines through inhibition of the DNA-binding activity of GATA-3.
Discussion
In this study, we demonstrate that LEF-1 physically associates with GATA-3 in cells of the T-cell lineage TG40, and that the HMG-box domain of LEF-1 is important in the association with GATA-3. Both zinc finger domains of GATA-3 appear to be required for the association with LEF-1. LEF-1 appears to inhibit the DNA binding of GATA-3, while suppressing the production of Th2 cytokines.
Several transcription factors, including repressor of GATA (ROG), friend of GATA (FOG), MAD homolog 3 (Smad3), spleen focus forming virus proviral integration oncogene spi1 (PU.1) and T-bet, have been reported to be associated with GATA-3 and regulate GATA-3 function.52–56 Smad3 acts as a co-activator of GATA-3, whereas ROG, FOG, PU.1 and T-bet appear to suppress GATA-3 function. Although the important regions of GATA-3 regarding the association with ROG and FOG have been determined, those for the association with PU.1 and T-bet remain unclear. ROG binds to the C-finger of GATA-3 and FOG binds to the N-finger, and control GATA-3 functions. In the current report, we found that both zinc fingers of GATA-3 are involved in the binding with LEF-1 (Fig. 1). This result may suggest that the association of LEF-1 with the zinc finger regions of GATA-3 is distinct from that of ROG or FOG.
The overexpression of ROG suppresses GATA-3-dependent transactivation52 and Th2 cell differentiation.17 PU.1 also suppresses Th2 cytokine production from the Th2 cell subpopulation through the inhibition of GATA-3 binding to the IL-4 VA enhancer site.55 An inhibitory effect of T-bet on GATA-3 binding at the IL-5 promoter has also been demonstrated.56 Interestingly, it was recently reported that GATA-3 could also interfere with the transactivation of the promoter of fucosyltransferase VII by T-bet.57 The functional consequence of the T-bet/GATA-3 interaction can be bidirectional, and these two opposing transcription factors can counteract each other. Like T-bet and PU.1, LEF-1 is a member of a family of transcription factors that directly recognize and bind to specific DNA sequences. Moreover, the DNA-binding domains of GATA-3 and LEF-1 are involved in their association (Figs 1–3). It is therefore likely that the binding of LEF-1 to GATA-3 inhibits the DNA-binding activity of GATA-3. In fact, an inhibitory effect of LEF-1 in the binding of GATA-3 to the IL-5 promoter was demonstrated (Fig. 4e).
The expression of LEF-1 was rapidly down-regulated in CD4 T cells after TCR-mediated stimulation (Fig. 4a). mRNA expression and the production of Th2 cytokines from in vitro-generated primary Th2 cells were strongly inhibited by the overexpression of LEF-1 (Fig. 4b,c). The TCR signal-mediated down-regulation of LEF-1 expression was inhibited by treatment with a MAP kinase/ERK kinase (MEK) inhibitor or a calcineurin inhibitor (M. Yamashita and T. Nakayama, unpublished observation). We previously reported that the activation of the ERK-MAPK and the Ca2+/calcinerurin signalling pathways upon TCR stimulation is required for the efficient development of Th2 cells.50,58 Although a more comprehensive investigation is required, the TCR-mediated down-regulation of LEF-1 may be required for the induction of Th2 function, i.e. robust Th2 cytokine production upon restimulation through the TCR.
In the absence of Wnt signals, LEF-1 associates with the Groucho family co-repressor protein, and represses the LEF-1-responsive genes.44 When the Wnt signalling pathway is activated, β-catenin is stabilized, binds to LEF-1 and transactivates the Wnt-responsive genes. In our experiments described in this report, we did not experimentally stimulate the Wnt signalling pathway. It therefore remains unclear which form of LEF-1 can bind to GATA-3. A lot of bands were detected in the precipitates (Fig. 1a), but we succeeded in the identification of only one band (Fig. 1, arrow). The identification of the other co-precipitated proteins may contribute to a better understanding of the GATA-3 complex, and the crosstalk between GATA-3 and the Wnt/β-catenin-mediated signalling pathways.
In summary, we identified LEF-1 as a component of the GATA-3 complex in T cells. The results of this study may help to elucidate a new network of transcription factors that control the activity of GATA-3 and the function of Th2 cells.
Acknowledgments
We thank Ms Kaoru Sugaya, Ms Hikari Asou, Ms Satoko Norikane and Mr Toshihiro Ito for their excellent technical assistance. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology (Japan) [Grants-in-Aid: for Scientific Research on Priority Areas #17016010; Scientific Research (B) #17390139, Scientific Research (C) #18590466, #19590491 and #19591609, Exploratory Research #19659121, and Young Scientists (Start-up) #18890046: Special Coordination Funds for Promoting Science and Technology, and Cancer Translational Research Project], the Ministry of Health, Labor and Welfare (Japan), The Japan Health Science Foundation, the Kanae Foundation, the Uehara Memorial Foundation, the Mochida Foundation, the Yasuda Medical Foundation, the Astellas Foundation and the Sagawa Foundation.
References
- 1.Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–73. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 2.Constant SL, Bottomly K. Induction of Th1 and Th2 CD4+ T cell responses: the alternative approaches. Annu Rev Immunol. 1997;15:297–322. doi: 10.1146/annurev.immunol.15.1.297. [DOI] [PubMed] [Google Scholar]
- 3.Seder RA, Paul WE. Acquisition of lymphokine-producing phenotype by CD4+ T cells. Annu Rev Immunol. 1994;12:635–73. doi: 10.1146/annurev.iy.12.040194.003223. [DOI] [PubMed] [Google Scholar]
- 4.Reiner SL, Locksley RM. The regulation of immunity to Leishmania major. Annu Rev Immunol. 1995;13:151–77. doi: 10.1146/annurev.iy.13.040195.001055. [DOI] [PubMed] [Google Scholar]
- 5.Reiner SL. Development in motion: helper T cells at work. Cell. 2007;129:33–6. doi: 10.1016/j.cell.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 6.Stockinger B, Veldhoen M. Differentiation and function of Th17 T cells. Curr Opin Immunol. 2007;19:281–6. doi: 10.1016/j.coi.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S, Radbruch A, Murphy KM. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12:27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- 8.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–96. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 9.Lee HJ, Takemoto N, Kurata H, Kamogawa Y, Miyatake S, O’Garra A, Arai N. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J Exp Med. 2000;192:105–15. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-γ production in CD4 and CD8 T cells. Science. 2002;295:338–42. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 11.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 12.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–23. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 13.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–56. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 14.Yamashita M, Ukai-Tadenuma M, Kimura M, Omori M, Inami M, Taniguchi M, Nakayama T. Identification of a conserved GATA3 response element upstream proximal from the interleukin-13 gene locus. J Biol Chem. 2002;277:42399–408. doi: 10.1074/jbc.M205876200. [DOI] [PubMed] [Google Scholar]
- 15.Zhou W, Chang S, Aune TM. Long-range histone acetylation of the Ifng gene is an essential feature of T cell differentiation. Proc Natl Acad Sci USA. 2004;101:2440–5. doi: 10.1073/pnas.0306002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–72. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- 17.Omori M, Yamashita M, Inami M, et al. CD8 T cell-specific downregulation of histone hyperacetylation and gene activation of the IL-4 gene locus by ROG, repressor of GATA. Immunity. 2003;19:281–94. doi: 10.1016/s1074-7613(03)00210-3. [DOI] [PubMed] [Google Scholar]
- 18.Inami M, Yamashita M, Tenda Y, et al. CD28 costimulation controls histone hyperacetylation of the interleukin5 gene locus in developing Th2 cells. J Biol Chem. 2004;279:23123–33. doi: 10.1074/jbc.M401248200. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto M, Ko LJ, Leonard MW, Beug H, Orkin SH, Engel JD. Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev. 1990;4:1650–62. doi: 10.1101/gad.4.10.1650. [DOI] [PubMed] [Google Scholar]
- 20.George KM, Leonard MW, Roth ME, Lieuw KH, Kioussis D, Grosveld F, Engel JD. Embryonic expression and cloning of the murine GATA-3 gene. Development. 1994;120:2673–86. doi: 10.1242/dev.120.9.2673. [DOI] [PubMed] [Google Scholar]
- 21.Hattori N, Kawamoto H, Fujimoto S, Kuno K, Katsura Y. Involvement of transcription factors TCF-1 and GATA-3 in the initiation of the earliest step of T cell development in the thymus. J Exp Med. 1996;184:1137–47. doi: 10.1084/jem.184.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–8. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 23.Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–75. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 24.Kurata H, Lee HJ, O’Garra A, Arai N. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity. 1999;11:677–88. doi: 10.1016/s1074-7613(00)80142-9. [DOI] [PubMed] [Google Scholar]
- 25.Yamashita M, Shinnakasu R, Asou H, et al. Ras-ERK MAPK cascade regulates GATA3 stability and Th2 differentiation through ubiquitin-proteasome pathway. J Biol Chem. 2005;280:29409–19. doi: 10.1074/jbc.M502333200. [DOI] [PubMed] [Google Scholar]
- 26.Hosokawa H, Kimura MY, Shinnakasu R, et al. Regulation of Th2 cell development by Polycomb group gene bmi-1 through the stabilization of GATA3. J Immunol. 2006;177:7656–64. doi: 10.4049/jimmunol.177.11.7656. [DOI] [PubMed] [Google Scholar]
- 27.Orkin SH. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80:575–81. [PubMed] [Google Scholar]
- 28.Shinnakasu R, Yamashita M, Shinoda K, Endo Y, Hosokawa H, Hasegawa A, Ikemizu S, Nakayama T. Critical YxKxHxxxRP motif in the C-terminal region of GATA3 for its DNA binding and function. J Immunol. 2006;177:5801–10. doi: 10.4049/jimmunol.177.9.5801. [DOI] [PubMed] [Google Scholar]
- 29.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function [corrected] Genes Dev. 1991;5:880–94. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 30.Waterman ML, Fischer WH, Jones KA. A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes Dev. 1991;5:656–69. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 31.van Noort M, Clevers H. TCF transcription factors, mediators of Wnt-signaling in development and cancer. Dev Biol. 2002;244:1–8. doi: 10.1006/dbio.2001.0566. [DOI] [PubMed] [Google Scholar]
- 32.Brannon M, Brown JD, Bates R, Kimelman D, Moon RT. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development. 1999;126:3159–70. doi: 10.1242/dev.126.14.3159. [DOI] [PubMed] [Google Scholar]
- 33.Hecht A, Stemmler MP. Identification of a promoter-specific transcriptional activation domain at the C terminus of the Wnt effector protein T-cell factor 4. J Biol Chem. 2003;278:3776–85. doi: 10.1074/jbc.M210081200. [DOI] [PubMed] [Google Scholar]
- 34.Atcha FA, Munguia JE, Li TW, Hovanes K, Waterman ML. A new beta-catenin-dependent activation domain in T cell factor. J Biol Chem. 2003;278:16169–75. doi: 10.1074/jbc.M213218200. [DOI] [PubMed] [Google Scholar]
- 35.Gradl D, Konig A, Wedlich D. Functional diversity of Xenopus lymphoid enhancer factor/T-cell factor transcription factors relies on combinations of activating and repressing elements. J Biol Chem. 2002;277:14159–71. doi: 10.1074/jbc.M107055200. [DOI] [PubMed] [Google Scholar]
- 36.Oosterwegel M, van de Wetering M, Timmerman J, Kruisbeek A, Destree O, Meijlink F, Clevers H. Differential expression of the HMG box factors TCF-1 and LEF-1 during murine embryogenesis. Development. 1993;118:439–48. doi: 10.1242/dev.118.2.439. [DOI] [PubMed] [Google Scholar]
- 37.van Genderen C, Okamura RM, Farinas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 38.Giese K, Grosschedl R. LEF-1 contains an activation domain that stimulates transcription only in a specific context of factor-binding sites. EMBO J. 1993;12:4667–76. doi: 10.1002/j.1460-2075.1993.tb06155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayall TP, Sheridan PL, Montminy MR, Jones KA. Distinct roles for P-CREB and LEF-1 in TCR alpha enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 1997;11:887–99. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- 40.Bruhn L, Munnerlyn A, Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes Dev. 1997;11:640–53. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 41.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 42.Eastman Q, Grosschedl R. Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr Opin Cell Biol. 1999;11:233–40. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 43.Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–20. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 44.Hsu SC, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol. 1998;18:4807–18. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levanon D, Goldstein RE, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci USA. 1998;95:11590–5. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimura M, Koseki Y, Yamashita M, et al. Regulation of Th2 cell differentiation by mel-18, a mammalian polycomb group gene. Immunity. 2001;15:275–87. doi: 10.1016/s1074-7613(01)00182-0. [DOI] [PubMed] [Google Scholar]
- 47.Watarai H, Inagaki Y, Kubota N, Fuju K, Nagafune J, Yamaguchi Y, Kadoya T. Proteomic approach to the identification of cell membrane proteins. Electrophoresis. 2000;21:460–4. doi: 10.1002/(SICI)1522-2683(20000101)21:2<460::AID-ELPS460>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 48.Krogan NJ, Cagney G, Yu H, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 49.Watarai H, Hinohara A, Nagafune J, Nakayama T, Taniguchi M, Yamaguchi Y. Plasma membrane-focused proteomics: dramatic changes in surface expression during the maturation of human dendritic cells. Proteomics. 2005;5:4001–11. doi: 10.1002/pmic.200401258. [DOI] [PubMed] [Google Scholar]
- 50.Yamashita M, Kimura M, Kubo M, Shimizu C, Tada T, Perlmutter RM, Nakayama T. T cell antigen receptor-mediated activation of the Ras/mitogen-activated protein kinase pathway controls interleukin 4 receptor function and type-2 helper T cell differentiation. Proc Natl Acad Sci USA. 1999;96:1024–9. doi: 10.1073/pnas.96.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamashita M, Hirahara K, Shinnakasu R, Hosokawa H, Norikane S, Kimura MY, Hasegawa A, Nakayama T. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity. 2006;24:611–22. doi: 10.1016/j.immuni.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 52.Miaw SC, Choi A, Yu E, Kishikawa H, Ho IC. ROG, repressor of GATA, regulates the expression of cytokine genes. Immunity. 2000;12:323–33. doi: 10.1016/s1074-7613(00)80185-5. [DOI] [PubMed] [Google Scholar]
- 53.Fox AH, Kowalski K, King GF, Mackay JP, Crossley M. Key residues characteristic of GATA N-fingers are recognized by FOG. J Biol Chem. 1998;273:33595–603. doi: 10.1074/jbc.273.50.33595. [DOI] [PubMed] [Google Scholar]
- 54.Blokzijl A, ten Dijke P, Ibanez CF. Physical and functional interaction between GATA-3 and Smad3 allows TGF-beta regulation of GATA target genes. Curr Biol. 2002;12:35–45. doi: 10.1016/s0960-9822(01)00623-6. [DOI] [PubMed] [Google Scholar]
- 55.Chang HC, Zhang S, Thieu VT, Slee RB, Bruns HA, Laribee RN, Klemsz MJ, Kaplan MH. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity. 2005;22:693–703. doi: 10.1016/j.immuni.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 56.Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–3. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- 57.Chen GY, Osada H, Santamaria-Babi LF, Kannagi R. Interaction of GATA-3/T-bet transcription factors regulates expression of sialyl Lewis X homing receptors on Th1/Th2 lymphocytes. Proc Natl Acad Sci USA. 2006;103:16894–9. doi: 10.1073/pnas.0607926103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamashita M, Katsumata M, Iwashima M, et al. T cell receptor-induced calcineurin activation regulates T helper type 2 cell development by modifying the interleukin 4 receptor signaling complex. J Exp Med. 2000;191:1869–79. doi: 10.1084/jem.191.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]