Abstract
We previously showed that Candida albicans orf19.4590, which we have renamed RFX2, expresses a protein that is reactive with antibodies in persons with candidiasis. In this study, we demonstrate that C. albicans RFX2 shares some functional redundancy with Saccharomyces cerevisiae RFX1. Complementation of an S. cerevisiae rfx1 mutant with C. albicans RFX2 partially restored UV susceptibility and the repression of DNA damage response genes. DNA damage- and UV-induced genes RAD6 and DDR48 were derepressed in a C. albicans rfx2 null mutant strain under basal conditions, and the mutant was significantly more resistant to UV irradiation, heat shock, and ethanol than wild-type strain SC5314. The rfx2 mutant was hyperfilamentous on solid media and constitutively expressed hypha-specific genes HWP1, ALS3, HYR1, ECE1, and CEK1. The mutant also demonstrated increased invasion of solid agar and significantly increased adherence to human buccal epithelial cells. During hematogenously disseminated candidiasis, mice infected with the mutant had a significantly delayed time to death compared to the wild type. During oropharyngeal candidiasis, mice infected with the mutant had significantly lower tissue burdens in the oral cavity and esophagus at 7 days and they were less likely to develop disseminated infections because of mucosal translocation. The data demonstrate that C. albicans Rfx2p regulates DNA damage responses, morphogenesis, and virulence.
Candida albicans is a versatile opportunistic pathogen that causes diverse diseases in humans, ranging from oropharyngeal candidiasis (OPC) and other mucosal infections to life-threatening disseminated bloodstream infections. Indeed, C. albicans is remarkable for its ability to cause invasive diseases of virtually all tissues and major organs. Several properties that contribute to candidal virulence have been characterized; these include adherence to host cells, secretion of hydrolytic enzymes, sequestration of iron, phenotypic switching, and the reversible transitioning from single-cell blastospores to forms with extended filaments (morphogenesis) (10). A large number of individual genes involved in these processes have been implicated in virulence through targeted disruption and testing of mutant strains in animal models. Rather than depending upon a dominant virulence factor, C. albicans achieves its success as a pathogen through coordinated expression of multiple genes as it senses and adapts to particular in vivo environments (47). As such, the regulation of biological processes important to the proliferation and survival of C. albicans cells within infected hosts has become an active area of investigation.
As a strategy to identify C. albicans genes that are expressed during the course of candidiasis in humans, we previously used pooled sera from human immunodeficiency virus-infected patients with active oropharyngeal or esophageal candidiasis to screen a C. albicans genomic DNA expression library (12, 36). We identified over 60 C. albicans genes that encoded proteins of diverse function that reacted with antibodies in the pooled sera (12, 36). We implicated several proteins identified by our screening in the regulation of yeast-hyphal morphogenesis and the pathogenesis of mucosal and/or disseminated candidiasis (3, 4, 12-14, 37). Among these previously unstudied regulators of candidal virulence were Not5p, a component of the CCR-NOT transcription-regulatory complex (13), Irs4p, an EH domain-containing protein that interacts with the 5′ phosphatase Inp51p and regulates phosphatidylinositol-(4,5)-bisphosphate levels (3, 4), and Set1p, a histone 3 lysine 4 methyltransferase (37). Interestingly, these proteins, like many identified by our screening, are known or predicted to localize to intracellular compartments, suggesting that they appear at the C. albicans cell surface at some point in the life cycle or that they are released from cells following cell death (13).
One of our previously unstudied genes corresponds to orf19.4590 in the Candida Genome Database (http://www.candidagenome.org). orf19.4590 encodes a protein of 1,112 amino acids that contains an RFX domain of approximately 103 amino acids at positions 427 to 530. This domain has 25.8% amino acid identity to the RFX domain of Saccharomyces cerevisiae Rfx1p (also known as Crt1p). RFX domains are unique winged-helix DNA binding domains that are conserved across eukaryotes (18, 20). Rfx1p, the sole member of the RFX protein family in S. cerevisiae, localizes transcriptional repressors to the promoters of DNA damage response genes like RNR2, -3, and -4 and HOG1 (25, 53). In response to DNA damage, Mec1p hyperphosphorylates Rad9p, which activates the protein kinase Rad53p (40, 49, 51). S. cerevisiae Rad53p is required for all transcriptional and cell cycle arrest responses (1, 52), the former of which are mediated, at least in part, by Rfx1p. When Rfx1p is phosphorylated by Rad53p, the transcriptional repressor complex is deactivated and DNA damage response genes are derepressed. As is true across eukaryotes (43), DNA checkpoint proteins are conserved in C. albicans. Moreover, they are essential for the filamentation of C. albicans cells in response to genotoxic stress (2, 45). Indeed, deactivation of the C. albicans Mec1-Rad53 pathway through deletion of RAD53 or RAD9 completely abolishes filamentous growth in response to DNA damage, including true hyphal growth (45). Conversely, activation of the DNA checkpoint by deletion of C. albicans RAD52, a gene responsible for the repair of double-stranded DNA breaks by homologous recombination, results in hyperfilamentous growth (2, 11, 15).
Based on these observations, we hypothesized that C. albicans orf19.4590 is involved in DNA damage responses and morphogenesis. Given the role of morphogenesis in candidal virulence, we further hypothesized that deletion of orf19.4590 would adversely affect the pathogenesis of mucosal and disseminated candidiasis in mice.
MATERIALS AND METHODS
Strains and growth condition.
C. albicans strains used or constructed in this study are described in Table 1. All strains were routinely grown in yeast extract-peptone-dextrose (YPD) medium (1% yeast extract, 1% Bacto peptone, 2% α-d-glucose) at 30°C unless otherwise noted. To induce hyphal formation in liquid media, C. albicans strains grown overnight on YPD agar were subcultured into liquid YPD supplemented with 5% fetal calf serum (FCS) at 37°C. To induce hyphal formation on solid media, overnight-grown C. albicans strains in YPD at 30°C were subcultured onto Spider medium, medium 199 (Gibco-BRL; adjusted to pH 7.5), and YPD medium supplemented with 5% FCS and grown at 37°C. A diploid S. cerevisiae rfx1 deletion strain (strain Hom14-D-1-34125; purchased from Open Biosystems, Huntsville, AL) was grown in YPD containing 200 μg/ml of G418.
TABLE 1.
Candida albicans and Saccharomyces cerevisiae strains used in this study
| Strain | Genotype or description | Reference or source |
|---|---|---|
| SC5314 | Clinical isolate | 23 |
| RFX2-1 | RFX2/rfx2Δ::SAT1-FLIP | This study |
| RFX2-2 | RFX2/rfx2Δ::FRT | This study |
| RFX2-3 | Rfx2Δ::SAT1-FLIP/rfx2Δ::FRT | This study |
| RFX2-4 (rfx2 mutant) | Rfx2Δ::FRT/rfx2Δ::FRT | This study |
| RFX2-5 (reinsertion) | RFX2::SAT1-FLIP/rfx2Δ::FRT | This study |
| RFX2-6 (reinsertion) | RFX2::FRTP/rfx2Δ::FRT | This study |
| BY4743 | Wild-type S. cerevisiae | Open Biosystems |
| Hom14D-1-34125 | S. cerevisiae rfx1 homozygous diploid mutant | Open Biosystems |
| Hom14D-1-34125 complemented with C. albicans orf19.4590 | Hom14D-1-34125 [yPB1-ADH1-orf19.4590] | This study |
| Hom14D-1-34125 complemented with S. cerevisiae RFX1 | Hom14D-1-34125 [yPB1-ADHp-ScRFX1] | This study |
Complementation study.
Genomic DNA of S. cerevisiae BY4743 and C. albicans SC5314 was used as templates for PCR. S. cerevisiae RFX1 and its downstream flanking sequence (999 bp downstream of the stop codon) were amplified with primers ScRFX1Exp-F (5′-CCGGTTCCAGATCTATGGTAATCTTCAAAGAACG-3′) and ScRFX1Exp-R (5′-CCGGTTCCAGATCTGGTGCTCTGTCTGAAAATGAC-3′). C. albicans orf19.4590, a 31-bp sequence upstream of the start codon, and a 716-bp sequence downstream from the stop codon were amplified with primers CaRFX1Exp-F (5′-CGCGGATCCTCCAAACTGTTCGTTGATTC-3′) and CaRFX1Exp-R (5′-CGCGGATCCAAGGAACGCGGGATAGTT-3′). BglII and BamHI restriction sites (underlined) were introduced into forward and reverse primers, respectively, of S. cerevisiae RFX1 and C. albicans orf19.4590. The PCR products were digested with BamHI or BglII and inserted into the BglII site of the C. albicans expression vector pYPB1-ADH1; this vector contains the C. albicans ADH1 promoter and terminator regions, an autonomously replicating sequence, C. albicans URA3 as a selectable marker, and S. cerevisiae 2μm sequences (kindly provided by Malcolm Whiteway). The insert sequence and orientation were confirmed by sequencing. The expression plasmid was then transformed into a diploid S. cerevisiae rfx1 null mutant (strain Hom14-D-1-34125) by high-efficiency yeast transformation using the lithium acetate/single-stranded carrier DNA/polyethylene glycol method (22). Ura+ transformants were selected using synthetic dextrose agar lacking uridine (0.67% yeast nitrogen base without amino acids, 0.077% uracil dropout supplement, 2% dextrose, and 1.5% agar). Vector yPB1-ADH1 alone was used as the control. Expression of S. cerevisiae RFX1 and C. albicans orf19.4590 was confirmed for the complemented strains and excluded for the control strain by reverse transcription-PCR (RT-PCR).
Construction of C. albicans RFX2 mutants.
For reasons elucidated in the Results and Discussion sections, we renamed orf19.4590 C. albicans RFX2. Disruptions of both alleles of C. albicans RFX2 were performed using the SAT1 flipper method (38). The proximal fragment (F1) at bp −898 to +346 relative to ATG of RFX2 was amplified by PCR using the primers 5′-CAAGTCACGTAGGGCCCCCTAAACTCAAACAACTTTGC and 5′-ATATACTCGAGGATCTAGCTCTGATGATCTG; the ApaI and XhoI restriction sites, respectively (underlined), were introduced. The distal gene fragment (F2) at bp +2557 to +3178 relative to the ATG of RFX2 was amplified using the primers 5′-TGGGCCATCTTCCGCGGACCACATCAATGGGTGAAG and 5′CTCATAGATTACACGAGCTCAGGGTTGGGTGTCATGTAGT, which contained the introduced SacII and SacI restriction sites (underlined). Following amplification, the fragments were digested with appropriate restriction enzymes and ligated sequentially into the plasmid pSFS2, resulting in pSFS2-RFX2. The disruption cassette was released by digesting pSFS2-RFX2 with ApaI and SacI, and it was transformed into C. albicans strain SC5314 by electroporation. Nourseothricin-resistant (NouR) transformants were selected on YPD plates containing 200 μg/ml of nourseothricin at 30°C.
After confirmation of disruption by PCR and Southern analysis, NouR colonies were grown in YPMal (1% yeast extract, 1% Bacto peptone, 2% maltose) at 30°C for 6 h. Thereafter, the culture was diluted, plated on YPD agar plates supplemented with 15 μg/ml nourseothricin, and incubated at 30°C. Smaller colonies were parallel streaked on YPD agar with 10 μg/ml nourseothricin and YPD agar to verify nourseothricin sensitivity. Genomic DNA was then isolated from several individual nourseothricin-sensitive (NouS) colonies, and excision of the cassette was verified by Southern blotting. To disrupt the second copy of RFX2, the NouS strain was transformed with pSFS2-RFX2. A screening process for nourseothricin susceptibility similar to that for the single RFX2 copy disruption was performed. The rfx2 null mutant strains were confirmed by PCR and Southern analysis.
To reinsert one copy of RFX2 at its own locus, the open reading frame was amplified from bp 117 upstream of the start codon to bp 465 downstream from the stop codon by PCR. The primers used were 5′-GCTACAGGTACCTGAGAGTGATCTAGAATAGTG and 5′-CAAGTCAACGTAGGGCCCGTGTGAGGGTATTATCGTGA, in which KpnI and ApaI sites, respectively (underlined), were induced. The amplified fragment and F2, described above, were sequentially cloned into pSFS2. The reinsertion cassette was released by KpnI/SacI digestion and used to transform a NouS rfx2 null mutant. The success of the reinsertion was confirmed by Southern blot analysis.
RT-PCR.
RNA was isolated using the RiboPure-Yeast kit (Ambion, Austin, TX). Contaminating chromosomal DNA was removed by treatment with DNase I and removal reagents provided in the RiboPure-Yeast kit. cDNA was made using the ImProm-II reverse transcription system (Promega, Madison, WI). PCR was performed on cDNA templates using primers designed from the sequences of the genes of interest (Table 2), under the following conditions: 1 min at 94°C, 1 min at 55°C, and 1 min at 68°C, preceded by denaturation for 5 min at 94°C and followed by a final extension cycle for 7 min at 68°C. The reactions were initially carried out at 30 cycles; for individual genes, additional cycles were performed to verify that expression ratios remained constant until maximal amplification intensity was achieved. The absence of genomic DNA contamination was controlled by including the constitutively expressed housekeeping gene EFB1 (elongation factor 1β gene) as an internal mRNA control (12). EFB1 contains an intron, such that PCR products from templates of genomic DNA and cDNA yield distinct product sizes of 891 and 526 bp, respectively. PCR products for the genes of interest were sequenced to verify that the desired C. albicans genes were amplified. Measurements of amplified band intensities were made with the ABI Prism 7700 SDS instrument (Applied Biosystems, Foster City, CA) based on ethidium bromide staining. We used EFB1 to normalize the transcript concentrations for each studied gene. EFB1 has been shown to be a useful internal standard for RT-PCR by our laboratory and others because its levels of expression in living C. albicans cells are similar in the conditions under which we performed our experiments (12, 41, 42). Furthermore, we showed by Northern blot analysis that the transcript levels of EFB1 quantitated against C. albicans ACT1 in the wild-type SC5314, rfx2 null mutant, and heterozygous mutant strains were similar (data not shown). All RT-PCR experiments were performed in triplicate on at least two separate days, and the average and standard deviation of the concentration of each band were calculated. The levels of expression by S. cerevisiae and C. albicans mutant strains were compared with those from the appropriate wild-type strains. The difference in levels of expression was considered significant if the mutant/wild-type ratio of expression was >2 and the difference was statistically significant (P value < 0.05).
TABLE 2.
Primers used for RT-PCR
| Species and gene | Primers | Source or reference |
|---|---|---|
| S. cerevisiae | ||
| RFX1 | 5′-ACTTCAAAGGCAGCTTTGCA-3′, 5′-TCAAAAGCGGATGTTCTGTG-3′ | This study |
| HUG1 | 5′-GCCTTAACCCAAAGCAAT-3′, 5′-TCTTACCAATGTCAGAAAGAC-3′ | This study |
| RNR3 | 5′-ATTGCCATGAAGGATGACTCT-3′, 5′-TGAAACCTCAACGAATGCTG-3′ | This study |
| EFB1 | 5′-TCTTTGGCTGACAAGTCATACATTG-3′, 5′-ATAGCAGCAATATCGGTAGATTGG-3′ | This study |
| C. albicans | ||
| RFX2 | 5′-GAGTACCGCCACCACTATA-3′, 5′-GCATTCCACATTACACCAAG-3′ | This study |
| HWP1 | 5′-ATGACTCCAGCTGGTTC-3′, 5′-TTAGATCAAGAATGCAGC-3′ | 12 |
| ECE1 | 5′-ACCTACTGTTCCTGCACCTCA-3′, 5′-CCGACAGTTTCAATGCTCTTT-3′ | This study |
| ALS3 | 5′-CACAATCCCCATCTGGTATT-3′, 5′-TGTGATCAAACCACATAACCA-3′ | This study |
| HYR1 | 5′-TTCTGGTTCTGGCTCTCAAA-3′, 5′-CCACCAGTAACAATAGATGAA-3′ | This study |
| RNR21 | 5′-GAGATGAAGGTTTACACACCG-3′, 5′-AAAGGTAAAAGCATCGGC-3′ | This study |
| RNR1 | 5′-GGGATTGGGATACATTAAAAC-3′, 5′-AGGACATGCGATACCTTTGG-3′ | This study |
| RAD6 | 5′-GACCTTCAGATACACCATTT-3′, 5′-TTCCTCCTCCTCATCATCA-3′ | This study |
| DDR48 | 5′-CGGTAAAGACGACGACAA-3′, 5′-CAGAAGATCCATAGGAGTCAC-3′ | This study |
| MEC1 | 5′-GTGGATTTGAGTGAACCAA-3′, 5′-CGATAAATCCCATCAACTC-3′ | This study |
| EFB1 | 5′-ATTGAACGAATTCTTGGCTGAC-3′, 5′-CATCTTCTTCAACAGCAGCTTG-3′ | 12 |
| ACT1 | 5′-ACTCTTCTGGTAGAACTACCG-3′, 5′-ACTTTCATAGAAGATGGAGAA-3′ | This study |
UV irradiation.
To test for susceptibility to UV radiation, overnight-grown yeast cells were diluted with fresh YPD to a concentration of 1,000 CFU/ml; 200 μl of the diluted culture was spread onto YPD agar plates. The plates were then irradiated at 10.0 mJ/cm2 (C. albicans) or 15 mJ/cm2 (S. cerevisiae) with a UV cross-linker (Spectrolinker XL-1000; Fisher Scientific, Pittsburgh, PA). Nonirradiated yeast cells were plated as controls. The plates were incubated at 30°C for 24 h for colony enumeration.
For RT-PCR experiments, C. albicans cells were grown in individual wells of a six-well plate (Costar) at 30°C with 200 rpm shaking until log phase. The supernatant was removed after centrifugation at 1,200 × g for 10 min. The uncovered six-well plate was irradiated at 10 mJ/cm2 using the UV cross-linker. Immediately after irradiation, 2 ml of 30°C-prewarmed YPD was added to each well. After 1- and 3-hour incubations at 30°C, the yeast cells were harvested for RNA extraction.
Susceptibility testing.
Log phase C. albicans cells were tested against hydroxyurea (10, 20, 30, 40, 50, and 60 mg/ml), methyl methanesulfonate (MMS; twofold dilution from 12 mM to 0.325 mM), bleomycin (twofold dilution from 16 μg/ml to 0.5 μg/ml), streptonigrin (twofold dilution from 1.6 μg/ml to 0.05 μg/ml), ethyl methanesulfonate (twofold dilution from 120 mM to 1.67 mM), H2O2 (17.6, 13.2, 8.8, 6.6, and 4.4 mM), NaCl (0.7 mM), and ethanol (10%). For heat shock experiments, log phase C. albicans cells were diluted 10-fold in YPD and placed in 1-ml Eppendorf tubes. The tubes were placed in a 48°C water bath for 3 min, and 100 μl of the samples was plated onto Sabouraud dextrose agar (SDA) plates for colony enumeration.
Agar invasion assay.
Overnight-grown Candida cells in YPD at 30°C were resuspended in fresh YPD to achieve a concentration at 106 CFU/ml. Two microliters of this inoculum was spotted onto the surfaces of YPD agar plates, which were then incubated for 48 h at 30°C. Cells that had not invaded the agar were washed away by rubbing the plate with a gloved finger while rinsing under running water. Cross-sectional slices of the agar-invasive growth were photographed.
Adherence to BECs.
Adherence to buccal epithelial cells (BECs) was measured as previously described (12, 39). Briefly, BECs pooled from three investigators were dispensed into 10 ml of phosphate-buffered saline (PBS), washed, and adjusted to a final concentration of 1 × 105 epithelial cells/ml PBS. Then, 0.5 ml of the epithelial cells was incubated in a glass tube with 0.5 ml of C. albicans cells at a concentration of 1 × 106 cells/ml in a shaking incubator at 35°C for 1 hour. Following incubation, the cells were vacuum filtered through prewet 20-mm-diameter polycarbonate filters with 10-μm pore size (Millipore, Billerica, MA) mounted on a filter manifold (Millipore, Bedford, MA). Each filter was washed 10 times with PBS to remove unattached Candida cells. The washed filters were then removed and pressed gently onto glass slides. The slides were air dried, heat fixed for 1 min, Gram stained, and examined by light microscopy. The Candida cells attached to 100 BECs were counted in multiple fields. Each experiment was performed in duplicate and repeated at least twice on two different days. Results for each strain were expressed as the mean percentage of adherence ± standard deviation (i.e., mean number of Candida cells/100 BECs). The difference in adherence to BECs between the wild-type and mutant strains was determined using Student's t test; a P value ≤0.05 was considered significant.
Murine model of OPC.
A murine model of OPC was developed as previously described (13) with some modifications. Seven-week-old male ICR mice (Harlan Sprague) with an approximate weight of 20 g were immunosuppressed with 200 mg/kg of body weight of cortisone acetate (Sigma Aldrich, St. Louis, MO) in saline with 0.1% Tween 80 administered subcutaneously on the day before inoculation and day 1 and day 4 after inoculation. Mice received tetracycline hydrochloride in their drinking water (0.5 mg/ml), starting the day before inoculation. For infection, the mice were first anesthetized by intraperitoneal injections with 140 mg/kg of pentobarbital sodium (Nembutal) solution (Abbott Laboratories, North Chicago, IL); this dose kept the mice well sedated for at least 3 h. After the mice were fully sedated, cotton wool balls (diameter, 3 mm) saturated with 50 μl of 1 × 108 CFU/ml of C. albicans were placed sublingually in the oral cavity for 2 h. Fifteen to 17 mice per group were used. The mice were euthanized by CO2 asphyxiation followed by cervical dislocation at 6, 24, and 72 h and on day 7 after infection. The esophagus, mandibular soft tissue, and tongue were dissected free of teeth and bone. The tissue was homogenized in saline, plated onto SDA containing ampicillin (100 μg/ml) and amikacin (60 μg/ml), and incubated at 30°C for 48 h. Tissue burden was enumerated by colony count. Interquartile values were used in the data analysis, and tissue burdens for mice infected with each strain were presented as mean log10 of tissue burden/gram of tissue ± standard deviation. The difference in tissue burden between mice infected with the wild-type or mutant strains was determined by Wilcoxon's test. P values of ≤0.05 were considered significant. For histopathology study, the tissues were fixed with formalin and embedded in paraffin, after which thin sections were prepared and stained with Gomori methenamine silver stain.
Murine model of disseminated candidiasis (DC).
Seven-week-old, male ICR mice (Harlan Sprague) were inoculated by intravenous injection of the lateral tail vein with 5 × 105 CFU of C. albicans strains in 0.2 ml of normal saline solution. Ten to 12 mice per group were used. Mice were followed until they were moribund, at which point they were sacrificed, or for 30 days. For tissue burden and histopathologic study, 12 mice per group were sacrificed at 6, 24, and 72 h postinfection, respectively, and the kidneys, livers, and spleens were obtained. For the tissue burden study, the kidneys, livers, and spleens were removed, weighed, and then homogenized in normal saline solution and plated on SDA containing ampicillin (100 μg/ml) and amikacin (60 μg/ml). Survival curves were calculated according to the Kaplan-Meier method using the PRISM program (GraphPad Software) and compared using the Newman-Keuls analysis. A P value of ≤0.05 was considered significant. As in OPC experiments, interquartile values were used in the data analysis. The tissue burdens were logarithmically transformed, and data were presented as mean log10 CFU/g tissue ± standard deviations; the differences in tissue burden between strains were calculated using Wilcoxon's test.
RESULTS
C. albicans orf19.4590 encodes an RFX domain-containing protein that complements an S. cerevisiae rfx1 null mutant strain.
In response to genotoxic insults, eukaryotes activate a conserved DNA damage response pathway that regulates the expression of genes such as those encoding the subunits of RNase reductase (RNR) (17, 56, 57). RNR catalyzes the rate-limiting step in the synthesis of deoxyribonucleotide triphophosphates, which are necessary for DNA replication and repair. S. cerevisiae RNR2, -3, and -4 and HUG1 (a gene of unknown function that is induced by hydroxyurea/UV light) are repressed under nonstressful conditions by S. cerevisiae Rfx1p, a downstream target of the DNA damage response pathway (52-54). As such, RNR genes and HUG1 are derepressed in an S. cerevisiae strain in which RFX1 is deleted (53).
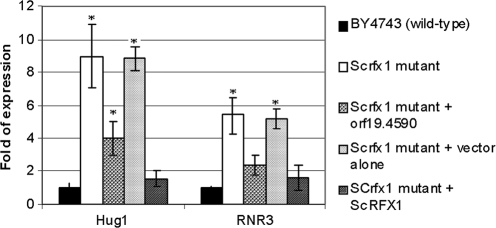
To determine if the protein encoded by C. albicans orf19.4590 is functionally related to S. cerevisiae Rfx1p, we measured expression of RNR3 and HUG1 in a diploid S. cerevisiae rfx1 null mutant strain (strain Hom14D-1-34152) transformed with a vector containing orf19.4590 and its flanking regions (Fig. 1). The S. cerevisiae rfx1 null mutant transformed with an S. cerevisiae RFX1 plasmid and the same mutant transformed with the vector alone served as positive and negative controls, respectively. In YPD medium at 30°C, the derepression of RNR3 and HUG1 in the null mutant was fully reversed in the transformants complemented with S. cerevisiae RFX1 (Fig. 1). In transformants complemented with orf19.4590, RNR3 and HUG1 derepression was partially reversed (Fig. 1). As anticipated, RNR3 and HUG1 derepression was unaffected in the S. cerevisiae rfx1 null mutant transformed with the vector alone. Experiments were repeated with independently created S. cerevisiae transformants with similar results. These findings suggest that orf19.4590 has some functional redundancy with S. cerevisiae RFX1.
FIG. 1.
HUG1 and RNR3 expression by an S. cerevisiae rfx1 null mutant complemented with C. albicans orf19.4590. Levels of HUG1 and RNR3 expression in wild-type S. cerevisiae strain BY4743, an S. cerevisiae rfx1 null mutant strain, and the S. cerevisiae rfx1 mutant complemented with C. albicans orf19.4590 were assessed by RT-PCR following growth to log phase in YPD at 30°C. The S. cerevisiae rfx1 mutants complemented with S. cerevisiae RFX1 or transformed with the vector alone were used as positive and negative controls, respectively. Data (means ± standard deviations) are expressed as the increases above the expression level of BY4743. The graph shows data from triplicate experiments. Asterisks denote statistical difference (P < 0.05) in the level of expression by the respective strains compared with the level of expression by BY4743.
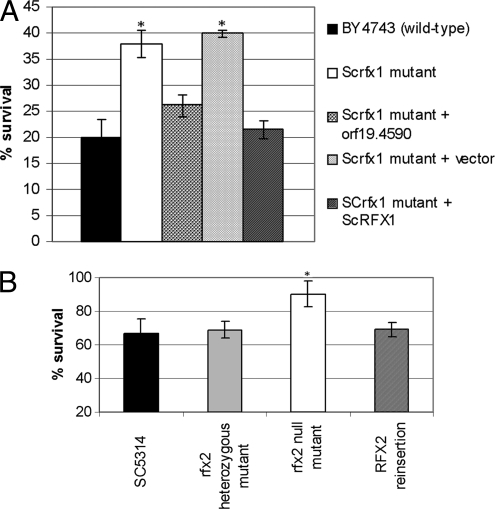
We next exposed the strains to UV radiation, which is known to induce S. cerevisiae RFX1 expression (33). In the presence of genotoxic insults, the S. cerevisiae DNA damage response pathway phosphorylates Rfx1p, which eliminates its repressor activity. The induction of RFX1 expression upon UV exposure is believed to ensure that ample nonphosphorylated Rfx1p is available to rapidly repress target gene transmission upon deactivation of the damage response pathway (33, 52). We exposed S. cerevisiae strains to 15 mJ/cm2 of UV radiation, followed by incubation in YPD at 30°C. As anticipated, S. cerevisiae RFX1 was upregulated by 13.1-fold ± 1.0-fold (data not shown). Deletion of RFX1 rendered S. cerevisiae significantly more resistant to UV killing (Fig. 2A). UV susceptibility was restored to levels consistent with the wild type in null mutant strains complemented with either orf19.4590 or S. cerevisiae RFX1 (Fig. 2A). The S. cerevisiae rfx1 null mutant transformed with the vector alone was indistinguishable from the null mutant.
FIG. 2.
Resistance of S. cerevisiae (A) and C. albicans (B) strains to UV irradiation. Log phase yeast cells freshly streaked onto a YPD agar plate were subjected to irradiation (15 mJ/cm2 for S. cerevisiae and 10 mJ/cm2 for C. albicans) using a UV cross-linker and then incubated at 30°C for 24 h for colony enumeration. The percent survival was calculated by the following formula: (number of colonies identified on a plate after UV exposure/number of colonies on a plate without UV exposure) × 100%. The experiments were performed in duplicate on two different days. The data presented are the mean percentages of survival ± standard deviations. Asterisks denote significant difference (P < 0.05) in survival of the respective strains compared with the survival of BY4743 (A) or SC5314 (B).
Taken together, the data suggest that C. albicans orf19.4590 can perform at least some of the functions of S. cerevisiae RFX1. Of note, a search of the C. albicans genome (http://www.candidagenome.org and http://genolist.pasteur.fr/CandidaDB) revealed two open reading frames that encode RFX domain-containing proteins (orf19.4590 and orf19.3865). In fact, the protein encoded by orf19.3865 more closely resembles S. cerevisiae Rfx1p in the DNA binding domain and over its full sequence (45.7% and 48.9% amino acid identity, respectively) than does the protein encoded by orf19.4590 (25.8% and 44.2% amino acid identity, respectively). For this reason, we will refer to orf19.4590 as C. albicans RFX2 hereafter, and we propose that the name C. albicans RFX1 be reserved for orf19.3865.
Deletion of C. albicans RFX2 results in increased resistance to UV killing, as well as the derepression of various stress-related genes.
To further study C. albicans RFX2, we independently constructed two sets of isogenic mutant strains in which one or both copies of the gene were disrupted and reinsertion strains in which a copy of the gene was reintroduced to the null mutant at the native locus. After verifying the desired disruptions and reinsertions by PCR amplifications and Southern analyses, we demonstrated that the mutant and reinsertion strains had growth rates similar to that of wild-type strain SC5314 in both liquid YPD and minimal media at 35°C (data not shown).
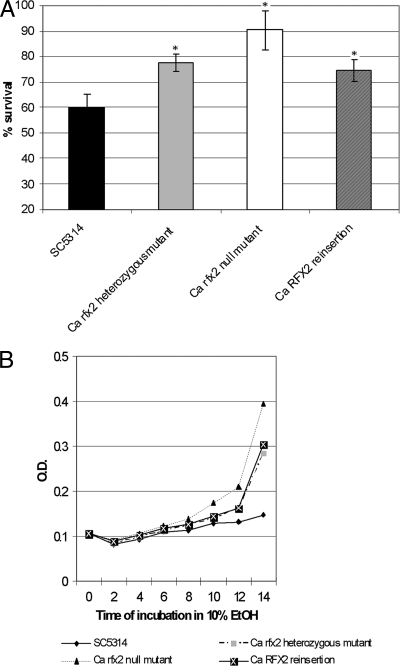
Next, we tested for susceptibility to ionizing radiation, hydroxyurea, and several radiomimetic drugs (bleomycin, MMS, streptonigrin, and ethyl methanesulfonate). Similar to the S. cerevisiae rfx1 null mutant and wild-type strains, the C. albicans rfx2 null mutant was more resistant to UV killing than SC5314 (Fig. 2B). The rfx2 heterozygous mutant and RFX2 reinsertion strains did not significantly differ from SC5314. We found no differences in the sensitivities of the rfx2 null mutant and SC5314 to hydroxyurea, bleomycin, MMS, streptonigrin, or ethyl methanesulfonate (data not shown). We also tested the susceptibility of the C. albicans strains to heat shock at 48°C and oxidative (ethanol, H2O2) and osmotic (NaCl) stresses. The rfx2 null mutant was significantly more resistant than SC5314 to heat shock and 10% ethanol (Fig. 3A and B) but equally susceptible to 0.7 mM NaCl and 13.2 mM H2O2 (data not shown). The rfx2 heterozygous mutant and RFX2 reinsertion strains were more resistant than SC5314 but more susceptible than the rfx2 null mutant to heat shock and 10% ethanol, results reflecting possible haploinsufficiency. As for each of the in vitro phenotypes presented below, independently created heterozygous mutant, null mutant, and reinsertion strains yielded similar results.
FIG. 3.
Effects of C. albicans RFX2 on resistance to heat shock (A) and ethanol (B). For the heat shock assay (A), log phase C. albicans cells were dispensed to 1-ml Eppendorf tubes and exposed to a 48°C water bath for 3 min. The cells were then cultured on SDA plates at 35°C for colony enumeration. For susceptibility to ethanol (EtOH) (B), log phase C. albicans cells were exposed to 10% ethanol at 35°C with shaking at 200 rpm. Hourly optical density (O.D.) readings at 600 nm were obtained up to 12 h. Both experiments were performed in triplicate. The data are presented as the means of all experiments. Asterisks denote significant difference (P < 0.05) in survival of the respective strains compared with the survival of SC5314.
In S. cerevisiae, various stress-associated genes are upregulated in response to UV radiation (26, 27). Based on these reports, we examined the expression of C. albicans RFX2, RNR1, RNR21, RAD6 (a gene induced by and required for resistance to UV radiation) (5, 31), and DDR48 (a gene encoding a Hog1p-induced immunogenic-stress-related protein and involved in DNA repair) (16). S. cerevisiae RNR1 and RNR2, -3, and -4 encode large and small subunits of RNR, respectively. In the C. albicans genome database, the corresponding genes are RNR1 and RNR21 (also referred to as RNR2). In YPD at 30°C, C. albicans RAD6 and DDR48 were significantly derepressed in the rfx2 null mutant compared to their expression by strain SC5314 (29.9- and 8.6-fold, respectively) (Table 3). RNR21 and RNR1 were derepressed only 1.9- and 1.3-fold, respectively, in the null mutant, differences of uncertain biological significance. The levels of RAD6 and DDR48 expression in the rfx2 heterozygous mutant and RFX2 reinsertion strains were intermediate to the levels in the null mutant and SC5314 (Table 3). The findings suggest that the expression of RAD6 and DDR48 is directly or indirectly repressed by RFX2.
TABLE 3.
Effects of C. albicans RFX2 on the expression of DNA damage genes
| Gene | Relative mRNA transcript levela in indicated strain
|
|||||||
|---|---|---|---|---|---|---|---|---|
| In YPD at 30°C
|
After UV radiation
|
|||||||
| SC5314 | rfx2 heterozygous mutant | rfx2 null mutant | rfx2 reinsertion strain | SC5314 | rfx2 heterozygous mutant | rfx2 null mutant | rfx2 reinsertion strain | |
| RNR21 | 1.00 ± 0.02 | 1.37 ± 0.03 | 1.86 ± 0.01 | 1.32 ± 0.02 | 2.19 ± 0.01* | 2.29 ± 0.02* | 2.43 ± 0.02* | 2.33 ± 0.03* |
| RNR1 | 1.00 ± 0.01 | 1.21 ± 0.04 | 1.30 ± 0.01 | 1.10 ± 0.01 | 1.07 ± 0.01 | 1.37 ± 0.00 | 1.78 ± 0.01 | 1.43 ± 0.01 |
| RAD6 | 1.00 ± 0.02 | 2.89 ± 0.37* | 29.92 ± 4.18* | 3.34 ± 0.81* | 4.52 ± 0.62* | 5.68 ± 0.48* | 27.51 ± 6.17* | 5.72 ± 0.70* |
| DDR48 | 1.00 ± 0.04 | 3.63 ± 0.03* | 8.61 ± 0.30* | 3.41 ± 0.02* | 4.93 ± 0.04* | 4.61 ± 0.30* | 9.38 ± 0.14* | 4.96 ± 0.06* |
| MEC1 | 1.02 ± 0.02 | 1.15 ± 0.01 | 1.01 ± 0.09 | 1.01 ± 0.02 | 4.28 ± 0.11* | 3.66 ± 0.04* | 3.84 ± 0.10* | 3.72 ± 0.18* |
Asterisks denote biological (increase of more than twofold) and statistical (P < 0.05) significance of the level of expression by the respective strains compared with the level of expression by SC5314.
Following exposure to UV radiation, C. albicans RFX2 was upregulated by 10.12-fold ± 0.25-fold in strain SC5314 (data not shown), a finding that was consistent with our earlier data for S. cerevisiae RFX1. RAD6 and DDR48 were also significantly upregulated in SC5314 after UV exposure (Table 3). Unlike SC5314, the rfx2 mutant was not able to significantly increase the expression of any of the genes in response to UV exposure, suggesting that the genes were already maximally derepressed.
S. cerevisiae MEC1 encodes a phosphoinositide kinase that functions upstream of Rad53p and Rfx1p as the major checkpoint in the DNA damage response pathway (25, 41). We demonstrated that, as anticipated, C. albicans MEC1 was induced in response to UV exposure (Table 3). The expression pattern was similar for the C. albicans rfx2 null mutant strain, demonstrating that RFX2 does not repress MEC1 and likely functions downstream in the damage response pathway.
The C. albicans rfx2 null mutant displays hyperfilamentous growth and overexpresses hypha-specific genes.
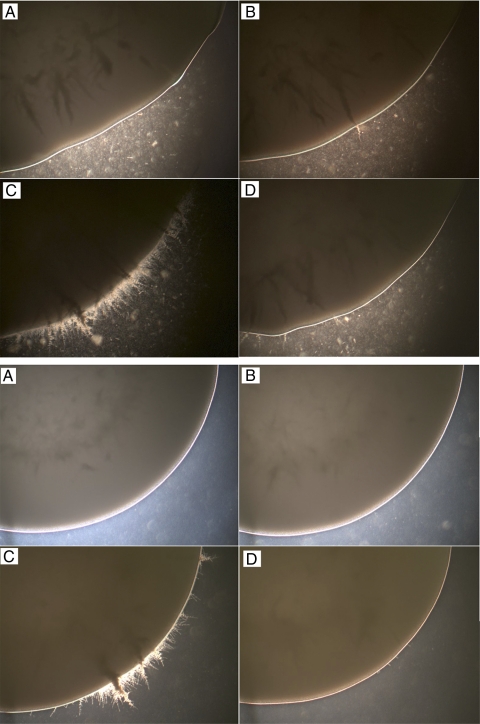
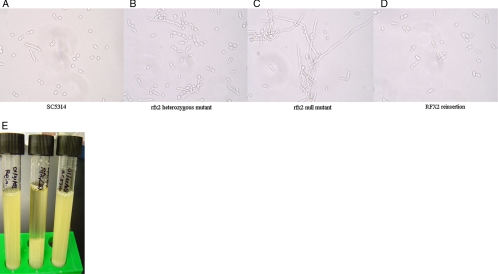
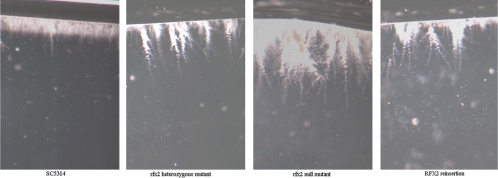
Having confirmed our hypothesis that RFX2 would repress the expression of DNA damage response genes, we evaluated the effects of gene deletion on morphogenesis. Colonies of SC5314, rfx2 heterozygous mutant, and RFX2 reinsertion strains were smooth under non-hypha-inducing conditions on solid agar (YPD at 30°C for 72 h). The colonies of the rfx2 null mutant, on the other hand, were wrinkled (data not shown). Examination of the colony margins revealed extensive filaments for the null mutant and no or minimal filaments for SC5314, heterozygous mutant, and reinsertion strains (Fig. 4). Similar results were obtained following growth on SDA medium at 30°C (Fig. 4). Under hypha-inducing conditions on solid agar (YPD medium supplemented with 5% FCS, medium 199, and Spider medium at 37°C), all strains demonstrated filamentous growth (data not shown). In YPD liquid medium incubated at 30°C, the strains were all in the yeast morphology and indistinguishable from one another. Under hypha-inducing conditions in liquid medium (YPD supplemented with 5% FCS at 37°C), the strains formed extensive hyphae, although those of the null mutant were 20 to 50% longer than those of the SC5314, rfx2 heterozygous mutant, and RFX2 reinsertion strains at the same time points (data not shown). Moreover, the majority of SC5314, heterozygous mutant, and reinsertion cells reverted to yeast forms following overnight growth in YPD plus 5% serum at 37°C, whereas the majority of null mutant cells remained in hyphal and pseudohyphal morphologies (Fig. 5). In addition to a hyperfilamentous phenotype, the rfx2 null mutant strain showed enhanced invasion into solid agar (Fig. 6). The invasive growth of rfx2 heterozygous mutant and RFX2 reinsertion strains was intermediate to that of SC5314 and that of the rfx2 null mutant.
FIG. 4.
Morphologies of C. albicans strains under non-hypha-inducing conditions on solid media. C. albicans cells grown overnight in YPD at 30°C were subcultured onto YPD agar (top) and SDA (bottom) and grown at 37°C for 72 h. (A) SC5314; (B) rfx2 heterozygous mutant; (C) rfx2 null mutant; (D) RFX2 reinsertion strain.
FIG. 5.
Morphologies of C. albicans strains after overnight growth. C. albicans strains were grown overnight in YPD supplemented with 5% serum at 37°C. Representative cell morphologies of C. albicans SC5314 and rfx2 heterozygous mutant, rfx2 null mutant, and RFX2 reinsertion strains are pictured. The null mutant existed predominantly as clumps of hyphae (pictured) and pseudohyphae (not shown). Following overnight growth, the cultures were transferred to test tubes and let stand for 15 min (E). As pictured, the filamentous cells of the null mutant precipitated at the bottom of the tube (tube 2). The blastoconidia of SC5314 (tube 1) and the RFX2 reinsertion strain (tube 3) remained in suspension. The tubes containing the rfx2 null mutant (not shown) resembled those containing C. albicans SC5314 and the RFX2 reinsertion strain.
FIG. 6.
Agar invasion by C. albicans strains. Overnight-grown C. albicans cells in YPD at 30°C were resuspended in fresh YPD and inoculated onto the surfaces of YPD agar plates, which were then incubated for 48 h at 30°C. Cells that did not invade the agar were washed away, and cross-sectional slices of the agar were photographed.
As a follow-up to the morphogenesis experiments, we evaluated the expression of hypha-specific genes using quantitative RT-PCR. Under conditions that typically promote growth as yeast (YPD liquid medium at 30°C), HWP1, ECE1, ALS3, HYR1, and CEK1 mRNA transcript levels in strain SC5314 were predictably low. mRNA transcript levels in the rfx2 null mutant strain were higher by 4.2-fold to 16.8-fold (Table 4), suggesting that Rfx2p either directly or indirectly repressed the hypha-specific genes under non-hypha-inducing conditions. Reintroducing RFX2 into a null mutant background reduced mRNA transcript levels of each gene to those observed for the rfx2 heterozygous mutant. Following exposure to UV radiation, HWP1, HYR1, and ECE1 were significantly induced in SC5314 (14.5-fold, 11.0-fold, and 4.9-fold, respectively), ALS3 was slightly induced (2.3-fold), and CEK1 was not significantly induced (1.8-fold) (Table 4). After UV exposure, the rfx2 null mutant was able to induce HWP1 and ECE1 above their derepressed basal levels but was not able to further induce ALS3, HYR1, and CEK1.
TABLE 4.
Effects of C. albicans RFX2 on the expression of hypha-specific genes
| Gene | Relative mRNA transcript levela in indicated strain
|
|||||||
|---|---|---|---|---|---|---|---|---|
| In YPD at 30°C
|
After UV radiation
|
|||||||
| SC5314 | rfx2 heterozygous mutant | rfx2 null mutant | RFX2 reinsertion strain | SC5314 | rfx2 heterozygous mutant | rfx2 null mutant | RFX2 reinsertion strain | |
| HWP1 | 1.02 ± 0.02 | 4.41 ± 0.2* | 14.43 ± 0.59* | 5.10 ± 2.32* | 14.55 ± 0.35* | 13.96 ± 2.50* | 33.30 ± 6.95* | 15.90 ± 2.75* |
| ECE1 | 0.96 ± 0.04 | 4.69 ± 1.53* | 16.82 ± 0.51* | 4.99 ± 2.50* | 4.86 ± 0.05* | 4.97 ± 3.16* | 29.26 ± 3.04* | 16.24 ± 2.56* |
| ALS3 | 1.00 ± 0.02 | 1.52 ± 0.01 | 5.18 ± 0.06* | 1.44 ± 0.01 | 2.31 ± 0.02* | 4.81 ± 0.07* | 8.70 ± 0.17* | 5.51 ± 0.21* |
| HYR1 | 1.01 ± 0.02 | 7.33 ± 2.15* | 13.34 ± 0.16* | 8.42 ± 0.40* | 11.04 ± 0.09* | 7.42 ± 0.36* | 14.28 ± 1.00* | 9.25 ± 1.43* |
| CEK1 | 1.00 ± 0.01 | 1.53 ± 0.97 | 4.21 ± 0.08* | 1.82 ± 0.30 | 1.84 ± 0.01 | 1.74 ± 0.02 | 3.32 ± 0.71* | 2.46 ± 0.32* |
Asterisks denote biological (increase of more than twofold) and statistical (P < 0.05) significance of the level of expression by the respective strains compared with the level of expression by SC5314.
The rfx2 null mutant demonstrates increased adherence to epithelial cells in vitro but decreased virulence during murine DC and OPC.
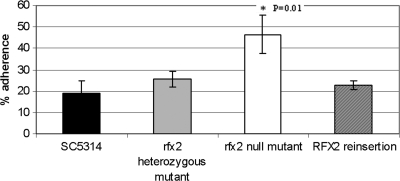
To assess the potential role of RFX2 in candidal virulence, we first measured the adherence of C. albicans strains to BECs in vitro. The rfx2 null mutant was significantly more adherent to BECs than SC5314, the rfx2 heterozygous mutant strain, or the RFX2 reinsertion strain (Fig. 7). The results were not ascribed to differences in cell morphology, as all strains were found to be blastoconidia by Gram staining (data not shown).
FIG. 7.
Adherence by C. albicans strains to BECs in vitro. C. albicans cells were coincubated with BECs for 60 min, as described in Materials and Methods. Percent adherence is defined as the mean number of C. albicans cells/100 BECs. Data are presented as means of all experiments ± standard deviations.
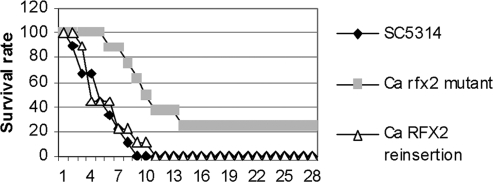
We next tested the strains in murine models of DC and OPC. For DC, groups of 10 to 12 ICR mice were infected via the lateral tail vein with 5 × 105 CFU/mouse. There was a significant delay in the time to death among mice infected with the rfx2 null mutant compared to those infected with SC5314 (mean of 17.5 ± 8.7 versus 6.3 ± 2.4 days, P = 0.002) (Fig. 8). All of the mice infected with SC5314 died by day 9, whereas 25% of mice infected with the null mutant were still alive on day 30. Results with the RFX2 reinsertion strain were similar to those with SC5314. In separate experiments, we measured candidal tissue burdens in the kidneys, livers, and spleens at 6, 24, and 72 h after intravenous infection with SC5314 or the rfx2 null mutant (12 mice per strain per time point) (Table 5). At 6 h, concentrations of the null mutant were slightly higher than those of SC5314 within the kidneys and liver (0.52 and 0.77 log10 CFU/g tissue, respectively). Concentrations within the spleen did not differ. At 24 h, there were no significant differences in tissue burdens within any of the organs. By 72 h, concentrations of SC5314 in the kidneys were 1.11 log10 CFU/g tissue higher than those of the rfx2 null mutant (P < 0.0001), while concentrations within the liver and spleen did not differ. Histopathologic examination of the kidneys at 72 h showed that both strains grew as mixtures of yeasts and hyphae (data not shown).
FIG. 8.
Effects of RFX2 on the survival of mice with HDC. Seven-week-old, male ICR mice (Harlan Sprague) were inoculated by intravenous injection of the lateral tail vein with 5 × 105 CFU of C. albicans strains and followed for 30 days.
TABLE 5.
C. albicans tissue burdens among mice with HDCa
| Organ and strain | Mean log10 CFU/g of tissueb ± SD at:
|
||
|---|---|---|---|
| 6 h | 24 h | 72 h | |
| Kidney | |||
| SC5314 | 4.22 ± 0.12 | 5.17 ± 0.27 | 5.59 ± 0.09 |
| rfx2 mutant | 4.74 ± 0.10 (0.03) | 5.11 ± 0.12 (NS) | 4.48 ± 0.14 (<0.0001) |
| Liver | |||
| SC5314 | 3.23 ± 0.05 | 3.03 ± 0.13 | 2.38 ± 0.31 |
| rfx2 mutant | 4.00 ± 0.04 (<0.0001) | 3.08 ± 0.14 (NS) | 2.56 ± 0.26 (NS) |
| Spleen | |||
| SC5314 | 3.94 ± 0.15 | 3.41 ± 0.66 | 3.06 ± 0.48 |
| rfx2 mutant | 4.11 ± 0.31 (NS) | 3.79 ± 0.32 (NS [P = 0.1]) | 2.92 ± 0.37 (NS) |
Mice were infected via lateral tail vein injection with 5 × 105 CFU/mouse. They were sacrificed at 6, 24, and 72 h. The kidneys, spleens, and livers were removed for CFU enumeration.
Significance (P values or NS [not significant]) for the wild type versus mutants is indicated in parentheses.
In the OPC model, mice were immunosuppressed with cortisone acetate and infected sublingually for 2 h with 5 ×106 CFU (15 to 17 mice per strain per time point). The candidal burdens within the tongues, buccal mucosa, and esophagi of mice infected with SC5314 at 6 and 24 h postinfection (3.45 ± 0.62 and 5.06 ± 0.40 log10 CFU/g tissue, respectively) were not different from the burdens due to the rfx2 null mutant (3.48 ± 0.45 and 5.03 ± 0.29 log10 CFU/g tissue, respectively). On the sixth day following infection, three mice infected with SC5314 died, with evidence of DC. For this reason, we sacrificed the remaining mice on day 7 and assessed the presence of C. albicans in the kidneys, livers, lungs, spleens, and preputial glands (the preputial glands, genital organs that secrete pheromones, often harbor significant concentrations of C. albicans during DC [S. Cheng, C. J. Clancy and M. H. Nguyen, unpublished data]). We found that 82.4% (14/17) of mice infected with SC5314 had evidence of DC (defined as the presence of C. albicans in any organ) on day 7, compared to only 26.7% (4/15) of mice infected with the rfx2 null mutant (P = 0.004). We then repeated the analysis of the OPC model by applying the same inoculum for 1 hour, including the RFX2 reinsertion strain in addition to SC5314 and the rfx2 null mutant (15 mice per strain per time point). The mice were observed for 7 days; no deaths were recorded. At 7 days postinfection, the mice infected with SC5314 had significantly higher tissue burdens within the tongue, buccal mucosa, and esophagus than mice infected with the null mutant (4.8 ± 0.5 versus 2.6 ± 0.8 log10 CFU/g tissue, respectively; P < 0.0001). There was no significant difference in tissue burdens of mice infected with SC5314 and the RFX2 reinsertion strain (data not shown). A histopathology study showed that the tongues and esophagi of mice infected with the rfx2 null mutant had minimal chronic inflammatory reaction compared with those of mice infected with SC5314. Both yeast and hyphal morphologies were observed in the tissues from mice infected with SC5314 (Fig. 9). A histopathology study of the tongues from mice infected with the rfx2 null mutant showed only rare hyphal elements, and no fungal elements were visualized on stains of the esophagus.
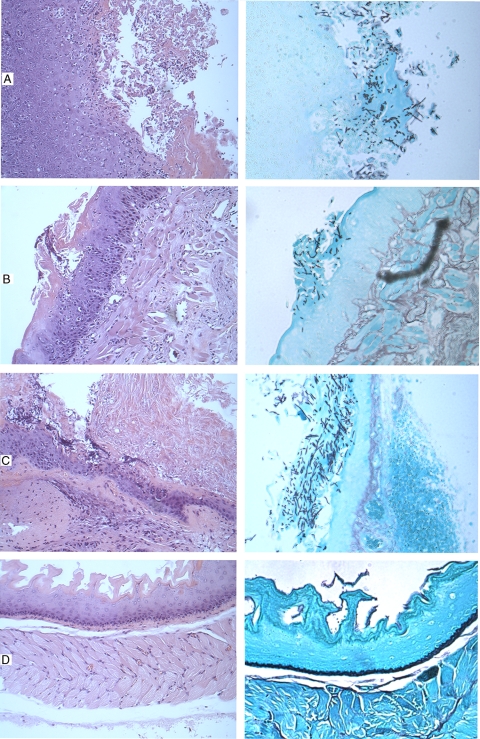
FIG. 9.
Histopathology of the tongues (A and B) and esophagi (C and D) of mice infected with C. albicans strains. Mice were infected sublingually with C. albicans SC5314 (A and C) or the rfx2 null mutant (B and D), as described in Materials and Methods. Organs were harvested after 7 days, processed, and stained with hematoxylin-eosin (left) and Gomori methenamine silver (right). In the tongues of mice infected with C. albicans SC5314 (A), an intense intraepithelial lymphocyte response is noted in the squamous epithelium, with associated hyperkeratosis and neutrophils. Fungal elements are extensive and comprise roughly equal measures of hyphae and yeasts. In the tongues of mice infected with the rfx2 null mutant (B), the epithelium has a mild degree of intraepithelial lymphocytosis and only focal keratosis. Fungal elements are less common and predominantly hyphae. In the esophagi of mice infected with C. albicans SC5314 (C), there is a moderate lymphocyte response. As in the tongue, there are extensive yeasts and hyphae. In the esophagi of mice infected with the rfx2 null mutant (D), no inflammation or fungal elements were detected.
DISCUSSION
Microbes suffer DNA damage due to a variety of stresses in the course of their interactions with infected hosts. As such, the regulation of DNA damage responses is an important determinant of successful adaptation to in vivo environments (19). In this study, we show that C. albicans RFX2 encodes an RFX domain-containing protein that represses the transcription of DNA damage response genes under nonstressful conditions. A C. albicans rfx2 null mutant exhibits hyperfilamentous and hyperinvasive growth, constitutive expression of hypha-specific genes, and attenuation of virulence during disseminated and mucosal candidiasis in mice. The data demonstrate that Rfx2p plays crucial roles in the regulation of DNA damage responses, morphogenesis, and virulence.
C. albicans RFX2 is one of two genes encoding RFX domain-containing proteins in the genome sequence database. The other is C. albicans orf19.3865, which encodes a protein that more closely resembles the sole RFX domain-containing protein of S. cerevisiae, Rfx1p. Given the sequence homology, we propose that orf19.3865 be named C. albicans RFX1, and we have named our gene RFX2. In this study, we demonstrate that C. albicans RFX2 has at least partial function redundancy with S. cerevisiae RFX1. In S. cerevisiae rfx1 null mutants, DNA damage response genes like RNR3 and HUG1 are derepressed and cells are rendered more resistant to UV killing. Transformation of an S. cerevisiae rfx1 mutant with a plasmid expressing C. albicans RFX2 significantly reduced expression of RNR3 and HUG1 and restored wild-type UV susceptibility. C. albicans wild-type strain SC5314 responded to UV exposure by inducing RFX2, RAD6 (a gene induced by and required for UV resistance), and DDR48 (a gene encoding a Hog1p-induced stress protein). Deletion of RFX2 resulted in significant derepression of RAD6 and DDR48 under nonstressful conditions. Moreover, the C. albicans rfx2 null mutant was more resistant than SC5314 to UV killing, 48°C heat shock, and 10% ethanol, suggesting that the basal derepression of DNA damage and UV-induced genes was protective. Taken together, the data indicate that C. albicans RFX2 can perform at least some of the functions of S. cerevisiae RFX1 and that the genes contribute to similar phenotypes.
UV exposure and other genotoxic insults to C. albicans result in filamentous growth (45). We demonstrate that Rfx2p is a crucial link between DNA damage responses and morphogenesis. As anticipated, exposure of wild-type C. albicans SC5314 to UV radiation induced the expression of hypha-specific genes HWP1, HYR1, ECE1, and, to a lesser extent, ALS3. Deletion of RFX2 resulted in the constitutive overexpression of these genes as well as CEK1. In addition, the rfx2 null mutant demonstrated hyperfilamentous growth on solid agar under non-hypha-inducing conditions. Along these lines, it is notable that genotoxic stress-induced filamentous growth by C. albicans is dependent upon intact DNA damage checkpoints (2, 45). Similar to S. cerevisiae Rfx1p, C. albicans Rfx2p is likely to function as a downstream effector of the highly conserved and well-characterized Mec1-Rad53 DNA checkpoint pathway.
The attenuated virulence of the rfx2 null mutant is consistent with reports of other hyperfilamentous C. albicans strains, including null mutants in which negative transcriptional regulators encoded by C. albicans TUP1, NRG1, RFG1, and SPT3 were disrupted (6-8, 28, 30, 34, 35). We showed that Rfx2p contributes to pathogenesis by assessing two distinct models of murine candidiasis. During hematogenously disseminated candidiasis (HDC), the rfx2 mutant caused significantly less overall mortality. The null mutant was also less likely than SC5314 to cause DC by mucosal translocation, as demonstrated in cortisone-treated mice that received 2-hour sublingual inoculations of C. albicans. When the inoculation time in these mice was shortened to 1 hour, the null mutant caused lower tissue burdens of infection and less inflammation within oral and esophageal mucosa than SC5314 after 7 days. It is not possible to discern from our data whether the attenuated virulence is a consequence of impaired DNA damage responses, dysregulated morphogenesis, some other process, or a combination of mechanisms. In general, it is unclear whether morphogenesis per se contributes to the pathogenesis of candidiasis rather than morphology-associated patterns of gene expression (24, 46, 48). In fact, pathways regulating C. albicans morphogenesis converge to influence the expression of genes encoding diverse virulence factors (29, 32). Nevertheless, it is notable that the rfx2 mutant not only exhibited derepressed filamentation but also failed to revert back to yeast morphology following overnight growth in liquid medium. To the extent that morphogenesis is a virulence determinant, therefore, the ability to switch back and forth between blastoconidial and filamentous growth might be more important to survival in different environments in vivo than merely the ability to form filaments.
It is interesting that the rfx2 null mutant was significantly more adherent than SC5314 to BECs in vitro and more invasive into solid agar. Although it has long been established that C. albicans hyphae are generally more adherent to host cells than conidia (39), our results in vitro cannot be attributed to the hyperfilamentous growth of the mutant since both strains were tested as yeasts. Indeed, Gram stains confirmed that SC5314 and the null mutant were morphologically similar during the in vitro assay. Rather, Rfx2p is likely to influence adhesion through its role in transcriptional repression. The derepression of HWP1 and ALS3, hypha-specific genes encoding adhesins (46, 55), under basal conditions associated with growth in the yeast morphology supports this hypothesis. The equivalent tissue burdens of SC5314 and the rfx2 null mutant after 6 h of murine OPC, however, highlight several important points: (i) adherence and penetration are not the sole determinants of pathogenicity, even at relatively early time points; (ii) in vitro assays cannot completely replicate complex in vivo systems; and (iii) mechanisms of pathogenesis differ at unique tissue sites.
The precise mechanisms by which Rfx2p contributes to the regulation of the interrelated processes of morphogenesis, virulence, and adherence will need to be elucidated in future studies. At least some of the cellular functions of Rfx2p are likely to be mediated through the transcriptional repressor Tup1p. Tup1p forms an evolutionarily conserved corepressor complex with Ssn6p, which is targeted to specific promoters through interactions with a variety of DNA binding proteins. S. cerevisiae Rfx1p targets Tup1p-Ssn6p to DNA damage response genes. As alluded to earlier, deletion of C. albicans TUP1 results in hyperfilamentous and invasive growth, derepression of hypha-specific genes, and attenuated virulence during murine candidiasis, similar to our observations with the rfx2 null mutant (6-9, 21, 34, 35, 44). Despite the phenotypic similarities, there are morphological differences between rfx2 and tup1 mutants. The rfx2 mutant grows as a mixture of blastospore and filamentous morphologies, whereas the tup1 mutant grows solely as filaments (35). In addition, rfx2 cells are able to form normal hyphal cells under a range of experimental conditions, unlike tup1 cells, which are fixed in a pseudohyphal morphology (35). In this regard, the rfx2 null mutant more closely resembles C. albicans strains with disruptions of NRG1, a gene encoding another DNA binding protein that targets TUP1 (6, 21, 34, 35). Interestingly, Nrg1p appears to regulate some C. albicans genes in a Tup1p-independent manner (34). It is likewise possible that Rfx2p regulates transcription by both Tup1p-dependent and -independent mechanisms.
Finally, our findings indicate that C. albicans Rfx1p cannot fully perform the cellular functions of Rfx2p in the absence of the latter protein. It is unclear at present whether the two RFX domain-containing proteins have similar, overlapping, or independent functions. In considering potential functions for Rfx1p and Rfx2p, it is worth noting that conserved DNA binding proteins like Nrg1p and Rfg1p (the homologue to S. cerevisiae Rox1p) have divergent cellular roles in C. albicans and S. cerevisiae (28, 34). In future studies, therefore, we will characterize the roles of C. albicans Rfx1p in DNA damage response, filamentation, and virulence, as well as determine if Rfx1p and Rfx2p contribute to cellular responses in S. cerevisiae not described.
Acknowledgments
Experiments were conducted in the laboratories of M. H. Nguyen and C. J. Clancy at the North Georgia/South Florida Veterans Health System and the University of Pittsburgh. We thank Joachim Morschhauser and Malcolm Whiteway for providing strains, plasmids, and cassettes used in this study.
Research was funded by an NIH Mycology Research Unit Program Project Award (5P01AI061537-02 to C.J.C. and M.H.N.) and supported by the VA Medical Research Service and the University of Florida.
Footnotes
Published ahead of print on 27 February 2009.
REFERENCES
- 1.Allen, J. B., Z. Zhou, W. Siede, E. C. Friedberg, and S. J. Elledge. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 82401-2415. [DOI] [PubMed] [Google Scholar]
- 2.Andaluz, E., T. Ciudad, J. Gómez-Raja, R. Calderone, and G. Larriba. 2006. Rad52 depletion in Candida albicans triggers both the DNA-damage checkpoint and filamentation accompanied by but independent of expression of hypha-specific genes. Mol. Microbiol. 591452-1472. [DOI] [PubMed] [Google Scholar]
- 3.Badrane, H., S. Cheng, M. H. Nguyen, H. Y. Jia, Z. Zhang, N. Weisner, and C. J. Clancy. 2005. Candida albicans IRS4 contributes to hyphal formation and virulence after the initial stages of disseminated candidiasis. Microbiology 1512923-2931. [DOI] [PubMed] [Google Scholar]
- 4.Badrane, H., M. H. Nguyen, S. Cheng, V. Kumar, H. Derendorf, and C. J. Clancy. 2008. The Candida albicans phosphatase Inp51p interacts with the EH domain protein Irs4p, regulates phosphatidylinositol-4,5-bisphosphate levels and influences hyphal formation, the cell integrity pathway and virulence. Microbiology 1543296-3308. [DOI] [PubMed] [Google Scholar]
- 5.Benton, M. G., S. Somasundaram, J. D. Glasner, and S. P. Palecek. 2006. Analyzing the dose-dependence of the Saccharomyces cerevisiae global transcriptional response to methyl methanesulfonate and ionizing radiation. BMC Genomics 7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277105-109. [DOI] [PubMed] [Google Scholar]
- 7.Braun, B. R., and A. D. Johnson. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 15557-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, B. R., D. Kadosh, and A. D. Johnson. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 204753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun, B. R., W. S. Head, M. X. Wang, and A. D. Johnson. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 15631-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calderone, R. A., and W. A. Fonzi. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9327-335. [DOI] [PubMed] [Google Scholar]
- 11.Reference deleted.
- 12.Chauhan, N., T. Ciudad, A. Rodríguez-Alejandre, G. Larriba, R. Calderone, and E. Andaluz. 2005. Virulence and karyotype analyses of rad52 mutants of Candida albicans: regeneration of a truncated chromosome of a reintegrant strain (rad52/RAD52) in the host. Infect. Immun. 738069-8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng, S., C. J. Clancy, M. A. Checkley, M. Handfield, J. D. Hillman, A. Progulske-Fox, A. S. Lewin, P. L. Fidel, and M. H. Nguyen. 2003. Identification of Candida albicans genes induced during thrush offers insight into pathogenesis. Mol. Microbiol. 481275-1288. [DOI] [PubMed] [Google Scholar]
- 14.Cheng, S., C. J. Clancy, M. A. Checkley, Z. Zhang, K. L. Wozniak, K. R. Seshan, H. Y. Jia, P. L. Fidel, Jr., G. Cole, and M. H. Nguyen. 2005. The role of Candida albicans NOT5 in virulence depends upon diverse host factors in vivo. Infect. Immun. 737190-7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng, S., M. H. Nguyen, Z. Zhang, H. Jia, M. Handfield, and C. J. Clancy. 2003. Evaluation of the roles of four Candida albicans genes in virulence by using gene disruption strains that express URA3 from the native locus. Infect. Immun. 716101-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciudad, T., E. Andaluz, O. Steinberg-Neifach, N. F. Lue, N. A. Gow, R. A. Calderone, and G. Larriba. 2004. Homologous recombination in Candida albicans: role of CaRad52p in DNA repair, integration of linear DNA fragments and telomere length. Mol. Microbiol. 531177-1194. [DOI] [PubMed] [Google Scholar]
- 17.Dib, L., P. Hayek, H. Sadek, B. Beyrouthy, and R. A. Khalaf. 2008. The Candida albicans Ddr48 protein is essential for filamentation, stress response, and confers partial antifungal drug resistance. Med. Sci. Monit. 14BR113-BR121. [PubMed] [Google Scholar]
- 18.Elledge, S. J., Z. Zhou, J. B. Allen, and T. A. Navas. 1993. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays 15333-339. [DOI] [PubMed] [Google Scholar]
- 19.Emery, P., B. Durand, B. Mach, and W. Reith. 1996. RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res. 24803-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu, Y., L. Pastushok, and W. Xiao. 2008. DNA damage-induced gene expression in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 32908-926. [DOI] [PubMed] [Google Scholar]
- 21.Gajiwala, K. S., H. Chen, F. Cornille, B. P. Roques, W. Reith, B. Mach, and S. K. Burley. 2000. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature 403916-921. [DOI] [PubMed] [Google Scholar]
- 22.García-Sánchez, S., A. L. Mavor, C. L. Russell, S. Argimon, P. Dennison, B. Enjalbert, and A. J. Brown. 2005. Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol. Biol. Cell 162913-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gietz, R. D., and R. H. Schiestl. 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 231-34. [DOI] [PubMed] [Google Scholar]
- 24.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198179-182. [DOI] [PubMed] [Google Scholar]
- 25.Gow, N. A. 2002. Candida albicans switches mates. Mol. Cell 10217-218. [DOI] [PubMed] [Google Scholar]
- 26.Huang, M., Z. Zhou, and S. J. Elledge. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94595-605. [DOI] [PubMed] [Google Scholar]
- 27.Jelinsky, S. A., P. Estep, G. M. Church, and L. D. Samson. 2000. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 208157-8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jelinsky, S. A., and L. D. Samson. 1999. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci. USA 961486-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadosh, D., and A. D. Johnson. 2001. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 212496-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane, S., C. Birse, S. Zhou, R. Matson, and H. Liu. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 27648988-48996. [DOI] [PubMed] [Google Scholar]
- 31.Laprade, L., V. L. Boyartchuk, W. F. Dietrich, and F. Winston. 2002. Spt3 plays opposite roles in filamentous growth in Saccharomyces cerevisiae and Candida albicans and is required for C. albicans virulence. Genetics 161509-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leng, P., P. E. Sudbery, and A. J. Brown. 2000. Rad6p represses yeast-hypha morphogenesis in the human fungal pathogen Candida albicans. Mol. Microbiol. 351264-1275. [DOI] [PubMed] [Google Scholar]
- 33.Liu, H. 2002. Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int. J. Med. Microbiol. 292299-311. [DOI] [PubMed] [Google Scholar]
- 34.Lubelsky, Y., N. Reuven, and Y. Shaul. 2005. Autorepression of Rfx1 gene expression: functional conservation from yeast to humans in response to DNA replication arrest. Mol. Cell. Biol. 2510665-10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murad, A. M., C. d'Enfert, C. Gaillardin, H. Tournu, F. Tekaia, D. Talibi, D. Marechal, V. Marchais, J. Cottin, and A. J. Brown. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42981-993. [DOI] [PubMed] [Google Scholar]
- 36.Murad, A. M., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 204742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen, M. H., S. Cheng, and C. J. Clancy. 2004. Assessment of Candida albicans genes expressed during infections as a tool to understand pathogenesis. Med. Mycol. 42293-304. [DOI] [PubMed] [Google Scholar]
- 38.Raman, S. B., M. H. Nguyen, Z. Zhang, S. Cheng, H. Y. Jia, N. Weisner, K. Iczkowski, and C. J. Clancy. 2006. Candida albicans SET1 encodes a histone 3 lysine 4 methyltransferase that contributes to the pathogenesis of invasive candidiasis. Mol. Microbiol. 60697-709. [DOI] [PubMed] [Google Scholar]
- 39.Reuss, O., A. Vik, R. Kolter, and J. Morschhäuser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341119-127. [DOI] [PubMed] [Google Scholar]
- 40.Samaranayake, L. P., and T. W. MacFarlane. 1982. Factors affecting the in-vitro adherence of the fungal oral pathogen Candida albicans to epithelial cells of human origin. Arch. Oral Biol. 27869-873. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271357-360. [DOI] [PubMed] [Google Scholar]
- 42.Schaller, M., H. C. Korting, W. Schäfer, J. Bastert, W. Chen, and B. Hube. 1999. Secreted aspartic proteinase (Sap) activity contributes to tissue damage in a model of human oral candidiasis. Mol. Microbiol. 34169-180. [DOI] [PubMed] [Google Scholar]
- 43.Schaller, M., W. Schafer, H. C. Korting, and B. Hube. 1998. Differential expression of secreted aspartyl proteinases in a model of human oral candidosis and in patient samples from the oral cavity. Mol. Microbiol. 29605-615. [DOI] [PubMed] [Google Scholar]
- 44.Segurado, M., and J. F. Diffley. 2008. Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev. 221816-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharkey, L. L., M. D. McNemar, S. M. Saporito-Irwin, P. S. Sypherd, and W. A. Fonzi. 1999. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J. Bacteriol. 1815273-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, Q. M., Y. M. Wang, X. D. Zheng, R. T. Lee, and Y. Wang. 2007. Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol. Biol. Cell 18815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staab, J. F., S. D. Bradway, P. L. Fidel, and P. Sundstrom. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 2831535-1538. [DOI] [PubMed] [Google Scholar]
- 48.Staib, P., S. Wirsching, A. Strauss, and J. Morschhäuser. 2001. Gene regulation and host adaptation mechanisms in Candida albicans. Int. J. Med. Microbiol. 291183-188. [DOI] [PubMed] [Google Scholar]
- 49.Sudbery, P., N. Gow, and J. Berman. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12317-324. [DOI] [PubMed] [Google Scholar]
- 50.Sun, Z., D. S. Fay, F. Marini, M. Foiani, and D. F. Stern. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10395-406. [DOI] [PubMed] [Google Scholar]
- 51.Toh, G. W., and N. F. Lowndes. 2003. Role of the Saccharomyces cerevisiae Rad9 protein in sensing and responding to DNA damage. Biochem. Soc. Trans. 31242-246. [DOI] [PubMed] [Google Scholar]
- 52.Weinert, T. A., G. L. Kiser, and L. H. Hartwell. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8652-665. [DOI] [PubMed] [Google Scholar]
- 53.Zaim, J., E. Speina, and A. M. Kierzek. 2005. Identification of new genes regulated by the Crt1 transcription factor, an effector of the DNA damage checkpoint pathway in Saccharomyces cerevisiae. J. Biol. Chem. 28028-37. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Z., and J. C. Reese. 2005. Molecular genetic analysis of the yeast repressor Rfx1/Crt1 reveals a novel two-step regulatory mechanism. Mol. Cell. Biol. 257399-7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao, X., S. H. Oh, G. Cheng, C. B. Green, J. A. Nuessen, K. Yeater, R. P. Leng, A. J. Brown, and L. L. Hoyer. 2004. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 1502415-2428. [DOI] [PubMed] [Google Scholar]
- 56.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408433-439. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, Z., and S. J. Elledge. 1992. Isolation of crt mutants constitutive for transcription of the DNA damage inducible gene RNR3 in Saccharomyces cerevisiae. Genetics 131851-866. [DOI] [PMC free article] [PubMed] [Google Scholar]