Abstract
As with other organisms with a completed genome sequence, opportunities for performing large-scale studies, such as expression and localization, on Toxoplasma gondii are now much more feasible. We present a system for tagging genes endogenously with yellow fluorescent protein (YFP) in a Δku80 strain. Ku80 is involved in DNA strand repair and nonhomologous DNA end joining; previous studies in other organisms have shown that in its absence, random integration is eliminated, allowing the insertion of constructs with homologous sequences into the proper loci. We generated a vector consisting of YFP and a dihydrofolate reductase-thymidylate synthase selectable marker. The YFP is preceded by a ligation-independent cloning (LIC) cassette, which allows the insertion of PCR products containing complementary LIC sequences. We demonstrated that the Δku80 strain is more effective and efficient in integrating the YFP-tagged constructs into the correct locus than wild-type strain RH. We then selected several hypothetical proteins that were identified by a proteomic screen of excreted-secreted antigens and that displayed microarray expression profiles similar to known micronemal proteins, with the thought that these could potentially be new proteins with roles in cell invasion. We localized these hypothetical proteins by YFP fluorescence and showed expression by immunoblotting. Our findings demonstrate that the combination of the Δku80 strain and the pYFP.LIC constructs reduces both the time and cost required to determine localization of a new gene of interest. This should allow the opportunity for performing larger-scale studies of novel T. gondii genes.
Toxoplasma gondii is an obligate intracellular protozoan in the phylum Apicomplexa that has garnered more intense study in recent decades. This is due in part to the genetic tractability and ease of growth of T. gondii and also because knowledge obtained for this parasite is often relevant to its kin, including other pathogens of medical and veterinary importance, such as Plasmodium falciparum, the parasite that causes malaria (11). The genome sequence of Toxoplasma was recently completed (10), opening the door to identifying novel genes involved in a variety of events, such as cell invasion, replication, gliding motility, metabolism, stage conversion, and virulence. With a large number of new genes of interest, it is essential to have tools that enable investigators to perform studies on a larger scale. One bottleneck for the analysis of novel genes is protein localization, since this generally necessitates time-consuming production of antibodies that often require additional affinity purification. Protein localization by ectopic expression of a tagged construct can also be problematic due to overexpression or mistiming of expression from a heterologous promoter. If genes could be tagged directly on the chromosome, this would better mimic natural expression and allow the use of fluorescent protein tags or standardized antibodies to monitor localization.
DNA double-strand breaks in mammalian cells are repaired via either the homologous recombination or nonhomologous end-joining (NHEJ) pathways, which differ in the requirement for homologous sequences versus ligation independent of DNA sequence homology. DNA breaks can be lethal to an organism if left unrepaired, resulting in genomic instability, sensitivity to DNA damage, and mutations leading to tumor development (16). Ku70 and Ku80 (Ku86 in higher eukaryotes) proteins form a heterodimeric complex that plays a pivotal role in NHEJ DNA repair. Ku70 and Ku80 orthologues are widely present in eukaryotes, having been identified in yeast, plants, invertebrates, and vertebrates (5). While the primary role of Ku proteins is in DNA repair, they have also been implicated in a range of other cellular activities, such as telomere maintenance, tumor suppression, gene transcription regulation, heat shock-induced responses, and apoptosis (15, 43). As observed by in vitro photo cross-linking of the Ku dimer and DNA, the Ku70 subunit preferentially contacts the major groove and Ku80 contacts the minor groove (48). Crystallography studies show that the Ku70/80 complex is in a 1:1 ratio in an open ring-shaped structure through which a DNA helix can pass (45). In a model of NHEJ, the Ku complex, which has a high affinity for DNA, binds to DNA termini and recruits the DNA-dependent protein kinase catalytic subunit, activating its kinase activity. This is followed by the recruitment of the DNA ligase IV and XRCC4 complex to the double-strand break, resulting in ligation of the DNA ends (5).
Neurospora crassa mutants deficient in Ku70 or Ku80 showed 100% homologous recombination, in contrast to the wild-type strain, which displayed only 21% homologous recombination (25). Deletions of either Ku70 or Ku80 in the fungal strain Sordaria macrospora or in Aspergillus spp. (3, 30, 40) have resulted in considerable improvements in gene targeting without notable growth impairment or other defects. The data on the effect of Ku70 or Ku80 disruption in mice or cell lines on susceptibility to DNA damaging agents are conflicting and unclear. Several studies have claimed an increased rate of mutation, chromosomal instability, and increased carcinogenesis (7, 12, 20, 26, 41), while other investigators observed a converse decrease in mutation frequency and chromosomal rearrangements (32, 44). It appears that the particular mouse strain or cell line used may play a role in this discrepancy.
We sought to exploit the potential improvement in homologous recombination in a T. gondii Ku80-disrupted strain to introduce a reporter protein at the 3′ end of a gene in order to localize the product within the parasite. We found that the Ku80-null parasite strain significantly improved gene targeting efficiency compared to wild-type parasites and is a useful tool for endogenous gene tagging. In a companion article, Fox, Bzik, and colleagues also show the utility of Δku80 parasites for creating targeted gene knockouts (9).
MATERIALS AND METHODS
Fusion PCR knockout (KO) construct for disruption of Ku80.
Genomic flanking sequences of the Ku80 gene (583.m05492) were obtained from the Toxodb database (www.toxodb.org; version 4.3). Primers were designed to amplify ∼4 kb of the 5′ and 3′ ends flanking the gene. The flanks were amplified to overlap on one end with the hypoxanthine xanthine guanine phosphoribosyltransferase (HXGPRT)-selectable marker cassette, and similarly, the HXGPRT marker cassette was amplified to contain Ku80 sequences on the ends (Fig. 1A).
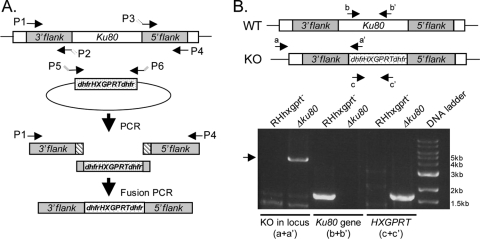
FIG. 1.
Targeted deletion of Ku80. (A) Generation of the Ku80 KO construct. Ku80 5′ and 3′ genomic flanks were amplified with overlaps to the dhfr-HXGPRT-dhfr selectable marker; the dhfr-HXGPRT-dhfr marker was amplified with overlaps to Ku80 flanks. A fusion PCR Δku80 product consisting of Ku80 flanks and the selectable marker was amplified. (B) Primer sets were used to evaluate the parental strain (RHhxgprt−) and Δku80 for replacement of the Ku80 gene with the selectable marker (a and a′), the presence or absence of the Ku80 gene (b and b′), and the presence or absence of the HXGPRT selectable marker (c and c′).
Primers to amplify the 3′ flank were P-1, 5′-GTCGACATGCATATGTTTTAGAGG-3′, and P-2, 5′-CCGCGGGCGGGTTTGAATGCAAGGTTTCGTGCTGcatgagtcgatatatctcgctagctataaatatatc-3′ (lowercase indicates HXGPRT sequences and uppercase indicates Ku80 gene sequences). Primers to amplify the 5′flank were P-3, 5′-gttctggcaggctacagtgacaccgcggtggGATACTCACTGTGGGTTGAGTTACAAG-3′, and P-4, 5′-ATGTAACTGTGCGCCTATCTACTTC-3′. Primers to amplify HXGPRT were P-5, 5′-GATATATTTATAGCTAGCGAGATATATCGACTCATGcagcacgaaaccttgcattcaaacccgcccgcgg-3′, and P-6, 5′-CTTGTAACTCAACCCACAGTGAGTATCccaccgcggtgtcactgtagcctgccagaac-3′.
RH genomic DNA was used as template for amplifying the 5′ and 3′ flanks, and pminiHXGPRT plasmid (6) was used as a template for amplifying the HXGPRT selectable marker. A fusion PCR product was then amplified using primers P1 and P4 and the three individual 5′, 3′, and HXGPRT fragments as templates.
Δku80 transfection and selection.
The RHhxgprt− (6) strain was cultured by passage in human foreskin fibroblasts in Dulbecco's modified Eagle's medium-HEPES-10% fetal bovine serum at 37°C and with 5% CO2. Parasites were filter purified, pelleted, and resuspended in cytomix buffer (2 mM EDTA, 120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4/KH2PO4, 25 mM HEPES, 5 mM MgCl2·6H2O; pH 7.6). Fifteen μg of the 3′Ku80-HXGPRT-5′Ku80 fusion PCR product was electroporated into 1 × 107 RHhxgprt− parasites using a Bio-Rad X-Cell electroporator (settings of 1.5kV, 25 mF, and no resistance). After overnight growth, transformants were placed under selection with 25 μg/ml mycophenolic acid and 50 μg/ml xanthine. Transformant pools were tested for the presence of a KO before cloning by limiting dilution under drug selection. Individual KO clones were replated in 96-well plates to ensure clonality. This strain was termed Δku80-HXG and is the strain used in all the experiments except those in Fig. 3D, below, which also included the Δku80-DHFR strain, which was generated by replacement of Ku80 with the dihydrofolate reductase-thymidylate synthase (DHFR-TS) selectable marker in RHhxgprt− parasites by using pyrimethamine selection.
FIG. 3.
Optimization and efficiency of gene tagging. (A) Agarose gel showing plasmids in the forms supercoiled (SC), linearized within the targeting sequence (LN), or nicked/open circle (OC). (B) Linearization of the vector is necessary for efficient integration. Only the LN construct integrated into the homologous loci, whereas the SC and OC constructs did not integrate during the 30-day experiment. (C) Linearization site and efficiency of integration. Two restriction enzyme digestion sites were used, cutting 350 or 750 into the 1-kb targeting sequence. Both linearized constructs were shown to be equally effective in integration into the endogenous ACP locus. (D) Δku80 favors homologous recombination. pCNA-YFP, ACP-YFP, and MIC3-YFP constructs with either HXGPRT or DHFR-TS selectable markers were transfected into RH or Δku80, and YFP-positive parasites were enumerated over time and are expressed as a percentage of the total population. Note that the y-axis scale of the MIC3-YFP graph is smaller than for the pCNA-YFP and ACP-YFP graphs.
Confirmation of Δku80 by PCR.
Primers within the HXGPRT selectable marker cassette, dhfr 5′ end 3.R (5′-GCGGGCGGGTTTGAATGCAAGGTTTCGTGC-3′) and outside the 3′ genomic flanking region, 3′Ku80.2730891.F (5′-CATACCTTCGAGTTGCTCTTCTTGTTGAC-3′), were used to detect the replacement of the Ku80 gene with HXGPRT. The absence of the Ku80 gene was confirmed with primers TgKu80.949.F (5′-TTCCTGATCCCCGTGTATGT-3′) and TgKu80.1748.R (5′-GACGCCGGATTGTAGTGTCT-3′); the presence of the HXGPRT selectable marker was confirmed with the primers TgKu80-5′dhfrHXGPRTdhfr-70bp.F (5′- GATATATTTATAGCTAGCGAGATATATCGACTCATGCAGCACGAAACCTTGCATTCAAACCCGCCCGCGG-3′) and 5′Ku80.2740775-3′dhfr.R (5′- CTTGTAACTCAACCCACAGTGAGTATCCCACCGCGGTGTCACTGTAGCCTGCCAGAAC-3′).
Southern blot analysis.
Digoxigenin (Roche)-labeled probes against HXGPRT were amplified from pminiHXGPRT using primers HXG.661.F (5′-GAGAACTTACTTCGGCGAG-3′) and HXG.978.R (5′-ATCGACTTCGGACCGACG-3′) and against Ku80 from genomic DNA using TgKu80.949.F (5′-TTCCTGATCCCCGTGTATGT-3′) and TgKu80.1748.R (5′-GACGCCGGATTGTAGTGTCT-3′. Genomic DNA was digested with XmnI or SacII for probing with HXGPRT and with XmnI or PstI for probing with Ku80.
pYFP.LIC.HXG and pYFP.LIC.DHFR vectors.
The YFP and DHFR-TS 3′-untranslated region (UTR) fragment was PCR amplified from the plasmid pTubYFPYFP-sagCAT (13), with a ligation-independent cloning (LIC) cassette 5′ to the start of YFP, and subcloned with KpnI and XhoI restriction enzyme ends into the pminiHXGPRT plasmid (6), generating the pYFP.LIC.HXG vector. The LIC cassette was introduced into the 5′ end of the YFP (35), with the following modifications: the LIC cassette (5′- CTGTACTTCCAATCCAATTTAATTAAAATTGGAAGTGGAGGACGG-3′) contains a unique PacI site, underlined in the sequence, and the stop codon in the original LIC cassette was removed to allow readthrough to the YFP. The HXGPRT cassette was then excised with HindIII and NotI and replaced with the DHFR-TS selectable marker cassette. The vector is a pBluescript backbone, and selection in Escherichia coli was performed with 100 μg/ml ampicillin.
Gene amplification, cloning, and transfection.
Genomic sequences were obtained from the Toxodb database (version 4.3) and primers were designed to amplify 1 to 4 kb of the 3′ ends of genes, which contained a unique restriction enzyme within the fragment. All forward primers contained the LIC sequence 5′-TACTTCCAATCCAATTTAATGC-3′ and all reverse primers contained the LIC sequence 5′-TCCTCCACTTCCAATTTTAGC-3′. The additional GC was added to the reverse primers to allow amplification of genes up to the second-to-last codon and for subsequent T4 DNA polymerase and dCTP treatment. Since a C is necessary for this treatment, this allowed the same LIC sequence to be used for all genes. In addition, this kept the YFP in frame with the gene after “ligation.” All reverse primers stopped just prior to the stop codon to allow readthrough to the downstream YFP. Both the pYFP.LIC.DHFR vector and the PCR products were treated with T4 DNA polymerase as described previously (35). Briefly, 0.2 pmol of PCR product was incubated in a 20-μl reaction mixture with 5 mM dithiothreitol (DTT), 4 mM dCTP, 1× T4 DNA polymerase buffer, and T4 DNA ligase and the following condition: 30 min at 22°C, 20 min at 75°C, and cooling on ice. Two μg of linearized vector DNA was incubated in a similar reaction mixture (60 μl), with dGTP in place of dCTP. Two μl of T4-treated PCR product and 1 μl of vector were incubated for 10 min at room temperature, 1 μl of 25 mM EDTA was added, and the mixture was incubated for another 5 min at room temperature before placing it on ice. Electrocompetent E. coli cells were transformed with 1 μl of reaction mix. Positive transformants were identified and confirmed by diagnostic restriction enzyme digests. For transfection, 2 to 20 μg of constructs was linearized within the region of homology, phenol-chloroform extracted, and ethanol precipitated. Nicked/open circle constructs were prepared by overnight digestion with EcoRI in the presence of 500 μg/ml of ethidium bromide at 26°C, followed by two phenol-chloroform and two chloroform-isoamyl alcohol extractions, as described previously (29). After overnight growth, transformants were selected with 2 μM pyrimethamine and kept under selection until the experiment was terminated.
Fluorescence and immunoblot analysis.
For localization, egressed parasites were inoculated onto eight-well chamber slides and fixed in 4% paraformaldehyde 24 h postinfection. YFP fluorescence was viewed directly on a Zeiss Axio inverted microscope with a YFP filter cube. For colocalization immunofluorescence, inoculated slides were fixed, permeabilized with 0.1% Triton X-100, stained with rabbit anti-MIC2 (1:250) (47) and mouse anti-green fluorescent protein (anti-GFP; 1:250; Clontech/BD) antibodies, followed by Alexa594-conjugated goat anti-rabbit and Alexa488-conjugated goat anti-mouse secondary antibodies (Invitrogen). 4′,6-diamidino-2-phenylindol (DAPI; 5 μg/ml final concentration) was added to visualize the nuclei.
For immunoblot analyses, parasites were isolated and lysed in 95°C sample buffer and separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. After semidry transfer to polyvinylidene difluoride membranes, blots were blocked, probed with mouse anti-GFP (1:1,000), washed, and probed with goat anti-mouse horseradish peroxidase-conjugated secondary antibody (Jackson Labs).
Plaque assay.
For transfection followed by plaque assay, 10 μg aliquots of linearized acyl carrier protein (ACP), microneme protein 3 (MIC3), and proliferating cell nuclear antigen (pCNA) in pYFP.LIC.DHFR were prepared and transfected into Δku80-HXG. Transfected parasites were diluted and 750 tachyzoites were inoculated into D150 petri dishes with pyrimethamine selection and left undisturbed for 7 days. Individual plaques were picked and expanded in 96-well and then 24-well plates, genomic DNA for PCR was extracted using Qiagen DNEasy columns, and proper integration into target loci was visualized by fluorescence. Integration of the construct into the endogenous DHFR-TS locus was detected using primers in the first and second exons of dhfr-ts: dhfr-ts.5′.268.F, 5′-GTTTCCTTTTTCTTCTGTTCGTTTC-3′, and dhfr-ts.896.R, 5′-GAATCCTTGTACTCTTCCTCCAGAAGG-3′.
RESULTS
Generation of a Δku80 strain.
Ku80 is a molecule found in a heterodimer with Ku70, and together they are involved in DNA repair via the NHEJ pathway. Deletion of Ku80 should result exclusively in homologous recombination, increasing the efficiency of genetic knockouts and incorporation of reporter proteins to endogenous loci. BLAST searches of the Toxoplasma genome database (version 4.3) using the Neurospora crassa Ku70 and Ku80 genes (mus-51 and mus-52; GenBank accession numbers AB177394 and AB177395, respectively) indicated the presence of single orthologues of Ku70 and Ku80 in T. gondii (with BLASTp values [Toxodb accession numbers] as follows: 2.6 × 1013 [50.m03211] and 4.9 × 107 [583.m05492], respectively). Both TgKu80 and TgKu70 have a canonical so-called Ku78 domain, which is the hallmark of this protein family. We focused on genetically disrupting TgKu80, using a fusion PCR-based method to replace the Ku80 gene with a selectable marker. To create the Ku80 KO (Δku80) strain, a fusion PCR KO construct was generated, consisting of 3′Ku80 and 5′Ku80 genomic flanks fused on either side of the HXGPRT selectable marker cassette (Fig. 1A). Once the individual PCR products were amplified, all three fragments were used as templates in a fusion PCR that resulted in a 3′Ku80-HXGPRT-5′Ku80 product. This KO construct was transfected into RHhxgprt− parasites and KO clones were identified by PCR using a forward primer annealing outside the 3′ flank of the KO construct and a reverse primer within the HXGPRT cassette. Clones were also tested by PCR for the absence of the Ku80 gene and the presence of the HXGPRT cassette. Parental RHhxgprt− parasites were included as a control. The results from one representative Δku80 clone are shown in Fig. 1B. Southern blot analysis was performed to confirm the presence of a single copy of the selectable marker and the absence of the Ku80 gene (data not shown). A similar method was used to generate a Δku80 strain with pyrimethamine selection in RHhxgprt− parasites (data not shown).
Construction of the pYFP.LIC.DHFR vector and LIC method.
To improve the efficiency of cloning individual genes into an expression vector, a plasmid was created that contained a LIC cassette (1, 35) upstream of an in-frame copy of YFP and a DHFR-TS selectable marker cassette (Fig. 2A). For insertion into the YFP vector, genes were amplified with primers that introduced LIC sequences at the 5′ and 3′ ends of the PCR products, homologous to the LIC sequences upstream of YFP. Restriction enzyme digestion with PacI linearized the vector, and subsequent T4 DNA polymerase treatment of both the vector and the PCR product exposed compatible sequences that annealed and were subsequently ligated after transformation of E. coli. All amplified genes were inserted into the YFP vector with this LIC method.
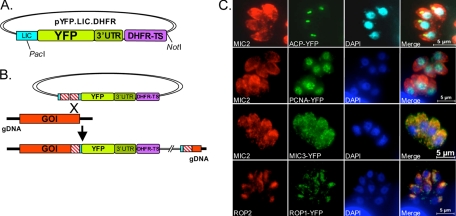
FIG. 2.
Endogenous gene tagging. (A) A pYFP.LIC.DHFR vector was generated that bore an upstream LIC cassette for cloning of genes of interest fused in frame to YFP, which was followed by the DHFR-TS 3′ UTR and a DHFR-TS cassette for selection of resistant parasites. (B) Schematic illustration of the single-crossover mechanism of integration of the YFP fusion protein to the 3′ end of a gene. (C) Positive control genes ACP (apicoplast), pCNA (nucleus), MIC3 (micronemes), and ROP1 (rhoptries) were cloned into the YFP expression vector and transfected into parasites, and localization was visualized by immunofluorescence with a GFP antibody. The DAPI staining in the ACP panel is pseudocolored with cyan to bring out the staining in the apicoplast. MIC2 staining was used to identify the micronemes and apical end of the parasite, and ROP2 staining was used to identify the rhoptries.
To exploit Δku80 parasites for gene tagging, we amplified the 3′ genomic region of a gene up to but not including the stop codon and cloned this fragment into the pYFP.LIC.DHFR vector, creating a fusion of the gene-of-interest fragment with YFP. By reciprocal recombination, the region of homology in the construct recombined with the homologous sequences on the chromosomal gene, introducing the YFP to the 3′ end of the endogenous gene (Fig. 2B). Studies in the yeast model have demonstrated that a plasmid linearized within a region of homology to chromosomal DNA increases the efficiency of homologous integration 10- to 1,000-fold; these and other studies formed the basis for the double-strand break repair model of recombination (27, 28, 39). In this model, the 3′ ends are processed to form 3′ single-stranded tails, which then invade the homologous duplex and priming DNA synthesis (review in reference 38). Because only a fragment (the 3′ region) of each gene is used for targeting and the YFP does not contain its own promoter for expression, YFP fluorescence in the expected pattern should only be seen upon homologous targeting and correct integration into the endogenous locus. For proof of principle, several genes that were previously tagged with reporter proteins and shown to target properly were examined: pCNA (31), ACP (46), MIC3 (37), and rhoptry protein 1 (ROP1) (36). As shown in Fig. 2C, all constructs targeted correctly to their respective organelles. Immunofluorescence dual staining was performed with anti-MIC2 antibody to visualize the apical end of the parasite or with ROP2 to localize the rhoptries.
It is worth noting that ACP and pCNA are essential genes (14, 46), indicating that chromosomal YFP tagging of these proteins did not overtly interfere with function. Having established that the YFP vector can successfully target an endogenous gene, we sought to determine whether the form of the transfected construct, i.e., supercoiled, linear, or nicked/open circle, affects the efficiency of integration. Using ACP and pCNA as easily identifiable YFP-tagged proteins in the parasites, we prepared constructs in all three forms (Fig. 3A). As expected based on the double-strand break model, only constructs linearized within the region of homology integrated into the target loci, while the supercoiled or nicked plasmids never integrated during the 35-day observation period with pyrimethamine selection (Fig. 3B). We also transfected constructs that were linearized downstream of the DHFR-TS selectable marker cassette with NotI (Fig. 2A) and did not observe integration after 30 days (data not shown). Given these results, it is necessary to amplify a region of the 3′ end of the gene that contains a unique restriction enzyme site to linearize the construct after cloning in the YFP vector. Next we examined whether the site of linearization affects integration. ACP-YFP was used since it contains two unique sites within the 1-kb targeting region and tagged ACP is easily identified within the parasite. Digests cutting 350 bp or 750 bp from the 3′ end of the ACP targeting sequence were transfected into parasites and examined by fluorescence for 33 days to determine the efficiency of integration (Fig. 3C). No apparent difference in the time required for integration of the two constructs was observed, indicating that linearization as close as 350 bp from the 3′ end of the gene of interest is sufficient for targeting if there are homologous sequences upstream of this cut site. The amount of transfected vector DNA required was also tested, and no difference was observed using 2.5 μg or 20 μg of DNA (data not shown).
To directly compare the efficiency of homologous targeting in Δku80 and wild-type parasites, as well as mycophenolic acid/xanthine (MPA/X) versus pyrimethamine selection, we transfected ACP, pCNA, and MIC3 in pYFP.LIC.DHFR into Δku80-HXG and RH strains and ACP, pCNA, and MIC3 in pYFP.LIC.HXG in Δku80-DHFR and RH and examined integration of the constructs over time by fluorescence. pCNA-YFP-positive parasites reached 90% of the population by 13 days posttransfection (dpt) in the Δku80-HXG strain and 40% in the Δku80-DHFR strain; both reached 100% within ∼20 dpt (Fig. 3D). In the RH strain, pCNA-YFP.DHFR-positive fluorescent parasites emerged much more slowly, reaching 12% by 18 dpt and ∼70% by 34 dpt. It is possible that given sufficient time in selection, all parasites would be fluorescent with transfection of this particular gene. In contrast, no fluorescent parasites were observed with pCNA-YFP.HXG in RH. ACP-YFP-positive parasites were detected in Δku80-HXG at 11 dpt and reached 100% by 27 dpt. ACP-YFP parasites in Δku80-DHFR were slower to emerge and reached ∼60% by 34 dpt. In RH, however, no ACP-YFP-positive fluorescent parasites were observed with the MPA/X selection, but a small percentage (4%) of YFP-positive parasites were observed in the pyrimethamine selection 34 dpt. Interestingly, MIC3-YFP-positive parasites reached ∼13% in Δku80-DHFR by 20 dpt but then dropped to 3% by 23 dpt and to only ∼1% by 34 dpt. No fluorescence was observed in the Δku80-HXG or RH transfections through the duration of the experiment (34 dpt). This indicates that (i) different genes have varied receptiveness to recombination and tagging, (ii) the Δku80 strain is more amenable to homologous recombination and gene tagging than the wild-type RH strain, and (iii) pyrimethamine selection appears to work better than MPA/X selection in some but not all cases.
To utilize the Δku80 strain and YFP tagging to investigate novel genes, we compiled a list of genes based on several criteria: (i) identification in a MudPIT/2DE proteomic screen of excreted-secreted antigen (ESA) products (49); (ii) evidence of a cell cycle expression profile consistent with a role in parasite invasion (M. Behnke and M. White, personal communication); (iii) the presence of domains previously identified in microneme proteins; and (iv) the presence of mass spectroscopy peptides in the Toxoplasma genome database. We present six examples of tagged genes that were confirmed to be expressed by both immunoblot analysis and by fluorescence. Table 1 shows all the genes tested, including the gene IDs, the predicted molecular masses of the proteins, and the localization of the protein by YFP. All primers used to amplify tested genes are provided in the Table S1 of the supplemental material. After cloning the genes into pYFP.LIC.DHFR and transfection/selection, transformant pools were examined for YFP expression. While expression of some YFP could be viewed directly by microscopy, many required staining with anti-GFP antibody to enhance the signal. The overall success rate of this method as measured by positive YFP signal after transfection and selection was 74%.
TABLE 1.
Genes chosen for YFP tagging of Δku80
| Toxodb gene IDa | Name | Mass (kDa)
|
Identified by: | No. of Toxodb MS peptidesb | Localization
|
||
|---|---|---|---|---|---|---|---|
| Predicted | Observed (with YFP) | Expected | Observed | ||||
| 55.m00019 | ACP | 19 | 50, 43 | Waller et al. (46) | 11 | Apicoplast | Apicoplast |
| 50.m00004 | pCNA | 35 | 60 | Guerini et al. (14) | 8 | Nucleus | Nucleus |
| 583.m00003 | ROP1 | 48 | 75 | Ossorio et al. (28a) | 24 | Rhoptries | Rhoptries |
| 641.m00002 | MIC3 | 38 | NAf | Zhou et al. (49) | 53 | Micronemes | Micronemes |
| 20.m03849 | PLP1 | 113 | 160 | Zhou et al. (49) | 16 | Micronemes | Micronemes |
| 65.m00002 | MIC5 | 26 | NA | Zhou et al. (49) | 21 | Micronemes | Subapical |
| 33.m00006 | M2AP | 35 | NA | Zhou et al. (49) | 72 | Micronemes | Subapical |
| 50.m00006 | HSP60 | 61 | NA | Toursel et al. (41a) | >100 | Mitochondria | No signal |
| 44.m00004 | IMC1 | 70 | NA | Mann et al. (23) | >100 | IMC | No signal |
| 44.m00009 | P30/SAG1 | 30 | NA | Kasper et al. (18a) | 35 | Surface | Anterior to nucleus |
| 645.m00037 | Cathepsin protease L | 47 | 70 | Putative MIC maturase | 2 | MVEc/lysosome | Anterior to nucleus puncta |
| 83.m01237 | Chitinase-like | 72 | 125 | Zhou et al. (49) | 11 | Micronemes | Micronemes |
| 83.m00006 | SPATR | 58 | 95 | Kawase et al. (19) | 3 | Apical | Micronemes |
| 20.m03858 | Hypothetical | 97 | 130 | Invasion cell cycle | 9 | Unknown | Micronemes |
| 20.m03958 | Toxolysin 4 | 256 | NA | Zhou et al. (49) | 26 | Unknown | Subapical |
| 49.m00054 | Hypothetical (dystroglycan dom.) | 101 | 140, 100 | Zhou et al. (49) | 6 | Unknown | Subapical |
| 55.m04865 | Hypothetical | 3 | Unknown | Subapical | |||
| 49.m03355 | Hypothetical | 12 | 40 | Invasion cell cycle | 14 | Unknown | Apical tip/conoid |
| 72.m00001 | Non-TM Ag [poly(ADP-ribose) glycohydrolase] | 62 | 85 | Invasion cell cycle | 49 | Unknown | PVg |
| 20.m03748 | C2 domain protein | 33 | 55 | Invasion cell cycle | 9 | Unknown | Cytosolic |
| 50.m00008 | Cathepsin protease B | 62 | NA | Que et al. (30a) | 5 | Rhoptries | No signal |
| 55.m04618 | Hypothetical (25 EGFd) | 172 | NA | MIC-like domains | 1 | Unknown | No signal |
| 8.m00176/8h | Hypothetical (4-PANe) | 39 | NA | Zhou et al. (49) | 2/12 | Unknown | No signal |
| 80.m02161 | Hypothetical | 48 | NA | Invasion cell cycle | >100 | Unknown | No signal |
As of October 2008.
Peptide(s) identified by mass spectroscopy according to Toxodb.
MVE, multivesicular endosome.
EGF, epidermal growth factor.
Also known as Apple domains.
NA, not applicable.
PV, parasitophorous vacuole.
Genes 8.m00176 and 8.m00178.
Perforin-like protein 1 (PLP1) is a novel microneme protein identified in a MudPIT analysis (49) that contains a MACPF pore-forming domain (18). PLP1 is predicted to be a 113-kDa protein (Table 1), although it migrates at 130 kDa on reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis. In PLP1-YFP parasites, the PLP1 band shifted up to ∼160 kDa (Fig. 4B), suggesting correct integration of the tagging construct at the PLP1 locus. Accordingly, antibodies against PLP1 and GFP showed an identical pattern, consistent with microneme staining (Fig. 4A, panel i).
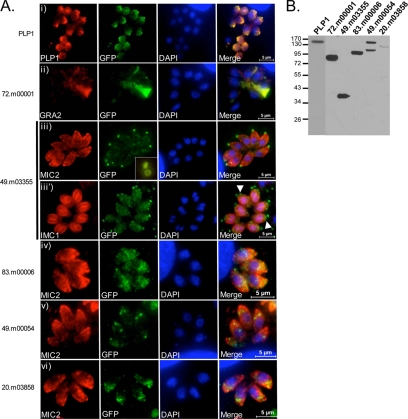
FIG. 4.
Localization of novel gene products by endogenous gene tagging. (A) YFP-tagged genes by immunofluorescence. (i) Microneme protein PLP1. (ii) 72.m00001, poly(ADP-ribose) glycohydrolase. (iii and iii′) 49.m03355 hypothetical protein. Arrowheads point to the YFP signal in apical poles of developing daughter cells. Inset: ring-like localization pattern. (iv) 83.m00006, SPATR-like protein. (v) 49.m00054, dystroglycan domain protein. (vi) 20.m03858 hypothetical protein. Scale bars were generated within the Zeiss Axio program. (B) Immunoblot of YFP-tagged genes probed with anti-GFP. Positions of molecular mass markers (in kDa) are indicated.
The 72.m00001 gene is annotated as a 62-kDa poly(ADP-ribose) glycohydrolase (PARG) family protein. YFP tagging of this protein showed localization in the parasitophorous vacuole and, in particular, an overlap with the intravacuolar membranous nanotubular network (42), as revealed by colocalization with the dense granule protein GRA2 (24) (Fig. 4A, panel ii). A single band at ∼80 kDa was observed by immunoblotting (Fig. 4B), consistent with the expected size of the YFP chimera.
Gene 49.m03355 is a hypothetical protein with a predicted molecular mass of 12 kDa. After endogenous tagging, 49.m03355-YFP lysates showed a single band at ∼38 kDa (Fig. 4B). This protein localized to a distinct spot at the anterior pole of the tachyzoite near where the conoid is positioned (Fig. 4A, panel iii). 49.m03355-YFP localized anterior to the inner membrane complex (IMC), as visualized with an antibody against IMC1. In dividing parasites, a discrete 49.m03355-YFP signal was detected just anterior of the developing IMC of the daughter cells (Fig. 4A, panel iii′). Rotation of a three-dimensional image obtained by optical sectioning of 49.m03355-YFP parasites revealed that the signal corresponded to a ring-like structure, possibly one of the polar rings associated with the conoid (Fig. 4A, panel iii, inset). Since this was an exceptionally small gene, the entire genomic fragment, including the endogenous promoter and upstream sequences, was amplified to include a unique restriction site. Thus, in this particular case, we cannot exclude the possibility that this construct integrated randomly and is expressing the YFP-tagged version ectopically as an extra copy of this gene.
83.m00006 is a hypothetical protein with similarity to Plasmodium falciparum SPATR (secreted protein with altered thrombospondin type I repeat domain), which is localized to the surface of sporozoites and the apical region of asexual erythrocytic stages (4). 83.m00006 was identified in a proteomic analysis of Ca2+-dependent secretion (19) and is predicted to encode a 58-kDa protein. A single ∼95-kDa band was observed in 83.m00006-YFP lysates (Fig. 4B). 83.m00006-YFP localized to the apical region, where it partially overlapped with MIC2 staining, indicating that it may reside to some extent in the micronemes (Fig. 4A, panel iv).
Parasites expressing the tagged hypothetical protein 49.m00054 showed punctate staining slightly posterior to the micronemes (Fig. 4A, panel v). In colocalizations with proM2AP, which occupies a site termed the VP1 compartment (16a), a fraction of 49.m00054 appeared to colocalize with proM2AP at certain stages of replication (data not shown). In other vacuoles, the 49.m00054-YFP signal was found posterior to proM2AP (data not shown). The predicted molecular mass of 49.m00054 is 101 kDa, and by immunoblot analysis using an anti-GFP antibody two bands were observed, at ∼100 kDa and ∼135 to 140 kDa (Fig. 4B). The higher molecular mass band is consistent with the addition of a 27-kDa YFP tag; the lower band may represent a processed form. No bands were observed in an induced ESA fraction (data not shown), suggesting that the tagged protein is likely not secreted.
20.m03858 is a hypothetical protein with no orthologues or putative conserved domains. It is highly expressed in RH (www.toxodb.org) and is predicted to be a 97-kDa transmembrane protein. 20.m03858-YFP is localized to the apical or subapical region of the parasite; some colocalization with MIC2 is observed (Fig. 4A, panel vi). A band of 130 kDa was seen by immunoblotting, implying integration of the YFP in the endogenous locus.
DISCUSSION
Completion of the Toxoplasma genome sequencing project and the availability of the Toxoplasma genome database unleashes the possibilities of studying large cohorts of genes predicted to play roles in particular cellular events. While gene disruption can be a powerful approach for determining protein function, deciphering the subcellular localization of a protein can also provide important clues to its role in a process or pathway. We present here a combination of approaches for improving the efficiency of determining protein localization. First, we generated a T. gondii strain lacking Ku80, which functions with Ku70 in NHEJ DNA repair. In the absence of Ku80, homologous recombination predominates. We have exploited this to introduce a YFP tag to genes at their endogenous loci. We also generated a YFP expression construct containing a LIC cassette for convenient cloning of genes of interest. The LIC method of cloning proved much more effective and efficient than conventional restriction enzyme-mediated subcloning, since a single restriction digest of the vector allows the cloning of any insert with complementary LIC sequences. This makes it an ideal method for high-throughput cloning of inserts.
Several proteins that have previously been tagged with GFP were used as positive controls for testing the effectiveness of this construct and system. ACP, pCNA, MIC3, and ROP1 all targeted properly to their respective organelles and, importantly, this analysis showed that essential genes, such as ACP and pCNA, are amenable to tagging. However, IMC1-YFP and HSP60-YFP failed to show expression, even after multiple attempts. One potential explanation is that the YFP is cleaved from the gene product, a likely scenario with IMC1 after the protein is incorporated into the inner membrane complex (23). Thus, a limitation of the system is that tagging and visualization of gene products that undergo C-terminal proteolysis or glycophosphatidyl inositol anchor addition are not feasible.
Homologous recombination generally requires either a double-strand break in one of the two DNA double helices (plasmid or genomic DNA) or a single-strand nick in both helices (2, 22). Early studies performed in yeast determined that plasmids containing a double-strand break in the region of homology to genomic sequences recombined with high efficiency after yeast transformation (28). Linearization of the integrating construct results in ends-in recombination, wherein the entire plasmid integrates into the genome at the region of homology and creates a duplication of the homologous sequence; a chromosomal break does not occur because the break or gap is repaired by gene conversion (reviewed in reference 38). In the absence of construct linearization, proper integration is dependent on the low probability of a chromosomal break at the targeted locus. After analyzing constructs in supercoiled, linear, or nicked/open circle forms, we confirmed that only constructs linearized within the gene fragment integrated into the homologous locus. Linearization of the construct near the selectable marker did not result in proper integration of YFP into the gene of interest, but it is possible that a proportion of these pyrimethamine-resistant parasites contained the DHFR-TS selectable cassette in the endogenous DHFR-TS locus by double-crossover gene replacement. This would result in YFP-negative, pyrimethamine-resistant parasites. Based on the findings of Fohl and Roos, however, these parasites have a fitness defect compared to those with either a normal copy or those with exogenous copies in addition to the endogenous gene (8). Thus, such parasites may only be seen in situations where tagging of the target locus results in a substantial growth defect.
Linearization within the gene fragment requires the presence of a unique restriction site, which may necessitate amplification of a larger fragment of DNA to include such a site. Fortunately, this was not a hindrance, since we found that LIC cloning of larger fragments (3 to 4kb) was equally efficient as that with shorter lengths (1 to 2 kb). In the case of 49.m03355, a particularly small gene of 240 bp, an additional 2.7 kb of upstream genomic DNA was amplified to include a unique restriction enzyme site. No other genes were present in the additional DNA amplified, but since the targeting sequence inserts by homologous recombination, any gene(s) present would nonetheless be reconstituted after integration.
The positive control YFP-tagged proteins targeted properly to their expected organelles, and many tagged hypothetical proteins showed a pattern consistent with being in apical invasion organelles (rhoptries and micronemes). However, additional genes selected for tagging either never showed YFP expression (55.m04618, 8.m00176, 8.m00178 80.m02161, and 50.m00008) or displayed a pattern that resembled that of mistargeting or retention (20.m03958 and 49.m00054) in a subapical location, which is presumably a staging ground for the apical invasion organelles (Table 1). Similar results have been seen with some exogenously GFP-tagged genes encoding secretory proteins (49). These gene products do not appear to have a feature in common, e.g., a transmembrane anchor, which could account for the failure of the fusion protein to reach the invasion organelles. Bands detected on immunoblots were the predicted sizes for the YFP-tagged proteins, implying that the proper genes were tagged. There are several possible scenarios: (i) these proteins may naturally occupy a subapical site; (ii) the precursor is observed in a subapical compartment and the YFP is cleaved from the mature protein prior to continuing on to the invasion organelles (as the case may be for 49.m00054); or (iii) the YFP tag hinders the protein from moving through the secretory pathway to its final destination. Distinguishing among the possibilities would require producing antibodies to establish the normal location of the protein. We are currently generating and testing additional constructs with smaller epitope tags, which may better facilitate the correct localization of the tagged gene product.
The genes chosen for tagging were based on their potential role in invasion, identified by the similarity of their expression profiles to other known invasion proteins or by their presence in an ESA proteomic screen. While we expected most of these genes to be in the invasion organelles, there were exceptions. For example, the 49.m03355 protein showed a distinct signal at the extreme apical tip of the parasite, with no overlap with microneme proteins AMA1 or MIC2 or with the inner membrane complex protein IMC1. The pattern is similar to TgCAM1 and TgCAM2 (17) which, like 49.m03355, are observed at the apical tip of mature parasites and in developing daughter cells. The resolution of light microscopy is insufficient to ascertain whether 49.m03355 is localized to the apical polar rings or the microtubules comprising the conoid body.
In addition to the YFP vector, we have also engineered constructs containing mCherry, tandem dimer Tomato (tdTomato), and cyan fluorescent reporter proteins (21, 33, 34). We speculate that low pH in the apical invasion organelles may hamper the YFP signal, and assessing additional fluorescent reporter proteins may improve signal detection. Since pCNA-mCherry and pCNA-tdTomato constructs integrated properly and are correctly expressed in the nucleus (data not shown), these vectors can now be used to try tagging genes that did not show fluorescence with YFP. Constructs encoding YFP and cyan fluorescent protein may also be utilized in fluorescent resonance energy transfer interaction studies of two chromosomally tagged proteins. Another application of the tagging method is to identify interacting partners in protein complexes. To this end, we have generated a tagging construct with a tandem affinity purification tag for isolation of protein complexes. The combination of these constructs, the LIC cloning method, and the Δku80 parasite strains, should prove to be useful tools for more rapid determination of protein localization and interacting partner proteins for the functional analyses of novel proteins.
Supplementary Material
Acknowledgments
This work was supported by a grant from the U.S. National Institutes of Health (AI046675 to V.B.C.).
We are grateful to Michael Behnke and Michael White for sharing data prior to publication. We thank Tracey Schultz for technical assistance and members of the Carruthers lab for helpful discussions, and we also thank David Bzik for constructive exchanges of ideas and suggestions.
Footnotes
Published ahead of print on 13 February 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Aslanidis, C., and P. J. de Jong. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 186069-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camerini-Otero, R. D., and P. Hsieh. 1995. Homologous recombination proteins in prokaryotes and eukaryotes. Annu. Rev. Genet. 29509-552. [DOI] [PubMed] [Google Scholar]
- 3.Chang, P. K. 2008. A highly efficient gene-targeting system for Aspergillus parasiticus. Lett. Appl. Microbiol. 46587-592. [DOI] [PubMed] [Google Scholar]
- 4.Chattopadhyay, R., D. Rathore, H. Fujioka, S. Kumar, P. de la Vega, D. Haynes, K. Moch, D. Fryauff, R. Wang, D. J. Carucci, and S. L. Hoffman. 2003. PfSPATR, a Plasmodium falciparum protein containing an altered thrombospondin type I repeat domain is expressed at several stages of the parasite life cycle and is the target of inhibitory antibodies. J. Biol. Chem. 27825977-25981. [DOI] [PubMed] [Google Scholar]
- 5.Critchlow, S. E., and S. P. Jackson. 1998. DNA end-joining: from yeast to man. Trends Biochem. Sci. 23394-398. [DOI] [PubMed] [Google Scholar]
- 6.Donald, R. G., D. Carter, B. Ullman, and D. S. Roos. 1996. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J. Biol. Chem. 27114010-14019. [DOI] [PubMed] [Google Scholar]
- 7.Errami, A., V. Smider, W. K. Rathmell, D. M. He, E. A. Hendrickson, M. Z. Zdzienicka, and G. Chu. 1996. Ku86 defines the genetic defect and restores X-ray resistance and V(D)J. recombination to complementation group 5 hamster cell mutants. Mol. Cell. Biol. 161519-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fohl, L. M., and D. S. Roos. 2003. Fitness effects of DHFR-TS mutations associated with pyrimethamine resistance in apicomplexan parasites. Mol. Microbiol. 501319-1327. [DOI] [PubMed] [Google Scholar]
- 9.Fox, B. A., J. G. Ristuccia, J. P. Gigley, and D. J. Bzik. 2009. Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryot. Cell 8520-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gajria, B., A. Bahl, J. Brestelli, J. Dommer, S. Fischer, X. Gao, M. Heiges, J. Iodice, J. C. Kissinger, A. J. Mackey, D. F. Pinney, D. S. Roos, C. J. Stoeckert, Jr., H. Wang, and B. P. Brunk. 2008. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 36D553-D556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenwood, B. M., D. A. Fidock, D. E. Kyle, S. H. Kappe, P. L. Alonso, F. H. Collins, and P. E. Duffy. 2008. Malaria: progress, perils, and prospects for eradication. J. Clin. Investig. 1181266-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu, Y., S. Jin, Y. Gao, D. T. Weaver, and F. W. Alt. 1997. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J. recombination. Proc. Natl. Acad. Sci. USA 948076-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gubbels, M. J., C. Li, and B. Striepen. 2003. High-throughput growth assay for Toxoplasma gondii using yellow fluorescent protein. Antimicrob. Agents Chemother. 47309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerini, M. N., M. S. Behnke, and M. W. White. 2005. Biochemical and genetic analysis of the distinct proliferating cell nuclear antigens of Toxoplasma gondii. Mol. Biochem. Parasitol. 14256-65. [DOI] [PubMed] [Google Scholar]
- 15.Gullo, C., M. Au, G. Feng, and G. Teoh. 2006. The biology of Ku and its potential oncogenic role in cancer. Biochim. Biophys. Acta 1765223-234. [DOI] [PubMed] [Google Scholar]
- 16.Hande, M. P. 2004. DNA repair factors and telomere-chromosome integrity in mammalian cells. Cytogenet. Genome Res. 104116-122. [DOI] [PubMed] [Google Scholar]
- 16a.Harper, J. M., M.-H. Huynh, I. Coppens, F. Parussini, S. Moreno, and V. B. Carruthers. 2006. A cleavable propeptide influences Toxoplasma infection by facilitating the trafficking and secretion of the TgMIC2.M2AP invasion complex. Mol. Biol. Cell 174551-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, K., J. Johnson, L. Florens, M. Fraunholz, S. Suravajjala, C. DiLullo, J. Yates, D. S. Roos, and J. M. Murray. 2006. Cytoskeletal components of an invasion machine: the apical complex of Toxoplasma gondii. PLoS Pathog. 2e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kafsack, B. F., J. D. Pena, I. Coppens, S. Ravindran, J. C. Boothroyd, and V. B. Carruthers. 2009. Rapid membrane disruption by a perforin-like protein facilitates parasite exit from host cells. Science 323530-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Kasper, L. H., J. H. Crabb, and E. R. Pfefferkorn. 1983. Purification of a major membrane protein of Toxoplasma gondii by immunoabsorption with a monoclonal antibody. J. Immunol. 1302407-2412. [PubMed] [Google Scholar]
- 19.Kawase, O., Y. Nishikawa, H. Bannai, H. Zhang, G. Zhang, S. Jin, E. G. Lee, and X. Xuan. 2007. Proteomic analysis of calcium-dependent secretion in Toxoplasma gondii. Proteomics 73718-3725. [DOI] [PubMed] [Google Scholar]
- 20.Little, J. B., H. Nagasawa, G. C. Li, and D. J. Chen. 2003. Involvement of the nonhomologous end joining DNA repair pathway in the bystander effect for chromosomal aberrations. Radiat. Res. 159262-267. [DOI] [PubMed] [Google Scholar]
- 21.Llopis, J., S. Westin, M. Ricote, Z. Wang, C. Y. Cho, R. Kurokawa, T. M. Mullen, D. W. Rose, M. G. Rosenfeld, R. Y. Tsien, and C. K. Glass. 2000. Ligand-dependent interactions of coactivators steroid receptor coactivator-1 and peroxisome proliferator-activated receptor binding protein with nuclear hormone receptors can be imaged in live cells and are required for transcription. Proc. Natl. Acad. Sci. USA 974363-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lloyd, R. G., and G. J. Sharples. 1992. Genetic analysis of recombination in prokaryotes. Curr. Opin. Genet. Dev. 2683-690. [DOI] [PubMed] [Google Scholar]
- 23.Mann, T., E. Gaskins, and C. Beckers. 2002. Proteolytic processing of TgIMC1 during maturation of the membrane skeleton of Toxoplasma gondii. J. Biol. Chem. 27741240-41246. [DOI] [PubMed] [Google Scholar]
- 24.Mercier, C., L. Lecordier, F. Darcy, D. Deslee, A. Murray, B. Tourvielle, P. Maes, A. Capron, and M. F. Cesbron-Delauw. 1993. Molecular characterization of a dense granule antigen (GRA2) associated with the network of the parasitophorous vacuole. Mol. Biochem. Parasitol. 5871-82. [DOI] [PubMed] [Google Scholar]
- 25.Ninomiya, Y., K. Suzuki, C. Ishii, and H. Inoue. 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 10112248-12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nussenzweig, A., K. Sokol, P. Burgman, L. Li, and G. C. Li. 1997. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival, and development. Proc. Natl. Acad. Sci. USA 9413588-13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orr-Weaver, T. L., and J. W. Szostak. 1983. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc. Natl. Acad. Sci. USA 804417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orr-Weaver, T. L., J. W. Szostak, and R. J. Rothstein. 1981. Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. USA 786354-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Ossorio, P. N., J. D. Schwartzman, and J. C. Boothroyd. 1992. A Toxoplasma gondii rhoptry protein associated with host cell penetration has an unusual charge assymetry. Mol. Biochem. Parasitol. 501-16. [DOI] [PubMed] [Google Scholar]
- 29.Pfannschmidt, C., and J. Langowski. 1998. Superhelix organization by DNA curvature as measured through site-specific labeling. J. Mol. Biol. 275601-611. [DOI] [PubMed] [Google Scholar]
- 30.Poggeler, S., and U. Kuck. 2006. Highly efficient generation of signal transduction knockout mutants using a fungal strain deficient in the mammalian ku70 ortholog. Gene 3781-10. [DOI] [PubMed] [Google Scholar]
- 30a.Que, X., A. Wunderlich, K. A. Joiner, and S. L. Reed. 2004. Toxopain-1 is critical for infection in a novel chicken embryo model of congenital toxoplasmosis. Infect. Immun. 722915-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radke, J. R., B. Striepen, M. N. Guerini, M. E. Jerome, D. S. Roos, and M. W. White. 2001. Defining the cell cycle for the tachyzoite stage of Toxoplasma gondii. Mol. Biochem. Parasitol. 115165-175. [DOI] [PubMed] [Google Scholar]
- 32.Rockwood, L. D., A. Nussenzweig, and S. Janz. 2003. Paradoxical decrease in mutant frequencies and chromosomal rearrangements in a transgenic lacZ reporter gene in Ku80 null mice deficient in DNA double strand break repair. Mutat. Res. 52951-58. [DOI] [PubMed] [Google Scholar]
- 33.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 221567-1572. [DOI] [PubMed] [Google Scholar]
- 34.Shu, X., N. C. Shaner, C. A. Yarbrough, R. Y. Tsien, and S. J. Remington. 2006. Novel chromophores and buried charges control color in mFruits. Biochemistry 459639-9647. [DOI] [PubMed] [Google Scholar]
- 35.Stols, L., M. Gu, L. Dieckman, R. Raffen, F. R. Collart, and M. I. Donnelly. 2002. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 258-15. [DOI] [PubMed] [Google Scholar]
- 36.Striepen, B., C. Y. He, M. Matrajt, D. Soldati, and D. S. Roos. 1998. Expression, selection, and organellar targeting of the green fluorescent protein in Toxoplasma gondii. Mol. Biochem. Parasitol. 92325-338. [DOI] [PubMed] [Google Scholar]
- 37.Striepen, B., D. Soldati, N. Garcia-Reguet, J. F. Dubremetz, and D. S. Roos. 2001. Targeting of soluble proteins to the rhoptries and micronemes in Toxoplasma gondii. Mol. Biochem. Parasitol. 11345-53. [DOI] [PubMed] [Google Scholar]
- 38.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein, and F. W. Stahl. 1983. The double-strand-break repair model for recombination. Cell 3325-35. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi, T., T. Masuda, and Y. Koyama. 2006. Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae. Mol. Genet. Genomics 275460-470. [DOI] [PubMed] [Google Scholar]
- 41.Teoh, N. C., Y. Y. Dan, K. Swisshelm, S. Lehman, J. H. Wright, J. Haque, Y. Gu, and N. Fausto. 2008. Defective DNA strand break repair causes chromosomal instability and accelerates liver carcinogenesis in mice. Hepatology 472078-2088. [DOI] [PubMed] [Google Scholar]
- 41a.Toursel, C., F. Dzierzinski, A. Bernigaud, M. Mortuaire, and S. Tomavo. 2000. Molecular cloning, organellar targeting and developmental expression of mitochondrial chaperone HSP60 in Toxoplasma gondii. Mol. Biochem. Parasitol. 111319-332. [DOI] [PubMed] [Google Scholar]
- 42.Travier, L., R. Mondragon, J. F. Dubremetz, K. Musset, M. Mondragon, S. Gonzalez, M. F. Cesbron-Delauw, and C. Mercier. 2008. Functional domains of the Toxoplasma GRA2 protein in the formation of the membranous nanotubular network of the parasitophorous vacuole. Int. J. Parasitol. 38757-773. [DOI] [PubMed] [Google Scholar]
- 43.Tuteja, R., and N. Tuteja. 2000. Ku autoantigen: a multifunctional DNA-binding protein. Crit. Rev. Biochem. Mol. Biol. 351-33. [DOI] [PubMed] [Google Scholar]
- 44.Uegaki, K., N. Adachi, S. So, S. Iiizumi, and H. Koyama. 2006. Heterozygous inactivation of human Ku70/Ku86 heterodimer does not affect cell growth, double-strand break repair, or genome integrity. DNA Repair (Amsterdam) 5303-311. [DOI] [PubMed] [Google Scholar]
- 45.Walker, J. R., R. A. Corpina, and J. Goldberg. 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412607-614. [DOI] [PubMed] [Google Scholar]
- 46.Waller, R. F., P. J. Keeling, R. G. Donald, B. Striepen, E. Handman, N. Lang-Unnasch, A. F. Cowman, G. S. Besra, D. S. Roos, and G. I. McFadden. 1998. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 9512352-12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan, K. L., V. B. Carruthers, L. D. Sibley, and J. W. Ajioka. 1997. Molecular characterization of an expressed sequence tag locus of Toxoplasma gondii encoding the micronemal protein MIC2. Mol. Biochem. Parasitol. 84203-214. [DOI] [PubMed] [Google Scholar]
- 48.Yoo, S., A. Kimzey, and W. S. Dynan. 1999. Photocross-linking of an oriented DNA repair complex. Ku bound at a single DNA end. J. Biol. Chem. 27420034-20039. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, X. W., B. F. Kafsack, R. N. Cole, P. Beckett, R. F. Shen, and V. B. Carruthers. 2005. The opportunistic pathogen Toxoplasma gondii deploys a diverse legion of invasion and survival proteins. J. Biol. Chem. 28034233-34244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.