Abstract
Activation of the unfolded protein response (UPR) in eukaryotes involves the splicing of an unconventional intron from the mRNA encoding the transcriptional activator of the pathway. In Saccharomyces cerevisiae a 252-nucleotide (nt) unconventional intron is spliced out of the transcript of HAC1, changing the 3′ end of the HAC1 open reading frame and relieving the transcript from a translational block in a single step. The translational block is caused by the base pairing of part of the unconventional intron with the 5′-untranslated region (5′UTR). In Aspergillus niger and other aspergilli, the unconventional intron in hacA mRNA is only 20 nt long. Since this intron is part of a stable stem-loop structure, base pairing with the 5′UTR, in contrast to the case with yeast HAC1, is not possible. However, analysis of the hacA mRNA revealed a GC-rich inverted repeat (18 base pairings). Upon the activation of the UPR, the 5′UTR of hacA mRNA is truncated by 230 nt, removing the left part of this inverted repeat. This implies a similar release of a translational block as in the case of S. cerevisiae HAC1 but in two steps. The mechanism behind the 5′ truncation, which does not take place in either yeast HAC1 or mammalian xbp1 mRNA, has been hitherto unknown. Here we show that during secretion stress in A. niger, hacA transcription starts from a new start site closer to the ATG, relieving the transcript from translational attenuation. This transcriptional switch is mediated by HacA itself and the unfolded protein response element 2 (UPRE2) in the hacA promoter.
In eukaryotic cells, the majority of secreted and membrane proteins are folded, assembled, and modified in the endoplasmic reticulum (ER). Chaperones and foldases assist in folding the newly synthesized proteins (4), and correctly folded proteins are transported to the Golgi compartment, where further modification takes place. The secretory proteins are transported in vesicles from the Golgi apparatus to the cell membrane, where the content of the vesicle is released into the medium. When the amount of newly synthesized proteins exceeds the folding capacity of the ER, or if folding in the ER is impaired by other means, the eukaryotic cell triggers an intracellular signaling pathway in order to counteract the resulting secretion stress. This response is known as the unfolded protein response (UPR) (11, 25, 31, 37) and leads to the increased synthesis of ER-resident foldases and chaperones as well as many other components of the secretory pathway (34). Genomic analysis of the UPR in Saccharomyces cerevisiae, Arabidopsis thaliana, Aspergillus nidulans, Trichoderma reesei, and Aspergillus niger all linked large sets of genes (several hundred) to the UPR, affecting all stages of the secretory pathway (2, 5, 16, 33, 34). The central activation step of the UPR involves the splicing of an unconventional intron from the mRNA encoding the transcriptional activator of the UPR. It is conserved from lower eukaryotes like yeasts to higher eukaryotes like mammalian cells and is mediated by Ire1p/IreA, a transmembrane kinase/RNase residing in the ER membrane. With its luminal domain, Ire1p/IreA senses the “folding state” of proteins in the ER, and when necessary, a signal is transmitted over the ER membrane by the activation of the RNase domain, which in turn processes the messenger encoding the transcriptional activator of the UPR.
In S. cerevisiae, the Ire1p-mediated splicing of a 252-nucleotide (nt) unconventional intron from HAC1 mRNA relieves the transcript from a translational block (12, 28). HAC1 encodes a basic leucine zipper-type transcription factor that activates its target genes by a direct interaction with upstream activating sequences. In mammalian cells, the HAC1 homologue XBP1 is activated in a similar way by the splicing of a 26-nt unconventional intron (15), whereas Trichoderma reesei hac1 and Aspergillus nidulans and A. niger hacA are activated by the splicing of a 20-nt unconventional intron (20, 29). To distinguish the unspliced hacA mRNA form the spliced form, they are referred to as hacAu (uninduced) and hacAi (induced), respectively.
Taking into account the magnitude of the UPR and the risk that the accumulation of unfolded proteins in the ER pose onto the cell, it seems plausible that natural selection has favored a strict controllability of the pathway. Although the UPR and its central activation step are conserved in all eukaryotic organisms, there are differences in the molecular mechanisms underlying the signaling pathway that give the different organisms different ways of control over the response. In mammalian cells, the UPR consists of three distinct pathways that operate in parallel and include general attenuation of translation, apoptosis, and increased folding and degradation of unfolded proteins (6). The mammalian UPR is triggered by at least three different ER stress sensors, all ER transmembrane proteins. This concerns (i) IRE1, a nuclease that splices the unconventional intron from XBP1 mRNA (38); (ii) ATF6, which migrates to the Golgi compartment in response to ER stress (in the Golgi compartment, its cytosolic domain, which encodes a transcription factor, is proteolytically released and relocated to the nucleus, where it upregulates its target genes) (9); and (iii) a protein kinase-like ER kinase (PERK) that phosphorylates translation initiation factor 2α in response to ER stress, resulting in a general inhibition of translation (8).
In contrast to mammalian cells, in S. cerevisiae, the only known sensor of the UPR is Ire1p. In addition to the on/off switch of yeast UPR, several other factors influencing the UPR circuit have been described. The transcription of HAC1 was shown to be autoregulated (23), and the translation of the unspliced HAC1u mRNA was blocked by long-range base pairing (28). Furthermore, a second ER-to-nucleus signaling pathway was found, which leads to an increased level of production of Hac1p combined with the production or activation of a putative UPR modulating factor (14). Both signals are required for super-UPR activation. Finally, Gcn4p and its activator, Gcn2p, were found to be required for the induction of the majority of the UPR target genes (26).
Integrated with the Ire1p pathway, these elements turn the UPR from a simple switch into a more complex response and give the yeast cell more fine-tuned control over the UPR.
The UPR in A. niger shares some characteristics with the S. cerevisiae response, but there are some differences. As saprophytic organisms, filamentous fungi are capable of secreting large amounts of proteins to recruit nutrients. This characteristic might have shaped the secretory pathway and have set specific demands to the stress responses, which might explain the differences with S. cerevisiae UPR.
First, in addition to the unconventional intron splicing, the 5′-untranslated regions (5′UTR) of the hacA transcripts of A. niger, A. nidulans, and T. reesei are truncated upon ER stress (20, 29). This has not been reported for yeast HAC1 or mammalian XBP1 homologues. Second, a novel type of transcriptional downregulation mechanism was described for filamentous fungi, which was termed repression under secretion stress (1, 24). Upon ER stress, it downregulates the transcription of some genes encoding secreted proteins, thereby lowering the burden on the ER. Third, three UPR elements (UPREs) are present in the hacA promoters of A. niger, A. nidulans, and A. fumigatus (19). These elements could play a role in the upregulation of hacA by the activator protein itself, which was experimentally shown for A. niger (20). The position of the UPREs in the hacA promoter in relation to start sites of the full-length and truncated hacA mRNAs could also implicate a role for them in the truncation of the hacA transcript.
Here we show the mechanism behind the truncation of the 5′UTR of the A. niger hacA mRNA and the involvement of the UPREs in the regulation of the response.
MATERIALS AND METHODS
Strains and transformations.
A. niger AB4.1 (cspA1 pyrG) and transformants thereof were grown on minimal medium plates containing (per liter) 6 g NaNO3, 1.5 g KH2PO4, 0.5 g MgSO4 · 7H2O, 0.5 g KCl, trace elements (35), 1.5% agar, and 20 g fructose as a carbon source (pH 6.0). A total of 5 mM uridine was added in case of auxotrophy. Protoplasts were transformed according to standard procedures (13). Agrobacterium tumefaciens-mediated transformation of A. niger was performed according to previously described methods (3). Transformation plates were incubated at 18°C.
Construction of an A. niger ΔhacA strain.
A 4.5-kb fragment containing hacA with 2 kb of its 5′-flanking region and 1.1-kb 3′-flanking region was PCR amplified from A. niger genomic DNA using primers P1 and P2 and cloned into pCR-Blunt II-TOPO (Invitrogen). The resulting plasmid, pHM58, was cut with BglII, filled in with Klenow fragment, and cut with SalI. This removed the major part of the hacA gene from the plasmid, which was replaced with the hygB cassette from pAN7-1 (GenBank accession number Z32698). For this purpose, pAN7-1 was cut with XbaI, filled in with Klenow fragment, and cut with XhoI. A 6.6-kb KpnI-XhoI fragment containing the hygB cassette flanked by the upstream and downstream regions of hacA was isolated from the resulting plasmid. The fragment was cloned between the left and right borders of A. tumefaciens plasmid pSDM14 (22). This resulted in plasmid pHM60, which was used for the replacement of hacA in A. niger by A. tumefaciens-mediated transformation.
Construction complementation plasmid.
A 3.2-kb FspI-BamHI fragment from plasmid pHM58 was ligated into the HincII-BamHI sites of pBluescriptSK(+), resulting in plasmid pHacA. For selective targeting of the plasmid to the pyrG locus of A. niger, the 3.9-kb XbaI fragment of pABpyrG*Not (kindly provided by Peter Punt, TNO Zeist, The Netherlands) was inserted into the SpeI site of pHacA, resulting in plasmid pHacA-pyrG*. The pyrG* gene contains a filled-in BamHI site disrupting the open reading frame (ORF). Only the homologous integration of the plasmid at the pyrG locus will restore uridine prototrophy, enabling the selective targeting of the construct to the pyrG locus.
Site-directed mutagenesis.
For purposes of mutagenesis of the elements present in the hacA promoter and 5′UTR of hacA mRNA, a 623-bp fragment containing these elements was PCR amplified from pHacA with primer pair P3 and P4 and cloned into pCR-BluntII-TOPO (Invitrogen), resulting in pCR-Blunt-PhacA. Site-specific mutations were introduced into the various elements using the QuikChange site-directed mutagenesis kit (Stratagene). The primers are listed in Table 1. The mutations were verified by sequencing performed with the BigDye sequencing kit (Applied Biosystems) and analyzed with the ABIPrism 3100 genetic analyzer (Applied Biosystems). A 493-bp BstEII-AspI fragment of pHacA was replaced with the corresponding but mutated fragment from plasmid pCR-Blunt-PhacA.
TABLE 1.
PCR oligonucleotide primers used in this studya
| Oligonucleotide | Sequence | Description |
|---|---|---|
| P1 | 5′-GGAGAACCAAGGTGCTATTG | −2,071-bp hacA start codon |
| P2 | 5′-GTTTAGGAGCCTTTACCTCC | +1,152-bp hacA stop codon |
| P3 | 5′-ATTTACCGTACGGTCAATTGGGGC | −503-bp hacA start codon |
| P4 | 5′-CCCTGCCTGTACTGACGAGTCATC | +120-bp hacA start codon |
| P5 | 5′-GGCCTGATCTGAACACGGACGCCTTTTAAAGAGT | UPRE1 |
| P6 | 5′-TCCCCCGTTATGACACGGACGCCTGTGTTCCTGT | UPRE2 |
| P7 | 5′-ATGGTTCTTAAGGACACCACTCCTTCTTGGCCCT | UPRE3 |
| P8 | 5′-TCTTTTTATTGTTCTCTGGTTCTTAAGGACACC | ATG uORF |
| P9 | 5′-CGACCTACATCACCGTCCTCCCAACGTCAGCGGTTAAGATAAGGCTCATAGTAAATCGATTG | 5′ splice site |
| P10 | 5′-TAGTGCCGTCCTCTGCGATCTTCA | Unspliceable intron us |
| P11 | 5′-TGAAGATCGCAGAGGACGGCACTA | Unspliceable intron ls |
| P12 | 5′-GCCTGGGTTAGCGCCCCCTGCAAGCCCCGTTATGACACGGTGGCCTGTGTTC | Inverted repeat 5′UTR |
| P13 | 5′-GACAGGTAATTCCTGCCCCCATGACTTTCTCTTCTTCACAGG | Inverted repeat hacA gene |
| P14 | 5′-TCCCCCGTTATGACACGGTGGCCTGTGTTCCTGT | −168-bp hacA start codon |
| P15 | 5′-TCAAACCGCTCAAGATTCGTTT | +1,253-bp hacA start codon |
| P16 | 5′-CGTCGAGAACGTCAAAGGCGAACCCGTC | For 5′RACE hacA |
| P17 | 5′-TTCGATGACAAGGATGTCCAGA | For RT-PCR bipA |
| P18 | 5′-GGGGATGAGCTTGGTCATGA | For RT-PCR bipA |
| P19 | 5′-ATTTAAATACCCTCTCCCATCGTCCTC | For RT-PCR pdiA |
| P20 | 5′-CCTCCTCGGCGGTGCAGTCAACCTTCAC | For RT-PCR pdiA |
Mutations are represented in boldface type.
The unconventional intron was removed by digesting pHacA with PstI followed by religation. The hacA variant with the unspliceable unconventional intron was constructed by ligation of the annealed oligonucleotide pair P10-P11 into PstI-digested plasmid pHacA. For mutation of the long-range inverted repeats, a 1,756-bp PCR fragment was generated in several successive PCR and assembly steps, which involved primers P3, P4, P12, P13, P14, and P15 (Table 1). The resulting fragment was cloned into pCR-BluntII-TOPO, and after sequencing, a 1,351-bp BstEII-BglII fragment was cloned into pHacA.
After introduction of the mutations into pHacA, the XbaI pyrG* fragment was cloned into the SpeI site of pHacA. Constructs having the pyrG* gene in the same orientation as that of wild-type plasmid pHacA-pyrG* were selected for transformation to A. niger ΔhacA.
Electrophoretic mobility shift assay (EMSA).
A total of 100 pmol oligonucleotide primers was annealed in 100 μl 10 mM Tris-HCl (pH 7.5) containing 100 mm NaCl by heating the mixtures to 95°C for 5 min and cooling them down to room temperature by placing the tubes on the work bench. A total of 1 pmol of annealed oligonucleotide was end labeled in a 20-μl reaction mixture with 5 μl [γ-32P]ATP and 20 units T4 polynucleotide kinase (New England Biolabs). The reaction mixture was incubated for 1 h at 37°C, and the radiolabeled oligonucleotide was purified on Biogel P30 spin columns (Bio-Rad). Binding reactions (20-μl mixtures) were performed using a buffer containing 25 mM Tris (pH 8.0), 5 mM dithiothreitol (DTT), 100 mM KCl, 4 mM spermidine, 0.1 μg/μl poly(dI-dC), 0.25% bovine serum albumin, 5% glycerol, and included 1 nM labeled probe, and 200 nM purified HacA protein (20). Binding was allowed to proceed on ice for 15 min. The reaction mixtures were loaded onto a 6% polyacrylamide gel and electrophoresed at 100 V in 0.25× TBE (89 mM Tris, 89 mM boric acid, 20 mM EDTA [pH 8.3]).
Southern blot analysis.
Genomic DNA for Southern blot analysis was isolated by adding 800 μl buffer (100 mM Tris [pH 8.0], 50 mM EDTA, 1 mM DTT, 35 mM sodium dodecyl sulfate) to 0.1 g of ground mycelium. The DNA was purified by phenol-chloroform extraction according to standard procedures (30). The DNA pellet was resuspended in H2O and incubated with DNase-free RNase (Roche Molecular Biochemicals). For each digest, 10 μg of genomic DNA was incubated for 6 h at 37°C with 20 U of an appropriate restriction enzyme. The reaction products were separated on 0.8% TBE agarose gels. Blotting and hybridization were done according to standard procedures (30).
Northern blot analysis.
For Northern blot analysis, cultures were grown overnight at 30°C on minimal medium supplemented with 0.2% (wt/vol) yeast extract. Aliquots of 0.5 g wet mycelium were transferred into 50 ml fresh medium, and the cultures were incubated for 3 h before provoking ER stress by the addition of either 20 μg/ml tunicamycin or 20 mM DTT. Equal amounts of either dimethyl sulfoxide or water were added to the control cultures. Samples from the stressed and unstressed cultures were taken 3 h after applying the stress agents. Since tunicamycin is a more specific agent to induce ER stress, Northern blots of these treatments are shown, where possible. For the analysis of the UPREs and hacA processing, the DTT blot is shown, as DTT resulted in much more pronounced hacA splicing.
For RNA isolation, mycelium was ground with 1-mm glass beads using the Fast Prep FP120 system (Bio 101 Savant), and total RNA was isolated using the RNeasy plant total RNA kit (Qiagen). In order to prevent the degradation of the RNA during the purification procedure, 1.5-ml screw-cap tubes were filled with approximately 0.5 ml 1-mm glass beads and 700 μl RLC buffer, which was provided with the RNeasy plant total RNA kit. The filled tubes were frozen in liquid nitrogen prior to the harvesting of the mycelia. The mycelium was harvested rapidly and dried thoroughly between tissue paper, and a small piece of mycelium (100 to 200 mg) was added to a prefrozen tube and refrozen in liquid nitrogen. The frozen tubes were than placed into the Fast Prep FP120 apparatus and run for four cycles of 20 s at a speed of 6.5, which allowed the thawing and simultaneous grinding of the mycelia. For Northern analysis, 10 μg of total RNA was separated on a 1.5% (wt/vol) agarose gel containing 10 mM phosphate buffer (pH 7) and blotted onto a Hybond N membrane (Amersham Pharmacia Biotech). Blotting and hybridization were done according to standard procedures (30). Probes were generated by labeling DNA fragments with deoxycytidine 5′-[α-32P]triphosphate using the Prime-It II labeling kit (Stratagene) and purified on Biogel P30 spin columns (Bio-Rad). Exposures were made using Curix HC-S Plus X-ray films (Agfa). Quantification of the Northern blot signals was done by phosphorimager analysis (Instant Imager; Packard).
5′RACE and RT-PCR.
For 5′ rapid amplification of cDNA ends (5′RACE) and reverse transcriptase PCR (RT-PCR), first-strand cDNA was made from 1 μg of total RNA using the Smart RACE cDNA amplification kit (Clontech). The 5′ end of the hacA transcript was amplified by PCR with primer P16 using cycling conditions recommended by the kit's supplier. For the transcription analysis of bipA and pdiA, cDNA was amplified using primer pairs P17-P18 and P19-P20, respectively.
Microscopy.
For microscopy, mycelia was grown between two glass slides in minimal medium supplemented with 0.2% yeast extract with and without the addition of 10 μg/ml tunicamycin. Images were taken using an Olympus BX51 microscope equipped with an Olympus DP11 digital camera.
RESULTS
The A. niger ΔhacA strain.
To assess the role of the hacA gene and the control mechanisms in the UPR of A. niger, we aimed to obtain a corresponding knockout strain. Recently, A. tumefaciens-mediated transformation was shown to be an efficient tool for gene replacement in Aspergillus awamori (18). Using this transformation procedure, the hacA gene of A. niger was successfully replaced with the hygromycin resistance cassette. The construct consisted of the hygromycin cassette from pAN7-1 flanked by 2 kb of the hacA 5′ upstream and 1.5 kb of the 3′ downstream regions, respectively.
Two types of colonies were present on the transformation plates. The majority of the colonies had a regular wild-type phenotype, whereas approximately 30% consisted of small nonsporulating yellow colonies. Southern analyses performed on both types of colonies showed the absence of the hacA gene in the aberrant colonies, whereas the construct was ectopically integrated in transformants having a wild-type phenotype.
Thus, the deletion of hacA results in a distinct phenotype. Growth is severely impaired, and sporulation is almost absent compared to the wild-type strain (Fig. 1A). On minimal medium plates as well as on plates with rich medium, such as complete medium (CM) and potato dextrose agar, the ΔhacA strain formed small compact colonies with only few conidia. The growth differences observed were analyzed further microscopically. Under normal growth conditions, parts of the hyphae of the ΔhacA strain are swollen compared to the wild type, and often, these swollen hyphae are branched at the tips, forming finger-like extensions as shown in Fig. 4B. In the presence of tunicamycin, growth of the ΔhacA strain was nearly abolished. The hyphae are shorter than those in the wild-type strain and have swollen irregular shapes.
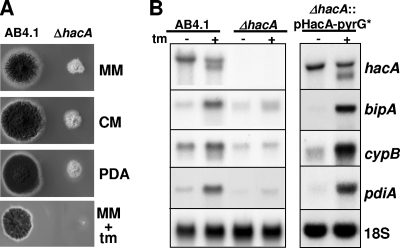
FIG. 1.
Phenotypic and transcriptional analysis of the A. niger ΔhacA strain. (A) Growth of the ΔhacA strain compared to that of wild-type strain AB4.1 on different agar media. MM, minimal medium; CM, MM supplemented with 0.5% yeast extract; PDA, potato dextrose agar; tm, 10 μg/ml tunicamycin. (B) Northern blot analysis showing the lack of a functional UPR in the ΔhacA strain and the restoration of the response by transforming the ΔhacA strain with plasmid pHacA-pyrG*.
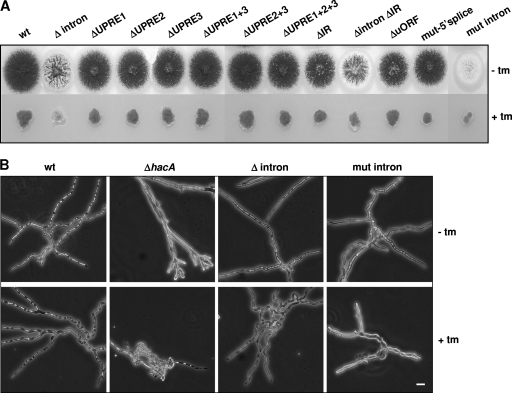
FIG. 4.
Phenotypes of the strains used in this study. (A) Growth of wild-type (wt) and mutant strains on CM plates in the absence (top row) or in the presence (bottom row) of 10 μg/ml tunicamycin (tm). (B) Microscope images of the wild-type strain, the hacA deletion strain (ΔhacA), the strain bearing the intronless hacA gene (Δintron), and the strain bearing the unspliceable hacA variant (mut-intron).
Thus, the application of ER stress in the form of 10 μg/ml tunicamycin severely affected the growth of the ΔhacA strain, indicating that a functional UPR is necessary for coping with ER stress.
Northern analysis showed the inability of the ΔhacA strain to activate the UPR (Fig. 1B). In a wild-type strain, the upregulation of the chaperone bipA and the foldases cypB and pdiA, due to ER stress, coincides with the processing of hacA mRNA. In the ΔhacA strain, however, ER stress does not trigger a comparable response. The basal transcript levels of bipA, cypB, and pdiA are low, and in response to tunicamycin treatment, only a slight, if any, upregulation was observed. In contrast, the retransformation of the ΔhacA strain with the wild-type hacA gene fully restored the transcriptional pattern of regulation typical for a wild-type strain.
The ΔhacA strain was used further as a background for studying the hacA gene expression mechanism.
Role of the small ORF and the putative 5′ splice site.
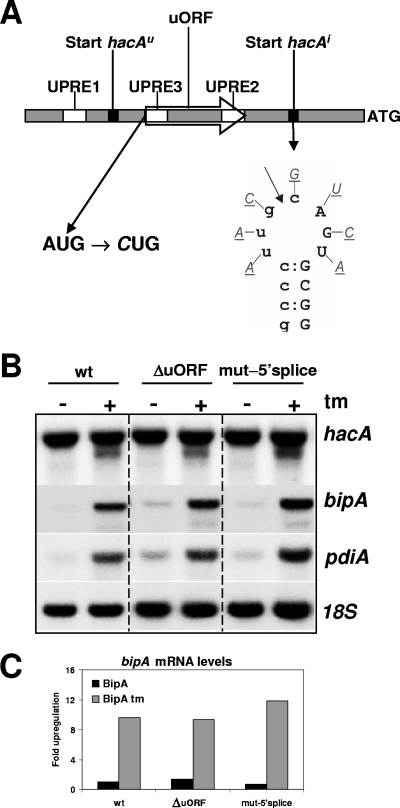
The 5′UTR of the full-length transcript of hacA contains a small ORF (uORF) of 44 codons (Fig. 2A), which is not present in the truncated hacA mRNA. We previously hypothesized that this region could be involved in a translational control mechanism of hacAu mRNA, since translational control is often associated with short ORFs, present in the 5′UTR of mRNAs (17). The S. cerevisiae transcription factor Gcn4p and the mammalian transcription factor ATF4 are translationally controlled by small uORFs (7, 10). To test this hypothesis, the ATG start codon of the uORF was mutated to CTG, and the mutated hacA construct was introduced into the ΔhacA strain. However, northern analyses of the ΔuORF strain and subsequent signal quantification did not show clear effects of the mutation on the transcription levels of the foldases and/or the truncation of hacA mRNA upon the application of ER stress compared to the wild type (Fig. 2B and C). Note that a second ATG is present in the reading frame of the 44-codon uORF of hacA. This ATG, however, is located 7 nt upstream of the central spacer nucleotide of UPRE2 (Table 1) and was therefore not mutated. As a result, an 8-codon-long uORF remains intact in the 5′UTR of the hacAu mRNA of this strain. Interestingly, this uORF shows remarkable sequence identity with two small uORFs present in the hacA transcripts of A. nidulans and A. fumigatus. The translated sequences of these uORFs of A. niger, A. nidulans, and A. fumigatus are MTRWPVFL, MTPWPLS, and MPTWLFL, respectively. Although this conservation could be the result solely of the fact that all three uORFs overlap with the UPRE2 sequence in each promoter, a function of the uORFs in translational attenuation cannot be excluded.
FIG. 2.
Mutational analysis of the uORF and the putative 5′ splice site, two elements present in the 5′UTR of hacAu mRNA. (A) Schematic overview of the 5′UTR of the hacA gene. ATG indicates the start codon of hacA, UPREs are indicated by open boxes, the starts of hacAu and hacAi mRNAs are indicated by black boxes, and the 44-codon uORF is indicated by an open arrow. The mutations introduced to disrupt the uORF and the putative 5′ splice site are indicated below. The putative IreA cutting site is indicated by an arrow. (B) Northern blot analysis showing the effect of ER stress imposed by tunicamycin (tm) on the transcription of hacA, bipA, and pdiA. 18S rRNA was used as a loading control. (C) Relative bipA levels in the wild type (wt), the ΔuORF strain, and the mut-5′ splice-site strain in the absence (black bars) and presence (gray bars) of 20 μg/ml tunicamycin.
Previous inspection of the sequence around the start site of hacAi mRNA revealed a strong sequence similarity with the intron borders of the unconventional intron (20). If this would lead to a similar secondary structure, as is the case for the unconventional intron borders, this structure could be a potential target for IreA and explain the mechanism behind the truncation of hacA. In contrast to the splicing of the unconventional intron, which involves an IreA-mediated cut in each of the two loops followed by the religation of the two exons by tRNA ligase (32), the hypothetical IreA-mediated 5′ truncation of hacA mRNA would involve only a cut at the recognition site. Although no RNA secondary structure incorporating the putative 5′ splice site in a stable stem-loop structure could be predicted, mutational analysis was performed to test whether or not this site would be a substrate for IreA. The seven nonpaired nucleotides in the putative loop were mutated from purines to pyrimidines, or vice versa, in order to destroy a putative recognition site for IreA (Fig. 2A). If IreA indeed would be involved, this strategy should lead to the inability to form the truncated hacA messenger. However, Northern analysis of a strain bearing such mutations did not show any effect of the mutations in the putative 5′ splice site on the processing of the hacA mRNA (Fig. 2B). Three hours after tunicamycin-induced stress, the truncated hacA mRNA was present at levels comparable to those of the wild-type strain. The upregulation of pdiA and bipA mRNA levels upon stress was also similar to the wild-type situation.
Role of UPREs in the hacA promoter.
Previously, we identified three UPREs in the hacA promoter by EMSA. The presence and conservation of these three boxes in the hacA promoters of A. niger, A. nidulans, and A. fumigatus suggest a function for all of them (19). Mutational analysis was carried out to determine the contribution of each of these elements to the transcriptional regulation of hacA. Mutations were introduced into the core sequences of the hacA UPREs. The loss of binding due to the mutations was confirmed in vitro by EMSAs with the HacA protein and 34-bp double-stranded oligonucleotides comprising the different mutated UPREs. Whereas two mutations (Fig. 3B) were sufficient to completely abolish binding of HacA to UPRE2 and UPRE3, the corresponding two mutations introduced into UPRE1 did not lead to a complete loss of binding. As a result, two additional mutations were introduced into UPRE1.
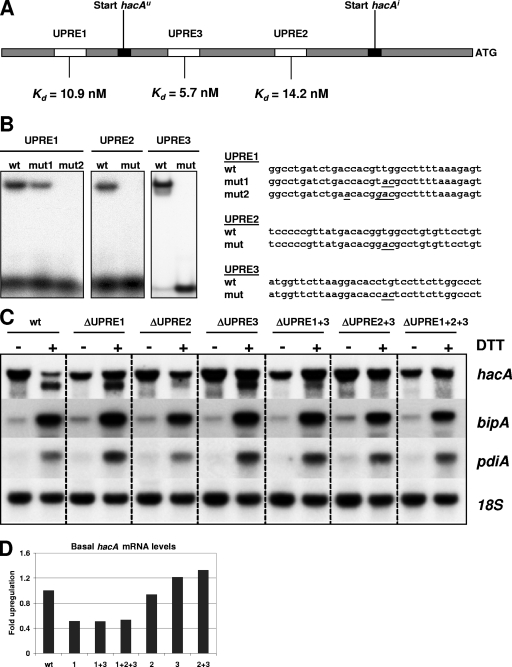
FIG. 3.
Role of the UPREs in regulation of hacA. (A) Schematic overview of the promoter region of hacA. The transcription start points of hacAu and hacAi are indicated by black boxes. The UPREs are represented by open boxes. The binding constant of each UPRE is shown at the bottom. (B) EMSAs showing the inability of HacA to bind to the mutated UPREs. The sequences of the 34-bp oligonucleotides representing the UPREs are shown, and mutations introduced into each UPRE to abolish binding by HacA are indicated in italics and are underlined. wt, wild type. (C) Northern analysis showing the effect of mutated UPREs on transcription levels of hacA, bipA, and pdiA. ER stress was imposed by the addition of 20 mM DTT to the medium. (D) Relative hacA mRNA levels in the different UPRE mutants under nonstress conditions.
None of the strains carrying mutations in the hacA UPREs showed phenotypic growth changes on a plate test (Fig. 4A), and these results were confirmed by light microscopy (data not shown). However, Northern analyses performed on the strains revealed an altered transcriptional pattern for hacA and the foldases.
When UPRE2 was mutated separately, in combination with UPRE3, or in combination with UPRE1 and UPRE3, the truncation of hacA mRNA did not occur anymore in response to ER stress (Fig. 3C). Although only the full-length transcript was formed in the UPRE2 mutants 3 h after DTT treatment, bipA and pdiA transcripts were upregulated to the same extent as in the wild-type strain, indicating that the truncation of hacA is not a necessity for the induction of the pathway. In contrast, the mutation of UPRE1 or UPRE3 separately did not lead to apparent changes in the splicing of hacA mRNA under ER stress conditions. However, a mutation of UPRE1 did result in lower basal levels of hacA mRNA (Fig. 3D). All three strains carrying mutations in UPRE1 had approximately half of the hacA mRNA levels compared to those of the wild type and the other mutant strains. This is in line with the finding that hacA is subject to autoregulation (20). Another observation is, however, more puzzling. In the wild-type strain, the amount of the full-length hacA messenger had dropped significantly 3 h after the addition of DTT to the culture. This effect was also observed for the mutants ΔuORF, mut-5′splice, and ΔIR (mutated inverted repeat) (see below) upon DTT stress (data not shown). However, except for ΔUPRE2, this effect was not observed for the other UPRE mutants. On the contrary, full-length hacA levels were slightly induced by ER stress in strains with a mutation in either UPRE1, UPRE3, or both elements. The observation that UPRE1 seems to repress the transcription of full-length hacA mRNA when HacAi is abundantly present implies a dual function for UPRE1. Under normal conditions, when the concentration of HacAi is low, UPRE1 functions as an activator site, whereas it functions as a repressor site during DTT stress. The underlying mechanism of this possible dual function of UPRE1 is unclear and will be the subject of further investigation.
The transcription start sites of the three single UPRE mutants were determined by 5′RACE on RNA samples of noninduced and DTT-induced cultures to check whether mutations in the UPREs had an effect on the exact start site of hacAu and hacAi mRNA. As in the wild type, in all three mutants (UPRE1, UPRE2, and UPRE3), the full-length hacA started at 303 nt upstream of the ATG. In DTT-stressed cultures of the UPRE1 and UPRE3 mutants, the transcription start site of the truncated hacA mRNA was found 73 nt upstream of the ATG, as in the wild type.
Long-range base pairing and translational block.
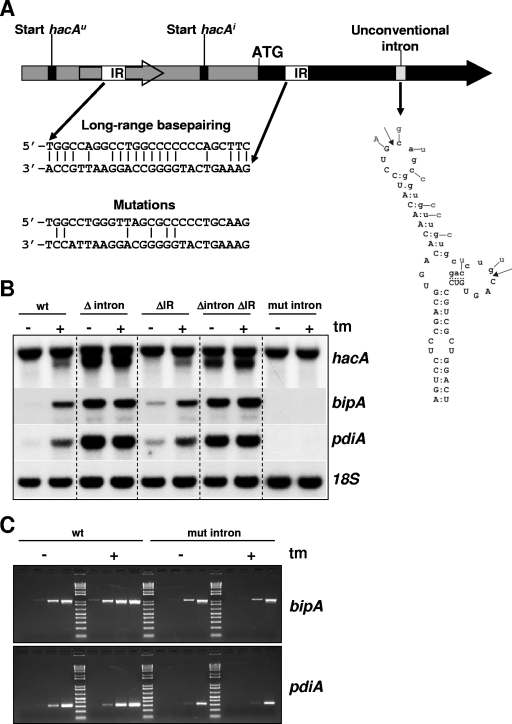
Long-range base pairing was shown to prevent the uninduced form of the yeast HAC1 homologue from being efficiently translated (28). Part of the 252-nt intron of yeast HAC1u mRNA forms a stable double-stranded structure by base pairing with the 5′UTR, thereby preventing the ribosomes from reading through. Unconventional intron splicing removes the 252-nt intron and with it the right half of the inverted repeat, thereby releasing HAC1u mRNA from its translation block.
An identical mechanism is not possible for A. niger hacAu mRNA. The unconventional intron of hacA is only 20 nt long, and a stable stem-loop structure is predicted for this sequence, which leaves no nucleotides available for a putative long-range base pairing with the 5′UTR. However, sequence analyses revealed a 26-nt-long inverted repeat in hacAu mRNA (Fig. 5A). This repeat contains 18 bp, 12 bp of which are GC pairs. In comparison, the yeast HAC1 inverted repeat stretches out over 19 nt and has 16 bp, 11 bp of which are GC pairs. The left half of the inverted repeat in A. niger hacA is located in the 44-codon uORF, and the right half is located in the hacA ORF close to the ATG. Truncation upon ER stress removes approximately 230 nt of the 5′UTR of hacA mRNA including the uORF and thereby the left half of the inverted repeat, possibly releasing hacA from a translational block analogous to the yeast hac1 mechanism.
FIG. 5.
Function of the unconventional intron and long-range base pairing in the regulation of the UPR. (A) Schematic overview indicating the unconventional intron and the location of the inverted repeats in relation to other elements present in hacA. The hacA ORF is indicated by a solid black arrow, and the 5′UTR is indicated by a gray box. The inverted repeats (IR) are depicted by open boxes, and the base pairing of the inverted repeats is shown at the bottom. The theoretically remaining base pairings after the introduction of silent mutations are also indicated. The structure of the unconventional intron is depicted by the stem-loop structure, and mutations introduced to prevent splicing by disrupting the structure are indicated. The intron sequence is shown in lowercase type, and cleavage sites are indicated by arrows. (B) Northern blot analysis showing the effect of the different mutations or deletions on the expression and truncation of hacA and on the transcript levels of bipA and pdiA. ER stress was imposed by treatment with tunicamycin (tm), and the gene coding for 18S rRNA was used as a control probe. (C) RT-PCR showing the transcript levels of bipA and pdiA in the wild-type (wt) and mut-intron strains. The four lanes under each condition correspond to 10, 15, 20, and 25 PCR cycles.
To determine the effect of the putative base pairing on the regulation of hacA, both half-sites of the inverted repeat were mutated. Since both half-sites are located in coding regions, only silent mutations were introduced to disrupt the putative base pairing (Fig. 5A). Although this strategy left 6 bp intact, it is likely that a stable base pairing of both half-sites is no longer possible. Analysis of the ΔIR strain by Northern blots showed that under normal growth conditions, approximately three-times-more bipA and pdiA transcripts were present than in the wild type (Fig. 5B), indicating higher basal levels of HacA in this strain. Under ER stress, the transcriptional pattern of hacA, bipA, and pdiA was similar to that of the wild-type strain.
The unconventional intron.
To study unconventional intron splicing, we aimed to construct an ΔireA strain. However, multiple attempts and techniques to construct such a strain all failed, presumably because of the lethality of the deletion. Therefore, the importance of the unconventional intron for the UPR was determined in the following two ways: by analysis of a strain carrying the hacA gene lacking the unconventional intron and by analysis of a strain having a hacA variant with an unspliceable unconventional intron. The removal of the unconventional intron from the hacA gene resulted in a constitutive UPR (Fig. 5B). Both the full-length and the truncated hacA transcripts were abundantly present under normal growth conditions, and the levels of bipA and pdiA were 19-fold and 13-fold higher, respectively, than levels measured in the wild-type strain under nonstress conditions.
The induction of ER stress by the addition of tunicamycin had no additional effect on the response in this strain, indicating that splicing of the unconventional intron is sufficient to fully induce the pathway. Northern blot analysis of a strain containing an unspliceable hacA variant (mut intron) revealed that the splicing of the unconventional intron and, as a result, HacAi is necessary to activate the downstream pathway (Fig. 5B). The truncation of the 5′-UTR of hacA mRNA in response to tunicamycin-induced stress and the accompanying upregulation of bipA and pdiA did not take place in this strain. bipA and pdiA transcripts could not even be detected on the Northern blot under both conditions. It was previously reported that the deletion of bipA is lethal for Aspergillus (27). Also, attempts to delete pdiA in A. niger were unsuccessful (21). Therefore, the apparent absence of bipA and pdiA transcripts on the Northern blot of the strain bearing the unspliceable hacA variant (mut-intron strain) is most likely a result of the limited exposure time of the X-ray film. RT-PCR performed on RNA from the mut-intron strain revealed that both bipA and pdiA transcripts were indeed present in this strain albeit at lower levels than those of the wild-type strain (Fig. 5C).
Remarkably, both the Δintron and mut-intron strains displayed clear phenotypes. When grown on agar plates, Δintron produced fewer conidia than did the wild-type strain, whereas sporulation was almost absent in strain mut-intron (Fig. 4A). Microscopic inspection of mut-intron grown in the presence of tunicamycin revealed a phenotype slightly comparable to that of the ΔhacA strain: poor growth and swollen hyphae (Fig. 4B).
Apparently, both HacAi and HacAu have physiological functions in A. niger, and the lack of either one results in the observed phenotype. The phenotype of the strain having the intronless variant of the hacA gene, and therefore capable of producing only the HacAi protein, is not merely a result of the permanent activation of the UPR. The overexpression of hacAi in a wild-type background, leading to a strain which constitutively produces the HacAi protein but is also capable of producing the HacAu protein from its endogenous hacA gene, did not lead to similar phenotypical changes (results not shown). This is in agreement with the previously reported observation that the amino acid sequence of the C-terminal parts of HacAu from different aspergilli are highly conserved (20). The precise role of HacAu in the fungus, however, is the subject of further investigation.
DISCUSSION
In eukaryotes, secretion stress leads to various responses, which aim at the homeostasis of ER functions. One of the secretion stress responses is the UPR, which is common to all eukaryotic cells. There are, however, differences in the molecular mechanisms underlying the UPR among different organisms. In this study, we describe some unique features of the A. niger UPR not reported for the S. cerevisiae and mammalian UPRs.
In both lower and higher eukaryotes, the activation of the UPR starts with the splicing of an unconventional intron from the hacA transcript, mediated by IreA. Splicing of the 20-nt intron from A. niger hacA mRNA is both necessary and sufficient to activate the UPR. A strain containing an unspliceable variant of hacA displayed poor growth and was unable to activate the pathway and upregulate the foldases in response to stress, whereas a strain containing an intronless variant of the hacA gene displayed a constitutive UPR (Fig. 5).
In addition to the rather drastic on/off switch of unconventional intron splicing, other regulatory elements are present in the promoter of the hacA gene and in the hacAu transcript, which provides the fungus additional regulatory control over the response.
Long-range base pairing in the hacA messenger prevents the efficient translation of the full-length transcript (Fig. 5). As opposed to the mechanism in S. cerevisiae, where pairing takes place between the 5′UTR and the unconventional intron, the 5′UTR of Aspergillus hacA mRNA forms a double-stranded structure with the coding region of the transcript, resulting in translational attenuation (Fig. 5). In the case of A. niger, the release of the translational block upon ER stress is exerted via a switch to an alternative transcription start located approximately 230 bp downstream of the usual transcription start site. This switch is dependent on UPRE2 and on HacA itself, thus confirming the autoregulation of hacA transcription, as demonstrated previously (20). UPRE2 is the HacA target most proximal to the ATG and located 81 bp upstream of the start site of hacAi mRNA. A mutation of UPRE2, either individually or in combination with UPRE1 and UPRE3, abolished the ability to synthesize the shorter transcript during ER stress (Fig. 3), indicating its importance for the transcriptional switch. The fact that the transcriptional switch depends on HacA itself was shown by two experiments. First, a strain containing the hacA gene without the unconventional intron, and therefore capable of producing only the active form of the HacA protein, produced both full-length and truncated hacA mRNAs under normal growth conditions (Fig. 5). Second, a strain carrying an unspliceable version of the hacA gene (cf. the mut-intron strain) (Fig. 5) and therefore unable to produce the active form of the HacA protein was unable to synthesize the truncated messenger in response to stress.
Whereas a clear function can be assigned to UPRE2, the roles of UPRE1 and UPRE3 in the regulation of hacA are somewhat more puzzling. On the whole, a mutation of UPRE1 and UPRE3 individually or together left the response and the accompanying truncation of hacA intact. However, these sites could provide an additional fine-tuning mechanism for hacA transcription. When UPRE1 was inactivated alone or in combination with other sites (ΔUPRE1, ΔUPRE1 plus ΔUPRE3, and ΔUPRE1 plus ΔUPRE2 plus ΔUPRE3), the basal level of hacA transcription dropped to approximately 50% compared to wild-type levels (Fig. 3D). In contrast, ER stress led to a slight upregulation of the full-length hacA transcript in those same mutants, with the most pronounced effect in the double mutant ΔUPRE1 plus ΔUPRE3 (Fig. 3C). Since UPRE1 is most distal to the ATG of the hacA gene and located 50 bp upstream of the transcription start point of hacAu (19), it seems plausible that UPRE1 affects the basal level of the full-length hacA messenger. The basal level of hacA transcription is, however, not dependent solely on HacA. A strain that was unable to synthesize HacAi (cf. mut intron) (Fig. 5) displayed hacA mRNA levels comparable to those of the wild-type strain under normal growth conditions. Most likely, another, as-yet-unknown factor plays an important role in the regulation of hacA. In mammalian cells, the hacA homologue XBP1 is upregulated by ATF6 in response to ER stress (15), and in S. cerevisiae, certain conditions can boost the accumulation of HAC1 mRNA via an Ire1p-independent pathway (14). A possible candidate involved in an additional regulation of A. niger hacA could be CpcA. CpcA is a homologue of S. cerevisiae Gcn4p, which acts as the central transcription factor in response to amino acid starvation (36). In addition, S. cerevisiae Gcn4p plays an essential role in the UPR and is responsible for the induction of a major part of the UPR targets during ER stress (26). Its mouse homologue, ATF4, has also been shown to be involved in secretion stress (7). The T. reesei and A. nidulans homologues cpc1 and cpcA, respectively, have also been linked to the UPR, as they have been shown to be upregulated in response to ER stress (2). The involvement of such a factor could explain the slight upregulation of the full-length hacA transcript observed upon stress in the UPRE1 mutant strains. The role of CpcA in the UPR and its effect, if any, on hacA transcriptional regulation, perhaps via an interaction with the UPRE1 target, and the controversial role of UPRE1 as an activator and a repressor site of hacA transcription remain to be established.
Whereas UPRE2 mediates the transcriptional activation of hacA upon stress, an opposite function could be attributed to UPRE3. Mutation analysis of UPRE3 suggests that this site could act as an HacA scavenger in order to prevent the transcriptional switch from being activated at low stress levels, at which full activation of the pathway is not necessary. The disruption of UPRE3 resulted in higher levels of hacA under both normal and ER stress conditions. Of the three UPREs present in the hacA promoter, UPRE3 possesses the highest affinity for HacA and stands apart from the other UPREs because of the missing spacer nucleotide between the two palindromic half-sites (19). Using polysomal fractionation, it has been shown that under nonstress conditions, the splicing of hacA still occurs but at a low level (5). The association of the spliced transcript with the polysomal fraction indicates that low levels of the HacAi protein will be synthesized under normal growth conditions, which is in agreement with our previous results from UPRE-promoter reporter constructs (19). UPRE3 could therefore have the function of a “threshold” switch; only when sufficient HacAi accumulates in the nucleus will the transcription of hacA shift to the new transcription start site, possibly initiated via the recognition of UPRE2. Although further analysis is needed, we suggest a mechanism in which UPRE3 intercepts HacAi when the protein comes into the vicinity of the promoter, thereby preventing the activation of transcription via UPRE1 or UPRE2. Only when the HacAi protein concentration reaches a certain threshold is the pathway further activated via the transcriptional switch to a new start of transcription followed by the consequent release from a translational attenuation checkpoint.
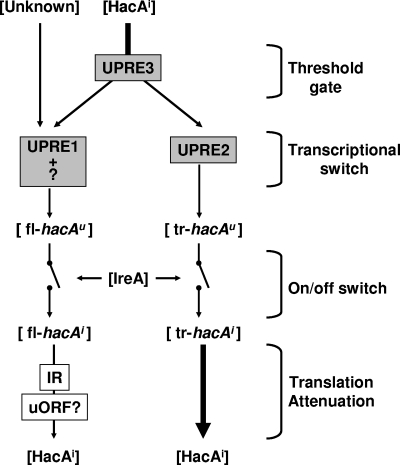
We unraveled several mechanisms behind the A. niger UPR, which offers the fungus controllability over and fine-tuning of the pathway. At the transcriptional level, activation occurs in two steps: intron splicing followed by a transcriptional switch to a new start site. At the translational level, long-range base pairing inhibits the efficient translation of the nontruncated hacA transcript. In addition to these mechanisms, our results indicate additional regulatory control of hacA transcription mediated via UPRE1 and HacA and most likely another transcription factor, possibly CpcA. UPRE3, finally, could function as a build-in safety mechanism to prevent unnecessary activation. The confirmation of the latter mechanisms and the integration into the regulation circuitry remains subject to further investigation. Figure 6 shows a schematic overview of our current working model. It implies that under normal growth conditions, low concentrations of active HacAi will be present in the cell as a result of the limited intron splicing that constitutively occurs in the fungal cell. UPRE3 acts as a threshold gate, keeping HacAi away from UPRE2 until the HacAi concentration reaches a certain level. The transcription of hacA at this stage is initiated mainly via UPRE1 and/or an additional as-yet-unknown promoter element and is mediated by HacAi and an unknown protein (possibly CpcA). The result is the transcription of full-length hacAu mRNA. IreA functions as the controller of an on/off switch to activate the UPR by splicing the hacAu mRNA. The resulting full-length hacAi transcript is, however, subject to translational attenuation caused by long-range base pairing and possibly the uORF. As a result, the production of the “active” HacAi protein is slow at this stage. Only during prolonged stress will enough HacAi accumulate to overcome the threshold value set by UPRE3 and will the transcriptional switch be activated. The transcriptional switch is then operated by HacAi, which will bind to the UPRE2 and initiate its own transcription from a new start site. This will result in the truncated hacAu transcript. This messenger lacks the translational block and can be efficiently translated. However, it still contains the unconventional intron, and only after activation by IreA will it contribute to the active pool of the HacAi protein and only then will the hacA-dependent part of the UPR be at its full strength.
FIG. 6.
Schematic overview of hacA regulation. The model depicts proteins and mRNAs between brackets indicating that their (active) concentration can vary. Elements present in the hacA promoter are in gray boxes, and elements present in the hacA mRNA are in white boxes. The splicing of full-length hacA (fl-hacAu) and truncated hacA (tr-hacAu) by IreA is depicted by the on/off switches. Translational attenuation is indicated by the thin arrow, compared to the arrow indicating nonattenuated translation. IR, inverted repeat.
As a saprophytic organism, A. niger secretes large amounts of enzymes into its environment to degrade complex polymeric substrates used as natural nutrients for the fungus. This characteristic might also have set special demands on the secretion machinery of the organism and on the complexity of the regulation of the UPR in comparison with, for example, S. cerevisiae. The activation of the UPR affects a large set of genes (2, 5, 33), and unnecessary or unnecessarily high-level activation of the UPR will be costly in terms of energy. Therefore, the presence of several subsequent checkpoints allow the fungal cell to optimally tune its secretion machinery to the environment.
Acknowledgments
This work was part of the Eurofung Program and was supported by EC grant no. QLRK3-00729.
Footnotes
Published ahead of print on 30 January 2009.
REFERENCES
- 1.Al Sheikh, H., A. J. Watson, G. A. Lacey, P. J. Punt, D. A. MacKenzie, D. J. Jeenes, T. Pakula, M. Penttila, M. J. Alcocer, and D. B. Archer. 2004. Endoplasmic reticulum stress leads to the selective transcriptional downregulation of the glucoamylase gene in Aspergillus niger. Mol. Microbiol. 531731-1742. [DOI] [PubMed] [Google Scholar]
- 2.Arvas, M., T. Pakula, K. Lanthaler, M. Saloheimo, M. Valkonen, T. Suortti, G. Robson, and M. Penttila. 2006. Common features and interesting differences in transcriptional responses to secretion stress in the fungi Trichoderma reesei and Saccharomyces cerevisiae. BMC Genomics 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Groot, M. J., P. Bundock, P. J. Hooykaas, and A. G. Beijersbergen. 1998. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16839-842. [DOI] [PubMed] [Google Scholar]
- 4.Gething, M. J., and J. Sambrook. 1992. Protein folding in the cell. Nature 35533-45. [DOI] [PubMed] [Google Scholar]
- 5.Guillemette, T., N. N. van Peij, T. Goosen, K. Lanthaler, G. D. Robson, C. A. van den Hondel, H. Stam, and D. B. Archer. 2007. Genomic analysis of the secretion stress response in the enzyme-producing cell factory Aspergillus niger. BMC Genomics 8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harding, H. P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18575-599. [DOI] [PubMed] [Google Scholar]
- 7.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 61099-1108. [DOI] [PubMed] [Google Scholar]
- 8.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397271-274. [DOI] [PubMed] [Google Scholar]
- 9.Haze, K., H. Yoshida, H. Yanagi, T. Yura, and K. Mori. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 103787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinnebusch, A. G. 1997. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J. Biol. Chem. 27221661-21664. [DOI] [PubMed] [Google Scholar]
- 11.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 131211-1233. [DOI] [PubMed] [Google Scholar]
- 12.Kawahara, T., H. Yanagi, T. Yura, and K. Mori. 1997. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell 81845-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kusters-van Someren, M. A., J. A. Harmsen, H. C. Kester, and J. Visser. 1991. Structure of the Aspergillus niger pelA gene and its expression in Aspergillus niger and Aspergillus nidulans. Curr. Genet. 20293-299. [DOI] [PubMed] [Google Scholar]
- 14.Leber, J. H., S. Bernales, and P. Walter. 2004. IRE1-independent gain control of the unfolded protein response. PLoS Biol. 2E235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, K., W. Tirasophon, X. Shen, M. Michalak, R. Prywes, T. Okada, H. Yoshida, K. Mori, and R. J. Kaufman. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16452-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez, I. M., and M. J. Chrispeels. 2003. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15561-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meijer, H. A., and A. A. Thomas. 2002. Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem. J. 3671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michielse, C. B., M. Arentshorst, A. F. Ram, and C. A. van den Hondel. 2005. Agrobacterium-mediated transformation leads to improved gene replacement efficiency in Aspergillus awamori. Fungal Genet. Biol. 429-19. [DOI] [PubMed] [Google Scholar]
- 19.Mulder, H. J., I. Nikolaev, and S. M. Madrid. 2006. HACA, the transcriptional activator of the unfolded protein response (UPR) in Aspergillus niger, binds to partly palindromic UPR elements of the consensus sequence 5′-CAN(G/A)NTGT/GCCT-3′. Fungal Genet. Biol. 43560-572. [DOI] [PubMed] [Google Scholar]
- 20.Mulder, H. J., M. Saloheimo, M. Penttila, and S. M. Madrid. 2004. The transcription factor HACA mediates the unfolded protein response in Aspergillus niger, and up-regulates its own transcription. Mol. Genet. Genomics 271130-140. [DOI] [PubMed] [Google Scholar]
- 21.Ngiam, C., D. J. Jeenes, P. J. Punt, C. A. van den Hondel, and D. B. Archer. 2000. Characterization of a foldase, protein disulfide isomerase A, in the protein secretory pathway of Aspergillus niger. Appl. Environ. Microbiol. 66775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Offringa, R., M. J. de Groot, H. J. Haagsman, M. P. Does, P. J. van den Elzen, and P. J. Hooykaas. 1990. Extrachromosomal homologous recombination and gene targeting in plant cells after Agrobacterium mediated transformation. EMBO J. 93077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogawa, N., and K. Mori. 2004. Autoregulation of the HAC1 gene is required for sustained activation of the yeast unfolded protein response. Genes Cells 995-104. [DOI] [PubMed] [Google Scholar]
- 24.Pakula, T. M., M. Laxell, A. Huuskonen, J. Uusitalo, M. Saloheimo, and M. Penttila. 2003. The effects of drugs inhibiting protein secretion in the filamentous fungus Trichoderma reesei. Evidence for down-regulation of genes that encode secreted proteins in the stressed cells. J. Biol. Chem. 27845011-45020. [DOI] [PubMed] [Google Scholar]
- 25.Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13349-355. [DOI] [PubMed] [Google Scholar]
- 26.Patil, C. K., H. Li, and P. Walter. 2004. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2E246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Punt, P. J., I. A. van Gemeren, J. Drint-Kuijvenhoven, J. G. Hessing, G. M. Muijlwijk-Harteveld, A. Beijersbergen, C. T. Verrips, and C. A. van den Hondel. 1998. Analysis of the role of the gene bipA, encoding the major endoplasmic reticulum chaperone protein in the secretion of homologous and heterologous proteins in black Aspergilli. Appl. Microbiol. Biotechnol. 50447-454. [DOI] [PubMed] [Google Scholar]
- 28.Ruegsegger, U., J. H. Leber, and P. Walter. 2001. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 107103-114. [DOI] [PubMed] [Google Scholar]
- 29.Saloheimo, M., M. Valkonen, and M. Penttila. 2003. Activation mechanisms of the HACI-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 471149-1161. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Sidrauski, C., R. Chapman, and P. Walter. 1998. The unfolded protein response: an intracellular signalling pathway with many surprising features. Trends Cell Biol. 8245-249. [DOI] [PubMed] [Google Scholar]
- 32.Sidrauski, C., J. S. Cox, and P. Walter. 1996. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87405-413. [DOI] [PubMed] [Google Scholar]
- 33.Sims, A. H., M. E. Gent, K. Lanthaler, N. S. Dunn-Coleman, S. G. Oliver, and G. D. Robson. 2005. Transcriptome analysis of recombinant protein secretion by Aspergillus nidulans and the unfolded-protein response in vivo. Appl. Environ. Microbiol. 712737-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101249-258. [DOI] [PubMed] [Google Scholar]
- 35.Vishniac, W., and M. Santer. 1957. The thiobacilli. Bacteriol. Rev. 21195-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wanke, C., S. Eckert, G. Albrecht, W. van Hartingsveldt, P. J. Punt, C. A. van den Hondel, and G. H. Braus. 1997. The Aspergillus niger GCN4 homologue, cpcA, is transcriptionally regulated and encodes an unusual leucine zipper. Mol. Microbiol. 2323-33. [DOI] [PubMed] [Google Scholar]
- 37.Welihinda, A. A., W. Tirasophon, and R. J. Kaufman. 1999. The cellular response to protein misfolding in the endoplasmic reticulum. Gene Expr. 7293-300. [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107881-891. [DOI] [PubMed] [Google Scholar]