Abstract
Each time Saccharomyces cerevisiae cells divide they ensure that both the mother and daughter cell inherit a vacuole by actively transporting a portion of the vacuole into the bud. As the mother cell begins budding, a tubular and vesicular segregation structure forms that is transported into the bud by the myosin V motor Myo2, which is bound to the vacuole-specific myosin receptor, Vac17 (41, 59, 70, 79). Upon arriving in the bud the segregation structure is resolved to found the daughter vacuole. The mechanism that regulates segregation structure resolution in a spatially dependent manner is unknown. In addition to resolving the segregation structure, Vac17 is degraded specifically in the bud to provide directionality to vacuole inheritance. It has been proposed that bud-specific degradation of Vac17 is promoted by proteins localized to or activated solely in the bud (77). The p21-activated kinases (PAKs) Cla4 and Ste20 are localized to and activated in the bud. Here we report that Cla4 is localized to the segregation structure just prior to segregation structure resolution, and cells lacking PAK function fail to resolve the segregation structure. Overexpression of either Cla4 or Ste20 inhibited vacuole inheritance and this inhibition was suppressed by the expression of nondegradable VAC17. Finally, PAK activity was required for Vac17 degradation in late M phase and CLA4 overexpression promoted Vac17 degradation. We propose that Cla4 and Ste20 are bud-specific proteins that play roles in both segregation structure resolution and the degradation of Vac17.
Each time a eukaryotic cell divides, it must ensure that both of the progeny cells contain a full complement of organelles. While high-copy-number organelles dispersed equally throughout the cell can rely primarily on a stochastic partitioning strategy, low-copy-number organelles localized to a specific region of the cell require ordered partitioning to ensure equal inheritance of organelles to each of the progeny (76). To better understand how ordered inheritance of low-copy-number organelles is regulated, we are studying organelle inheritance in the budding yeast Saccharomyces cerevisiae, focusing specifically on vacuole inheritance.
The budding yeast vacuole is analogous to the mammalian lysosome and serves as a site of macromolecular degradation. The vacuole is the primary storage site for amino acids and phosphates, is involved in the endocytic pathway, and serves as the site for both pH and osmoregulation (46). Under normal physiological conditions the yeast vacuole is a low-copy-number organelle containing on average one to three lobes and requires ordered transport to ensure its inheritance by the daughter cell (65).
At the time of bud emergence, proteins necessary for PtdIns(3,5)P2 production are required to form the tubular and vesicular structure called the segregation structure (7, 8). Transport of the segregation structure into the bud requires attachment of the vacuole to the myosin V motor, Myo2 (12, 13, 36). Attachment between the vacuole and myosin requires the vacuole-specific Myo2 receptor Vac17, which binds both Myo2 and Vac8 directly (41, 70). Vac8 is a vacuole membrane protein and is inserted into the vacuole membrane by its myristoylation and palmitoylation (56, 75). Once attached to myosin, vacuoles, like many other organelles in budding yeast, are transported into the bud along polarized actin cables (3, 6, 10, 27, 53, 78). Upon entering the bud, the segregation structure is resolved, probably by fusion of the tubules and vesicles, to found the daughter vacuole (17, 32, 33, 47, 86).
Directionality in vacuole transport is achieved by spatial degradation of Vac17 in a bud-specific manner. The vacuole inheritance mutants vac8 and myo2-2 (a point mutant within Myo2 that specifically disrupts the Myo2-Vac17 interaction) fail to transport Vac17 into the bud, and Vac17 accumulates to high levels. Bud-specific degradation of Vac17 requires a PEST sequence within Vac17. Cells expressing VAC17 without the PEST sequence (vac17ΔPEST) accumulate Vac17 to high levels and the vacuole is transported back to the mother bud neck, where the actin cytoskeleton is polarized, prior to cytokinesis (41, 70).
Vac17 degradation is regulated in a spatially dependent manner. It has been proposed that the spatial degradation of Vac17 solely in the bud is promoted by factors specifically localized to and/or activated in the bud (77). However, these factors have not yet been identified. The fusion of tubules and vesicles leading to the formation of a separate daughter vacuole does not take place until after entering the bud. As proposed for Vac17 degradation, there may be factors in the bud that promote vacuole fusion to help resolve the segregation structure only after entering the bud.
Budding yeast contain two main p21-activated kinases (PAKs), Cla4 and Ste20. Regions in the N-terminal halves of the PAKs interact with the kinase domain in the C-terminal half of the PAKs, leading to autoinhibition (4, 5, 48). Autoinhibition is relieved when PAKs bind to activated GTP-bound Cdc42 through the p21 binding domain (PBD) located within the N-terminal half of the protein (5, 18, 69). The adaptor protein Bem1 facilitates the activation of Cdc42 in the bud by binding both Cdc42 and its guanine nucleotide exchange factor (GEF), Cdc24 (43, 50, 58, 87). Bem1 contains two SH3 domains, and the second SH3 domain interacts with proline-rich domains in Ste20 and probably Cla4 (9, 31, 82). Cla4 and Ste20 are localized to and activated in the bud (39, 40, 48, 57). In this report we provide evidence that Cla4 and Ste20 regulate vacuole inheritance. PAK function is required to resolve the segregation structure and for Vac17 degradation prior to cytokinesis.
MATERIALS AND METHODS
Plasmid and strain construction.
Yeast strains and sources are listed in Table 1. The yeast strains used in this study are all derivatives of W303 except for the CLA4-GFP strains (S288c derivatives). CLA4-GFP strains were created by C-terminal tagging of CLA4 as described previously (52) and obtained from other sources, and they all showed the same localization pattern (39, 40). Yeast strains were constructed by genetic crosses followed by tetrad dissection or by transformation using the lithium acetate method (11). ProA and the adjacent HIS3 and URA3 markers were amplified by PCR and integrated to generate VAC17-ProA as described previously (1). A GAL-cla4-K594R strain was made by site-directed mutagenesis of pBB135 (4) as described previously (89). Plasmid sources and construction methods are listed in Table 2 and DNA manipulations were performed as described previously (64).
TABLE 1.
Strains
| Strain | Genotype | Source |
|---|---|---|
| Piatti2711 | MATaura3::4X URA3::GAL1-CLA4t swe1::LEU2 (W303) | 15 |
| KN3591 | MATacla4::LEU2 (W303) | 18 |
| KN3621 | MATaste20::URA3 (W303) | 18 |
| KN4580 | MATabar1::hisG cla4::LEU2 YCp-TRP1-cla4-75 ste20::URA3 | 18 |
| W303-1A | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 (W303) | 71 |
| YCB91 | MATahis3::GAL-cla4-K594R-HIS3 (W303) | This study |
| YCB94 | MATaste20::KanMX6-pGAL-STE20-GFP-TRP1(k.l) (W303) | This study |
| YCH1028 | MATacdc15-2 CLA4-GFP-HIS3 (S288c) | This study |
| YCH1665 | MATavac8::kanMX4 CLA4-GFP-HIS3 (S288c) | This study |
| YCH1752 | MATamyo2-2 CLA4-GFP-HIS3 (S288c) | This study |
| YCH3250 | MATaCLA4-GFP-HIS3 (S288c) | This study |
| YCH4737 | MATalte1::KanMX6 (W303) | This study |
| YCH4774 | MATaVAC17-ProA-HIS3-URA3 (W303) | This study |
| YCH4811 | MATapRS316 (W303) | This study |
| YCH4837 | MATapRS314-GAL-GST-STE20 (W303) | This study |
| YCH4843 | MATapRS314-GAL-GST-STE20 VAC17-ProA-HIS3-URA3 (W303) | This study |
| YCH4852 | MATacdc14-1 VAC17-ProA-HIS3-URA3 (W303) | This study |
| YCH4862 | MATacdc15-2 VAC17-ProA-HIS3-URA3 (W303) | This study |
| YCH4869 | MATadbf2-2 VAC17-ProA-HIS3-URA3 (W303) | This study |
| YCH4882 | MATahis3::GAL-CLA4-HIS3 VAC17-ProA-HIS3-URA3 (W303) | This study |
| YCH4893 | MATaura3::4X URA3::GAL1-CLA4t swe1::LEU2 VAC17-ProA-HIS3-URA3 (W303) | This study |
| YCH4894 | MATaura3::4X URA3::GAL1-CLA4t VAC17-ProA-HIS3-URA3 (W303) | This study |
| YCH4934 | His3::GAL-CLA4-HIS3 lte1::KanMX6 VAC17-ProA-HIS3-URA3 (W303) | This study |
| YCH4974 | MATahis3::GAL-CLA4-HIS3 vac17::kanMX4 pRS414-pVAC17-VAC17 (W303) | This study |
| YCH4975 | MATahis3::GAL-CLA4-HIS3 vac17::kanxMX4 pRS414-pVAC17-vac17ΔPEST (W303) | This study |
| YCH4981 | MATapRS316-GAL-GST-STE20 (W303) | This study |
| YCH4982 | MATapRS316-GAL-GST-ste20-K649R (W303) | This study |
| YCH4992 | MATapRS316-GST-STE20 vac17::kanMX4 pRS414-pVAC17-VAC17 (W303) | This study |
| YCH4993 | MATapRS316-GST-STE20 vac17::kanMX4 pRS414-pVAC17-vac17ΔPEST (W303) | This study |
| YCH5162 | MATaPDS1-HA-LEU2::leu2 VAC17-ProA-HIS3 (W303) | This study |
| YCH5286 | bub2::HIS3MX6 ura3-52::GAL-BUB2-URA3 lte1::KanMX6 ste20::TRP1 VAC17-ProA-HIS3-URA3 (W303) | This study |
| YCH5295 | ura3::GAL-GFP-CLA4-URA3 (W303) | This study |
| YMG694 | MATabar1-1 his3::GAL-CLA4-HIS3 (W303) | 31 |
TABLE 2.
Plasmids
| Plasmid | Description | Source |
|---|---|---|
| pCH1915 | pRS414-pVAC17-VAC17; made by insertion of 1.94-kb BamHI-SalI fragment of pVAC17 (70) into the BamHI-SalI site of pRS414 (68) | This study |
| pCH1916 | pRS414-pVAC17-vac17ΔPEST; made by insertion of 1.75-kb BamHI-SalI fragment of pVAC17(Δ97-259) (70) into the BamHI-SalI site of pRS414 (68) | This study |
| pCH1924 | pRS313-pVAC17-VAC17; made by insertion of 1.94-kb BamHI-SalI fragment of pVAC17 (70) into the BamHI-SalI site of pRS313 (68) | This study |
| pCH1925 | pRS313-pVAC17-vac17ΔPEST; made by insertion of 1.75-kb BamHI-SalI fragment of pVAC17(Δ97-259) (70) into the BamHI-SalI site of pRS414 (68) | This study |
| pCH1926 | pRS314-GAL-GST-STE20; made by insertion of a 4.53-kb SacII-KpnI fragment of pRD20-STE20-ATG (57) into the SacII-KpnI site of pRS314 (68) | This study |
| pRD20-STE20-ATG | pRS316-GST-STE20 | 57 |
| pRD20-STE20-K649R | pRS316-GST-ste20-K649R | 57 |
| pRS316 | pRS316 | 68 |
Microscopy.
Images were acquired using two microscopes. All images, with the exception of those for Fig. 1A and 2, below, were taken using a microscope (BX60; Olympus) with a UPlanApo 100×, numerical aperture (NA) 1.30 oil immersion objective (Olympus) and a DAGE ISIT-68 camera and using NIH image 1.62 (Wayne Rasband). Images for Fig. 1A and 2 were acquired using a microscope (BX50; Olympus) with a UPlanF1 100×, NA 1.30 oil immersion objective (Olympus) and a CoolSNAP HQ camera (Photometrics). Images were collected using MetaVue version 4.6 (Molecular Devices). Green fluorescent protein (GFP) was visualized using an X-cite 120 UV lamp and a Chroma filter set. Within each experiment, all images were collected and scaled identically. Images were processed with Photoshop 9.0 software (Adobe). Video microscopy was performed with mid-log-phase cells placed on a drop of 2% agarose in yeast extract-peptone-dextrose (YPD) and flattened with an uncoated glass slide as described previously (74). Cells were visualized every 3 to 5 minutes. Small, medium, and large budded cells were grouped into size categories as described previously (63). Cell outline color codes were as follows: red, bud contains segregation structure; blue, bud has inherited vacuole material from mother cell; yellow, medium or large bud lacking inherited vacuole material from mother cell. The scale bars are 2.5 μm throughout all figures.
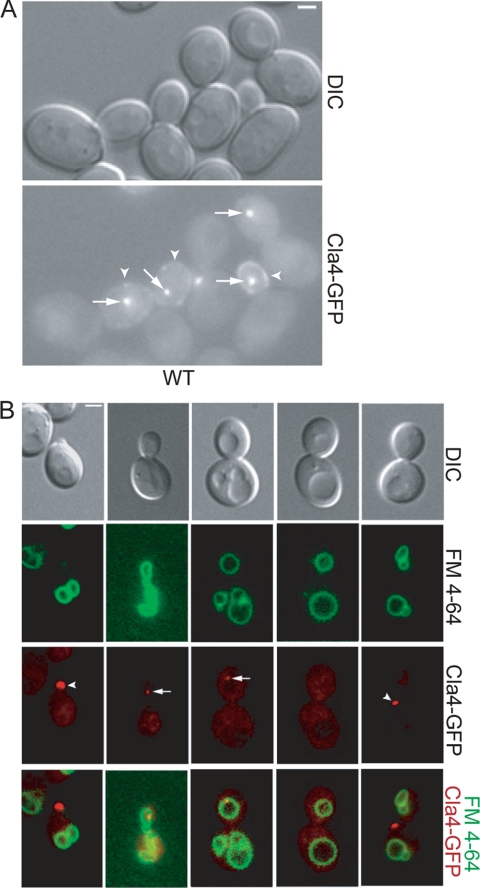
FIG. 1.
Cla4 localizes with the vacuole. (A) Cla4-GFP was visualized in asynchronous wild-type (YCH3250) cells. Arrows indicate perivacuolar Cla4-GFP localization, and arrowheads indicate cortex localization. (B) CLA4-GFP (YCH3250) cells were FM 4-64 stained and examined by fluorescence microscopy.
FIG. 2.
Cla4 requires vacuole inheritance for vacuole-associated punctate structure localization and remains on the daughter vacuole until late M phase. (A) Asynchronous cultures of FM 4-64-stained Cla4-GFP (YCH3250) were examined by fluorescence microscopy and each cell was categorized as small, medium, or large budded (>200 cells were counted for each bud size). Each cell was examined for both vacuole morphology (green) and Cla4-GFP localization to a punctate spot (red), and all cells fell into one of the categories shown. The percentages of small, medium, or large budded cells with the indicated vacuole morphology and punctate Cla4 localization are listed. (B) CLA4-GFP (YCH3250) localization was visualized in cells of various bud sizes and in cells arrested in S phase with hydroxyurea (HU), at G2/M with nocodazole (NZ), or at mitotic exit by arresting cdc15-2 (YCH1028). Values indicate the percentages of cells with the displayed Cla4-GFP localization. (C) Cla4-GFP was visualized in vac8 (YCH1665) and myo2-2 (YCH1752) cells. Arrowheads indicate Cla4-GFP localization at the cortex. (D) Cla4-GFP and FM 4-64-stained vacuole localization was quantified in small, medium, and large budded wild-type (YCH3250), vac8 (YCH1665), and myo2-2 (YCH1752) cells (>200 cells were counted for each bud size).
In vivo labeling of vacuoles and nuclei staining.
To visualize vacuoles, yeast cells were concentrated and incubated for 1 h with N-(3-triethylammoniumpropyl)-4(6(4(diethylamino)phenyl)hexatrienyl) pyridium dibromide (FM 4-64; Molecular Probes) at a final concentration of 16.5 μM (73). Cells were then washed with appropriate medium and grown for ≥3 h before being viewed by fluorescence microscopy. Vacuole inheritance was quantified by looking for the presence of FM 4-64-stained vacuole in large budded cells with >200 cells counted for each experiment. Nuclei were visualized by fixation in 70% ethanol followed by staining with 4′,6-diamidino-2-phenylindole (DAPI) at 0.1 mg/ml.
SDS-PAGE and Western blot analysis.
Yeast protein levels and gel mobility were assayed by trichloroacetic acid precipitation followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 7.5%) and Western blot analysis using rabbit anti-mouse immunoglobulin G (1:1,000; Promega), mouse anti-hemagglutinin (16B12; 1:1,000; Covance), goat anti-actin (1:2,000), and mouse anti-GFP (1:1,000) antibodies.
Vac17-ProA immunoprecipitation and phosphatase treatment.
Pelleted cells where resuspended in lysis buffer (20 mM Tris-HCl [pH 6.5], 5 mM MgCl2, 2% Triton X-100, 150 mM NaCl, 1× protease inhibitor [Roche], 1 mM phenylmethylsulfonyl fluoride, 5 mM EDTA, 50 mM NaF, 10 mM Na4P2O7, 0.5 mM NaVO4, and 1× phosphatase inhibitor [Calbiochem]). Cells were lysed using a French press, and lysates were incubated with 300 μl of immunoglobulin G-Sepharose beads (Amersham Biosciences) and washed with wash buffer (20 mM Tris-HCl [pH 6.8], 5 mM MgCl2, 2% Triton X-100, 150 mM NaCl). Immunoprecipitates were then split and treated with λ-protein phosphatase (New England Biolabs) and phosphatase inhibitor (Calbiochem) as indicated.
Cell cycle arrests.
Cell cycle arrests were performed by the addition α-factor (ZymoResearch), hydroxyurea (Sigma), or nocodazole (Sigma) at final concentrations of 3 μM, 200 mM, or 15 μg/ml, respectively, or by shifting temperature-sensitive mutants from the permissive temperature (23°C) to the nonpermissive temperature (37°C) for 3 hours. Conditions for growth and release of synchronous cultures from arrest by α-factor were described previously (14). Centrifugal elutriation was performed in a Beckman-Coulter type J centrifuge as recommended by the manufacturer and as described previously (44). Fractions with greater than 98% unbudded cells were pooled and used for each experiment.
Other methods.
YP medium containing 1% yeast extract and 2% Bacto peptone was used with addition of glucose (YPD), raffinose (YPR), and/or galactose (YPG) as carbon sources at a 2% final concentration. Alternately, strains with plasmids were grown on synthetic medium (SC) lacking tryptophan or histidine to maintain plasmids.
RESULTS
Cla4 localizes to the vacuole segregation structure.
In addition to its reported localization at the bud cortex, we found that Cla4 localized to a non-cortex-localized punctate structure within the buds of small, medium, and some large budded cells (Fig. 1A). Biochemical studies indicate that Cla4 is enriched on isolated vacuoles (25). Therefore, we postulated that the Cla4 punctate structure might colocalize with the vacuole. To determine if Cla4 colocalized with the vacuole, CLA4-GFP strains were FM 4-64 stained. We found that in 100% of cells in which the punctate Cla4-GFP structure was visualized that it colocalized with the segregation structure or the daughter vacuole (Fig. 1B).
Further quantification of the vacuole morphology and Cla4 localization to the vacuole showed that Cla4 localized to the vacuole almost immediately upon entry of the segregation structure into the bud, as only a very small percentage of cells with a segregation structure in the bud did not colocalize with Cla4 (Fig. 2A). Most cells in which Cla4 colocalized with the vacuole had resolved the segregation structure and formed a daughter vacuole (Fig. 2A). Cla4 persisted on the vacuole until late M phase and cells that arrested in S phase or G2/M, but not at mitotic exit, had Cla4 spots in the bud (Fig. 2A and B).
The machinery required to form and transport the segregation structure to the bud consists of a myosin V motor, Myo2, a vacuole receptor, Vac17, and a vacuole-associated protein, Vac8. We observed that cells that did not form vacuole segregation structures, including vac8 and myo2-2 mutants, lacked a Cla4 spot in the mother or bud (Fig. 2C and D). Thus, Cla4 localization at the vacuole was dependent on vacuole partitioning. Cla4 leaves the vacuole late in mitosis and localizes to the mother bud neck prior to cell separation (data not shown).
PAKs are required for resolution of the vacuole segregation structure.
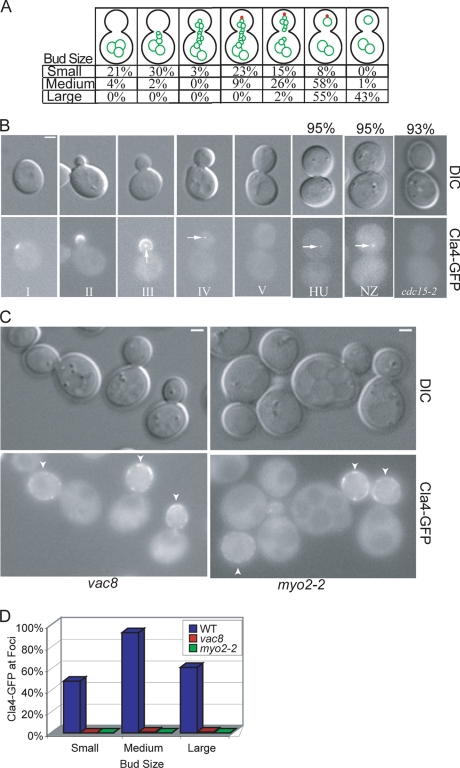
Cla4 accumulation at the incipient bud site early in the cell cycle and its localization at the vacuole segregation structure prior to daughter vacuole formation places Cla4 in the right place at the right time to play a role in resolving the segregation structure. Cla4 is proposed to have overlapping functions with the PAK Ste20, as cla4 and ste20 single mutants are viable but the cla4 ste20 double mutant is synthetically lethal (18). Consistent with these observations, we found that cla4 and ste20 single mutants formed daughter vacuoles (Fig. 3A).
FIG. 3.
Cells lacking PAK function form but do not resolve segregation structures. (A) Mid-logarithmic-phase FM 4-64-strained wild-type (W303-1A), cla4 (KN3591), and ste20 (KN3621) strains were examined by fluorescence microscopy. (B to D) Asynchronous FM 4-64-stained wild-type (W303-1A) and cla4-75 ste20 (KN4580) cells were elutriated to obtain a uniform population of G1 cells, grown at 37°C, and examined at the indicated times by fluorescence microscopy. Values in panels B and C are the percentages of cells with a daughter vacuole.
In order to characterize cells lacking both Cla4 and Ste20 function we used a cla4-75 ste20 strain with mutant cla4 and ste20 kept alive by plasmid-based expression of a temperature-sensitive cla4-75 allele (18). When grown at the permissive temperature, vacuoles in the cla4-75 ste20 cells were similar to vacuoles in ste20 cells (data not shown). After cells were synchronized by centrifugal elutriation in G1 and released at the restrictive temperature, the cla4-75 ste20 double mutant cells initially were similar to wild-type cells and formed a segregation structure that was directed toward the bud tip with similar kinetics as wild-type cells (Fig. 3B and C). Strikingly, after 120 min, 98% of all cla4-75 ste20 cells had a persistent segregation structure (Fig. 3C) and after 240 min 95% of all cells arrested with an intact segregation structure (Fig. 3D). This was in sharp contrast to wild-type cells that resolved their segregation structures soon after they entered the bud (Fig. 3B). The failure of the cla4-75 ste20 cells to resolve the segregation structure is a novel phenotype.
Cells overexpressing CLA4 have a vacuole inheritance defect.
Video microscopy of FM 4-64-stained wild-type and GAL-CLA4 cells was performed to determine if CLA4 overexpression inhibited vacuole inheritance. Wild-type (data not shown) and GAL-CLA4 cells were grown on YP plus 2% galactose medium. Unlike wild-type cells, in which segregation structures were frequently observed and 100% of cells inherited a vacuole (data not shown), segregation structures were not formed in cells overexpressing CLA4 (Fig. 4A).
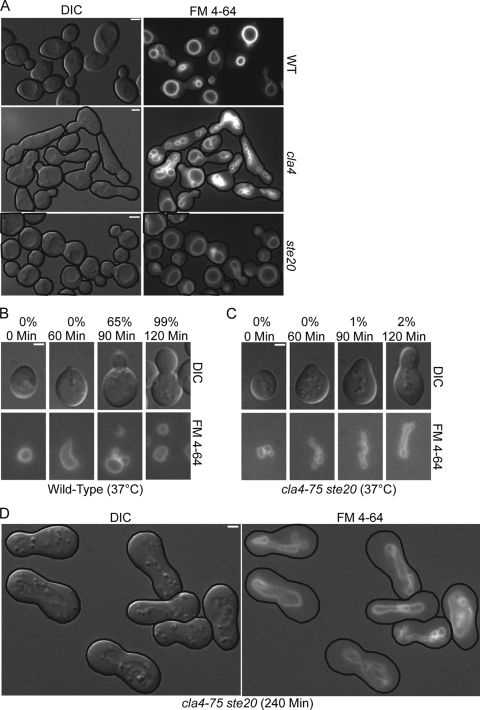
FIG. 4.
Cells overexpressing CLA4 or STE20 have a vacuole inheritance defect. (A) Video microscopy and pedigree analysis of FM 4-64-stained GAL-CLA4 (YMG694) cells was performed on cells grown in YP plus 2% galactose. (B) FM 4-64-stained GAL-CLA4 (YMG694) cells were grown in YP plus 2% raffinose to mid-logarithmic stage. At time zero 2% galactose was added to half the culture and the presence of segregation structures in small and medium budded cells and of daughter vacuoles in large budded cells was examined. Representative pictures at the 6-h time point are shown. (C) Wild-type (W303-1A), GAL-CLA4 (YMG694), and GAL-cla4-K594R (YCB91) cells were treated as for panel B, and the percentage of large budded cells with a discrete daughter vacuole was quantified by fluorescence microscopy (>200 large budded cells were counted for each strain). (D) Mid-logarithmic-phase GAL-GFP-CLA4 (YCH5295) cells were grown in YP plus 2% raffinose. At the zero time point 2% galactose was added to induce GFP-CLA4 overexpression, samples were taken every hour, and Western blot analysis was performed. (E and F) FM 4-64-stained pRS316 (YCH4811), p316-GAL-GST-STE20 (YCH4981), and pRS316-GAL-GST-ste20-K649R (YCH4982) cells were grown in SC-URA plus 2% raffinose to the mid-logarithmic stage and treated as for panels B and C. Representative pictures at the 0- and 5-h time points are shown for GAL-STE20 cells. (G) Mid-logarithmic-phase GAL-STE20-GFP (YCB94) cells were treated as for panel D. Pedigree analysis: M, mother; d1 to -3, primary, secondary, and tertiary daughters.
To further explore the ability of CLA4 to inhibit vacuole inheritance, FM 4-64-stained wild-type, GAL-CLA4, and GAL-cla4-K594R (a kinase dead version of Cla4 [37]) cells were grown in YP plus 2% raffinose to mid-log phase, at which time 2% galactose was added and samples were examined every 2 hours. The presence of a daughter vacuole in large budded cells was scored for each sample. Segregation structures were present in wild-type and GAL-cla4-K594R small and medium budded cells and 100% of all large budded cells contained a daughter vacuole in the presence or absence of galactose throughout the time course of the experiment (Fig. 4C). GAL-CLA4 cells grown under noninducing conditions had similar vacuole inheritance percentages as wild-type cells (data not shown). In contrast, GAL-CLA4 cells grown under inducing conditions exhibited a decrease in vacuole inheritance as early as 2 h and by 6 h postinduction only 2% of large budded cells contained a daughter vacuole (Fig. 4B and C).
Cells overexpressing STE20 have a vacuole inheritance defect.
Cla4 and Ste20 play many unique and overlapping roles in the cell (35, 45). We therefore hypothesized that STE20-overexpressing cells might also have a vacuole inheritance defect. Cultures of FM 4-64-stained wild-type, GAL-STE20, and GAL-STE20-K649R (a kinase dead version of Ste20 [2]) cells were grown to mid-log phase, at which time 2% galactose was added at the zero time point. Under inducing conditions 100% of all large budded wild-type and GAL-ste20-K649R cells contained a daughter vacuole throughout the time course of the experiment (Fig. 4F). GAL-STE20 cells grown under inducing conditions had decreasing amounts of vacuole inheritance as early as 2 h, and by 6 h postinduction only 24% of large-budded cells had a discrete daughter vacuole (Fig. 4E and F). Because PAK function was required for the resolution of the segregation structure and overexpression of PAKs prevented segregation structure formation, we propose that PAKs regulate vacuole inheritance by promoting segregation structure resolution in the bud.
vac17ΔPEST expression suppresses the vacuole inheritance defect of CLA4- or STE20-overexpressing cells.
Vac17 is degraded in a bud-specific manner, but degradation is not necessary for resolution of the segregation structure (41, 70). It has been proposed that Vac17 degradation is due to bud-specific proteins that promote Vac17 degradation (77). An alternate, but not mutually exclusive, interpretation of our overexpression results is that the PAKs are the bud-specific factors that promote Vac17 degradation and that overexpression of CLA4 or STE20 leads to premature destruction of Vac17. We hypothesized that CLA4 or STE20 overexpression caused a vacuole inheritance defect by promoting the premature degradation of Vac17. If this model were correct then we would have expected that expression of the nondegradable VAC17 (vac17ΔPEST) should suppress the vacuole inheritance defect caused by CLA4 or STE20 overexpression.
FM 4-64-stained GAL-CLA4 vac17 and GAL-STE20 vac17 cells containing a plasmid with VAC17 or vac17ΔPEST were examined for vacuole inheritance. As expected 100% of all GAL-CLA4 and GAL-STE20 large budded cells contained a daughter vacuole in the presence of VAC17 or vac17ΔPEST under noninducing conditions. Under inducing conditions GAL-CLA4 and GAL-STE20 cells containing VAC17 exhibited vacuole inheritance defects after 6 h of overexpression. However, expression of vac17ΔPEST fully suppressed the vacuole inheritance defect in GAL-CLA4 and GAL-STE20 cells under inducing conditions (Table 3).
TABLE 3.
vac17ΔPEST expression suppresses the vacuole inheritance defect of CLA4- or STE20-overexpressing cells
| Genotype | % Vacuole inheritance of large budded cells |
|---|---|
| GAL-CLA4 vac17 pVAC17 (−GAL) | 100 |
| GAL-CLA4 vac17 pvac17ΔPEST (−GAL) | 100 |
| GAL-CLA4 vac17 pVAC17 (+GAL) | 2 |
| GAL-CLA4 vac17 pvac17ΔPEST (+GAL) | 100 |
| GAL-STE20 vac17 pVAC17 (−GAL) | 100 |
| GAL-STE20 vac17 pvac17ΔPEST (−GAL) | 100 |
| GAL-STE20 vac17 pVAC17 (+GAL) | 19 |
| GAL-STE20 vac17 pvac17ΔPEST (+GAL) | 100 |
Vac17 is a phosphoprotein, and CLA4 overexpression causes a decrease in Vac17 levels.
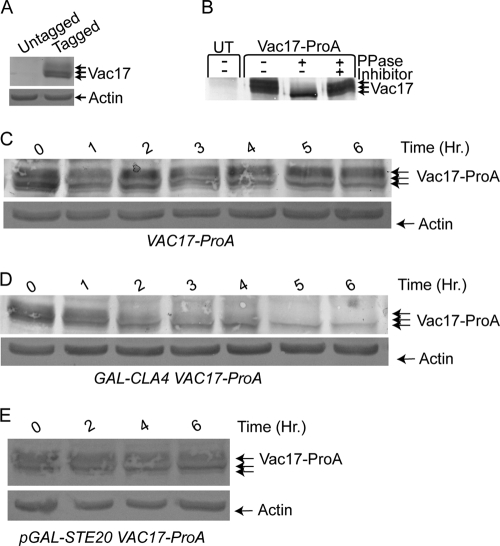
Vac17 levels oscillate during the cell cycle. Vac17 levels increase as cells bud and decrease prior to cytokinesis (70). We examined Vac17 levels throughout the cell cycle to further characterize when Vac17 was degraded. We tagged Vac17 with a ProA tag. Vacuole inheritance was assayed in VAC17-ProA cells and the kinetics of vacuole inheritance were identical to wild-type cells, suggesting that Vac17-ProA was functional (data not shown). Vac17-ProA levels were then examined in asynchronous cells by SDS-PAGE followed by Western blot analysis. We found that Vac17 ran as multiple bands, suggesting the possibility that Vac17-ProA is a phosphoprotein (Fig. 5A). Vac17-ProA was immunoprecipitated and treated with λ-phosphatase or λ-phosphatase plus phosphatase inhibitor. Vac17-ProA treated with λ-phosphatase collapsed down to a single band and phosphatase inhibitors blocked this collapse (Fig. 5B).
FIG. 5.
Vac17 is a phosphoprotein and CLA4-overexpressing cells exhibit a decrease in Vac17 levels. (A) Vac17-ProA and actin protein levels were examined by Western blot analysis in untagged (W303-1A) and VAC17-ProA (YCH4774) cells. (B) Vac17-ProA was immunoprecipitated from protein extracts of 4X GAL-CLA4t swe1Δ VAC17-ProA (YCH4893) or untagged (UT) 4X GAL-CLA4t swe1 (Piatti2711) strains grown on YP plus 2% galactose medium for 6 h. Immunoprecipitated material was split and treated with buffer alone, λ-phosphatase, or λ-phosphatase plus phosphatase inhibitors. (C and D) Mid-logarithmic-phase VAC17-ProA (YCH4774) and GAL-CLA4 VAC17-ProA (YCH4882) cells were grown in YP plus 2% raffinose. At the zero time point 2% galactose was added to induce CLA4 overexpression, samples were taken every hour, and Western blot analysis was performed. (E) Mid-logarithmic-phase GAL-STE20 VAC17-ProA (YCH4843) cells were grown in SC-TRP plus 2% raffinose. At the zero time point 2% galactose was added to induce STE20 overexpression, samples were taken every hour, and Western blot analysis was performed.
We next determined if PAK overexpression caused Vac17 degradation. Asynchronous cultures of wild-type and GAL-CLA4 cells were grown to mid-log phase, 2% galactose was added at the zero time point, and samples were taken every hour for examination of Vac17-ProA levels. Over the 6-h time course of the experiment, Vac17 remained constant in wild-type cells (Fig. 5C). However, Vac17-ProA levels decreased in GAL-CLA4 cells over the 6-h time course. A reduction in Vac17-ProA levels was observed as early as 2 h, with the phosphorylated Vac17-ProA disappearing quickest and Vac17-ProA levels reached very low levels at the 6-hour time point (Fig. 5D). Importantly, the decrease in Vac17-ProA levels occurred with similar kinetics to vacuole inheritance defects (Fig. 4C). Therefore, CLA4-overexpressing cells have a severe vacuole inheritance defect like the class I vacuole inheritance mutants vac8 and myo2-2, which fail to inherit vacuoles. However, unlike vac8 and myo2-2 cells, CLA4-overexpressing cells do not accumulate Vac17 to high levels like other class I vacuole inheritance mutants (70), but instead Vac17 levels drop after CLA4 overexpression is initiated, which may be due to either Vac17 turnover or a decrease in synthesis. Thus, we concluded that CLA4 overexpression causes a decrease in Vac17 protein levels. It is currently unknown if this decrease is due to a decrease in Vac17 synthesis or turnover.
Additionally, the ability of STE20 to promote Vac17-ProA degradation was examined. Asynchronous cultures of GAL-STE20 were grown under inducing conditions, and while vacuole inheritance defects (data not shown) were seen as shown in Fig. 4D and E, Vac17-ProA levels remained constant throughout the 6-hour time course of the experiment (Fig. 5E).
Vac17 degradation in late M phase requires PAK function.
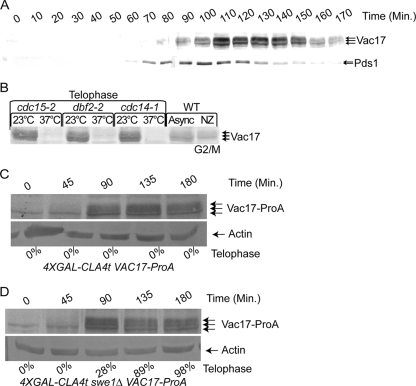
Vac17 levels oscillate during the cell cycle. Vac17 levels increase as cells bud and decrease prior to cytokinesis (70). We examined Vac17 levels throughout the cell cycle to further characterize when Vac17 was degraded. VAC17-ProA PDS1-HA cells were arrested in G1 with α-factor and released. Samples were taken every 10 min, and Vac17 and Pds1 protein levels were examined throughout a single cell cycle. Pds1 is degraded as cells enter anaphase, and Vac17 degradation occurred after this point (Fig. 6A) (16).
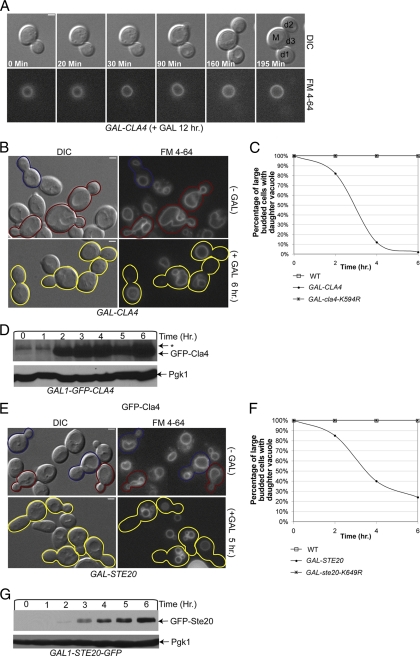
FIG. 6.
PAK function is required for Vac17-ProA degradation in late M phase. (A) VAC17-ProA PDS1-HA (YCH5162) cells were arrested in G1 with α-factor and released, samples were taken every 10 min, and Vac17-ProA and Pds1-HA were examined by Western blot analysis. (B) Vac17-ProA levels were assayed by Western blotting in cdc15-2 (YCH4862), dbf2-2 (YCH4869), and cdc14-1 (YCH4852) cells grown at 23°C or arrested at telophase in 37°C medium. (C and D) 4XGAL-CLA4t VAC17-ProA (YCH4894) and 4XGAL-CLA4t swe1 VAC17-ProA (YCH4893) cells were α-factor arrested and released, and samples were taken at the indicated times and subjected to Western blot analysis. Additionally, the percentage of telophase cells was determined by DAPI staining and the number of cells with mother and bud nuclei staining was scored.
Because Vac17 degradation occurred after anaphase onset, we examined if mitotic exit was required for Vac17 degradation. CDC15, DBF2, and CDC14 are required for mitotic exit, and temperature-sensitive mutants for these genes arrest prior to mitotic exit (19, 54). Vac17-ProA levels were examined in cdc15-2, dbf2-2, and cdc14-1 cells grown at the permissive and nonpermissive temperatures to arrest cells at mitotic exit. Phosphorylated Vac17-ProA was common in cells grown at the nonpermissive temperature. However, cells arrested at mitotic exit only contained a small amount of what appeared to be unphosphorylated Vac17 (Fig. 6B). This suggested that Vac17 was destroyed after anaphase entry and before mitotic exit during late M phase.
If PAKs are required for Vac17 degradation, then in the absence of Cla4 and Ste20 activity Vac17-ProA should not be degraded. A CLA4 allele with a truncated C terminus (CLA4t) when overexpressed causes Cla4 and Ste20 to delocalize from the bud cortex and cells display similar defect as seen in cells lacking PAKs (15). 4X GAL-CLA4t VAC17-ProA cells were arrested in G1 and released, and Vac17 levels and mitotic progression were examined. As in wild-type cells (Fig. 6C), Vac17-ProA is phosphorylated and cells arrest at G2/M in a SWE1-dependent manner (Fig. 6C). Since Vac17 is stable at G2/M, the same experiment was performed in the absence of SWE1, a situation in which cells progress to late M phase (15) when Vac17 protein levels are low (Fig. 6D). We observed that even though cells progressed to late M, Vac17-ProA levels remained high with large amounts of phosphorylated Vac17 present (Fig. 6D). Thus, we concluded that active PAKs were required for Vac17 degradation in late M.
Cla4 requires Lte1 to modulate Vac17 protein levels.
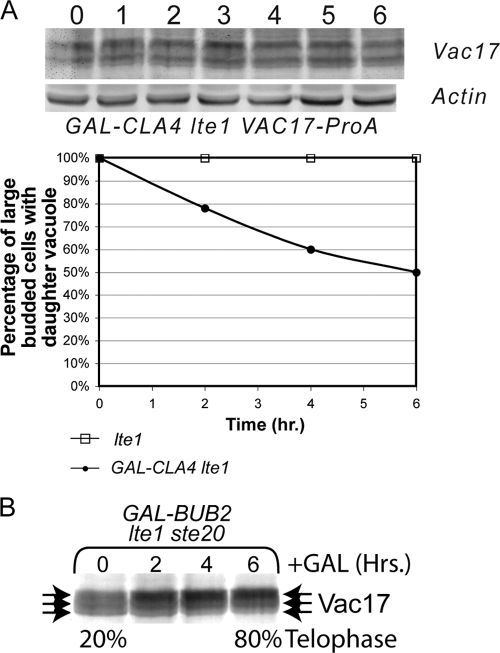
Because Vac17 is phosphorylated in the absence of PAK activity (Fig. 6C and D), we hypothesized that Cla4 may act indirectly to affect Vac17 protein levels. Cla4 plays a role in the proper localization and activation of the guanine nucleotide exchange factor Lte1 in the bud (37, 38, 66, 67, 81, 88). Asynchronous cultures of wild-type and GAL-CLA4 lte1 cells were grown to mid-log phase, 2% galactose was added at the zero time point, and samples were taken every hour for examination of Vac17-ProA levels. Over the 6-h time course of the experiment, Vac17 remained constant (Fig. 7A). Because PAK activity was required to degrade Vac17 in late M phase, we looked in cells lacking both STE20 and LTE1. The lte1 and ste20 mutants show synthetic lethality, but this lethality is suppressed by deletion of BUB2. We looked at Vac17 protein levels in bub2 GAL-BUB2 lte1 ste20 cells (37) grown under noninducing conditions (2% raffinose) to which 2% galactose was added at the zero time point. As previously reported (37) these cells arrest in late M phase but Vac17-ProA is not degraded (Fig. 7B). Thus, we concluded that Cla4 acts indirectly through Lte1 to affect Vac17 protein levels.
FIG. 7.
LTE1 is required for degradation of Vac17 in CLA4-overexpressing cells. (A) Mid-logarithmic-phase lte1 (YCH4737) and GAL-CLA4 lte1 VAC17-ProA (YCH4934) cells were grown in YP plus 2% raffinose and FM 4-64 stained. At the zero time point 2% galactose was added to induce CLA4 overexpression, samples were taken every hour, and Western blot analysis was performed and vacuole inheritance was quantified. (B) Mid-logarithmic-phase GAL-BUB2 bub2 lte1 ste20 Vac17-ProA (YCH5286) cells were grown in YP plus 2% raffinose, 2% glucose was added at time zero, and samples were taken at the indicated times and subjected to Western blot analysis. Additionally, the percentage of telophase cells was determined by DAPI staining and scoring the number of cells with mother and bud nuclei staining.
DISCUSSION
To ensure that the daughter cell contains a functioning vacuole before cell separation, a portion of the vacuole is transported into the bud. To facilitate transport the segregation structure is formed and partitioned into the bud along actin cables by the vacuole-specific myosin receptor Vac17. The segregation structure is transient and resolved to form a discrete bud vacuole soon after entering the bud. Based on the rate of vacuole movement and the distance of travel, segregation structures persist for as little as 30 s during the cell cycle (77). How the segregation structure is resolved in a spatially dependent manner, only after entering the bud, is currently unknown. In addition to resolving the segregation structure, Vac17 is also destroyed in the bud to ensure that the vacuole is not transported to the mother bud neck after the actin cytoskeleton rearranges to the mother bud neck. The mechanism of bud-specific Vac17 degradation is currently unknown. In this paper we provide evidence that Cla4 and Ste20, which are localized and activated specifically in the bud, act in the bud to promote both segregation structure resolution and Vac17 degradation.
How does the cell promote resolution of the segregation structure in a spatially dependent manner only after it has entered the bud? We found that that Cla4 localizes to the vacuole at the right place and at the right time to promote segregation structure resolution (Fig. 1 and 2). This, however, is not the first report of Cla4 localization to the vacuole, as other investigators have previously identified Cla4 on isolated vacuoles (25). However, our findings add substantially to the understanding of the timing of Cla4 localization to the vacuole. Cla4-GFP localized to the cortex of small, medium, and some large budded cells as previously reported (39, 40, 60). Additionally, Cla4-GFP localized to segregation structures when they entered the bud. Almost every segregation structure that entered the bud and no segregation structures that had not entered the bud had Cla4-GFP localized to them (Fig. 2A). Cla4-GFP localization to the vacuole is dependent on the vacuole transport machinery, Vac8 and Myo2 (Fig. 2C and D), further suggesting that Cla4-GFP localization to the vacuole is dependent on transport of vacuoles into the bud.
In further support of a role for Cla4 in segregation structure resolution, PAK function is required for the resolution of the segregation structure (Fig. 3C and D). We propose that Cla4 localizes to the segregation structure, where it promotes segregation structure resolution. Ste20 may promote resolution of the segregation structure when it comes into contact with Ste20 at the bud cortex or Ste20 may be localized to the vacuole at low levels below the detection limit of fluorescence microscopy.
How do Cla4 and Ste20 promote the resolution of the segregation structure? Our current research does not answer this question. However, a possible mechanism(s) already exists within the extant literature. Segregation structure formation is promoted by the vacuole fission machinery and the inhibition of vacuole fusion machinery (7, 8, 21, 28, 29, 47, 62). Since the production of the segregation structure requires the fission machinery and inhibition of fusion, it has be postulated that upregulation of fusion and/or the downregulation of fission promotes segregation structure resolution (17, 32, 33, 78, 80, 86). Intriguingly, Cla4 and Ste20 play roles in vacuole fusion by acting downstream of Cdc42 to promote and actin polymerization, which is required for the docking stage of vacuole fusion (23-26, 49, 55, 83, 84). Intriguingly, cla4 mutants, but not ste20 mutants, have fragmented vacuoles, suggesting that Cla4 may play a larger role than Ste20 in vacuole fusion (25, 65).
In further support of this model, overexpression of either CLA4 or STE20 led to vacuole inheritance defects (Fig. 4). Overexpression of the PAKs may have led to Cla4 and Ste20 localization in the mother. In support of this, GFP-CLA4 when overexpressed was easily seen in both the mother and daughter cytoplasm (see Fig. S1 in the supplemental material). Overepressed Cla4 and Ste20 could potentially be activated in the mother vacuole, where Cdc42 is constitutively localized (25, 55, 61). In further support of this possibility, the scaffolding protein Bem1 that brings together Cdc42 with its GEF Cdc24 (72, 90) is localized to the vacuole, where it plays a role with Cdc42 to promote homotypic vacuole fusion (34, 85). Cla4 and Ste20 bind directly to the second SH3 domain of Bem1 (9, 31, 51, 82), therefore bringing it into proximity with Cdc42 and allowing potential PAK activation. Further research by other labs will be necessary to determine the molecular mechanism by which Cla4 and Ste20 promote segregation structure resolution.
After the segregation structure has been resolved in the bud, the vacuole transport complex is disassembled and Vac17 is degraded to ensure that the vacuole is not transported back to the mother bud neck. We propose a model in which Cla4 and Ste20 localization and activation in the bud mark the bud as the destination for Vac17 degradation. Upon entry of the segregation structure into the bud, the vacuole transport complex comes into contact with activated Cla4 and Ste20, or proteins activated by Cla4 and Ste20 phosphorylation, which primes Vac17 for degradation. It is important to note that segregation structure resolution does not require Vac17 degradation, as vac17ΔPEST mutants readily form daughter vacuoles (70). Thus, the role of PAKs in regulation of Vac17 levels is distinct from its role in segregation structure resolution. In this study we have provided multiple experimental lines of evidence supporting this model that PAKs regulate Vac17 protein level. First, cells overexpressing CLA4 have decreased Vac17 levels (Fig. 5D) and the timing of this decrease coincides temporally with the vacuole inheritance defect (Fig. 4C). Second, PAK function is required for Vac17 degradation in late M phase (Fig. 6D). Third, expression of vac17ΔPEST suppressed the vacuole inheritance defects of CLA4- and STE20-overexpressing cells (Table 3). We propose that PAKs play a second role in regulating vacuole inheritance by regulating Vac17 protein levels. It is currently unknown if PAK regulation of Vac17 is due to a decrease in Vac17 synthesis or turnover.
Potentially inconsistent with this hypothesis is our finding that STE20 overexpression did not cause a decrease in Vac17 levels (Fig. 5E). However, this result must be taken in context with other vacuole inheritance mutants, such as vac8 and myo2-2, that accumulate Vac17 to high levels (70). GAL-STE20 cells have a vacuole inheritance defect but do not accumulate Vac17 to high levels (Fig. 5E). The presence of low levels of Vac17 in STE20-overexpressing cells is also consistent with our findings that around 18% of the cells still inherit vacuoles (Fig. 4E and F) and with a lesser effect of Ste20 in comparison to Cla4 throughout our experiments. Alternately, downstream targets on which Ste20 acts to promote a decrease in Vac17 may only be available in the daughter and therefore STE20 overexpression would not act to regulate Vac17 levels in the mother. Despite the possibility that PAKs may regulate vacuole inheritance through vacuole fusion, our data suggest that PAKs also regulate Vac17 protein levels. Our finding that PAK-deficient cells do not degrade Vac17 during late M phase (Fig. 6D), a time in which Vac17 is usually degraded, argue for a model in which PAKs are the daughter-specific factors that promote Vac17 degradation. Also in agreement with this model, expression of vac17ΔPEST suppressed the vacuole inheritance defects of cells overexpressing CLA4 or STE20 (Table 3). However, our interpretation of the vac17ΔPEST experiment should be only accepted cautiously because the Vac17ΔPEST protein level is abundant. The high levels of Vac17ΔPEST may simply suppress a defect that is not directly due to Vac17 turnover.
How might Cla4 and Ste20 promote a decrease in Vac17 protein levels? The kinase activities of Cla4 and Ste20 are required for the vacuole inheritance defect seen in STE20-overexpressing cells (Fig. 4C and F). Therefore, Cla4 and Ste20 might directly phosphorylate Vac17 priming it for destruction. While possible, our findings suggest this is not the case. Vac17 is phosphorylated in cells overexpressing CLA4t, suggesting that PAKs regulation may be indirect. Furthermore, the finding that LTE1 is required for Vac17 degradation in CLA4-overexpressing cells (Fig. 7A) and that in the absence of STE20 that LTE1 was required for Vac17 degradation in late M phase (Fig. 7B) suggest that Cla4 acts indirectly to regulate Vac17. Further research is therefore required to understand how PAKs regulate vacuole inheritance and Vac17 protein level.
The study of segregation structures in yeast has been a daunting task due to its transient existence. Based on the rate of vacuole movement and the distance of travel, segregation structures can be around for as little as 30 s during the cell cycle (77). During this 30 s a complex set of reactions has been proposed to occur, including the production of PtdIns(3,5)P2 (77), the recruitment of PtdIns(3,5)P2 binding proteins (20, 22, 30, 42), formation of the segregation structure, transport of the segregation structure into the bud, and fusion to resolve the segregation structure. The very transient nature of the segregation structure and the impossibility of reliably synchronizing a population with intact segregation structures have made further analysis of the segregation structure difficult if not impossible. The identification of vacuole segregation structure resolution mutants cla4-75 ste20 and 4X GAL-CLA4t may provide a much-needed tool to further study these steps in vacuole inheritance.
Supplementary Material
Footnotes
Published ahead of print on 13 February 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Aitchison, J. D., M. P. Rout, M. Marelli, G. Blobel, and R. W. Wozniak. 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 1311133-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ash, J., C. Wu, R. Larocque, M. Jamal, W. Stevens, M. Osborne, D. Y. Thomas, and M. Whiteway. 2003. Genetic analysis of the interface between Cdc42p and the CRIB domain of Ste20p in Saccharomyces cerevisiae. Genetics 1639-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr, F. A. 2002. Inheritance of the endoplasmic reticulum and Golgi apparatus. Curr. Opin. Cell Biol. 14496-499. [DOI] [PubMed] [Google Scholar]
- 4.Benton, B. K., A. Tinkelenberg, I. Gonzalez, and F. R. Cross. 1997. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol. Cell. Biol. 175067-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bokoch, G. M. 2003. Biology of the p21-activated kinases. Annu. Rev. Biochem. 72743-781. [DOI] [PubMed] [Google Scholar]
- 6.Boldogh, I. R., K. L. Fehrenbacher, H. C. Yang, and L. A. Pon. 2005. Mitochondrial movement and inheritance in budding yeast. Gene 35428-36. [DOI] [PubMed] [Google Scholar]
- 7.Bonangelino, C. J., N. L. Catlett, and L. S. Weisman. 1997. Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol. Cell. Biol. 176847-6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonangelino, C. J., J. J. Nau, J. E. Duex, M. Brinkman, A. E. Wurmser, J. D. Gary, S. D. Emr, and L. S. Weisman. 2002. Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J. Cell Biol. 1561015-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bose, I., J. E. Irazoqui, J. J. Moskow, E. S. Bardes, T. R. Zyla, and D. J. Lew. 2001. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J. Biol. Chem. 2767176-7186. [DOI] [PubMed] [Google Scholar]
- 10.Bretscher, A. 2003. Polarized growth and organelle segregation in yeast: the tracks, motors, and receptors. J. Cell Biol. 160811-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Catlett, N. L., and L. S. Weisman. 1998. The terminal tail region of a yeast myosin-V mediates its attachment to vacuole membranes and sites of polarized growth. Proc. Natl. Acad. Sci. USA 9514799-14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catlett, N. L., and L. S. Weisman. 2000. Divide and multiply: organelle partitioning in yeast. Curr. Opin. Cell Biol. 12509-516. [DOI] [PubMed] [Google Scholar]
- 14.Cheng, L., L. Hunke, and C. F. Hardy. 1998. Cell cycle regulation of the Saccharomyces cerevisiae Polo-like kinase cdc5p. Mol. Cell. Biol. 187360-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiroli, E., R. Fraschini, A. Beretta, M. Tonelli, G. Lucchini, and S. Piatti. 2003. Budding yeast PAK kinases regulate mitotic exit by two different mechanisms. J. Cell Biol. 160857-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen-Fix, O., J. M. Peters, M. W. Kirschner, and D. Koshland. 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 103081-3093. [DOI] [PubMed] [Google Scholar]
- 17.Conradt, B., J. Shaw, T. Vida, S. Emr, and W. Wickner. 1992. In vitro reactions of vacuole inheritance in Saccharomyces cerevisiae. J. Cell Biol. 1191469-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cvrckova, F., C. De Virgilio, E. Manser, J. R. Pringle, and K. Nasmyth. 1995. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 91817-1830. [DOI] [PubMed] [Google Scholar]
- 19.de Bettignies, G., and L. H. Johnston. 2003. The mitotic exit network. Curr. Biol. 13R301. [DOI] [PubMed] [Google Scholar]
- 20.Dove, S. K., R. C. Piper, R. K. McEwen, J. W. Yu, M. C. King, D. C. Hughes, J. Thuring, A. B. Holmes, F. T. Cooke, R. H. Michell, P. J. Parker, and M. A. Lemmon. 2004. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 231922-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Efe, J. A., R. J. Botelho, and S. D. Emr. 2005. The Fab1 phosphatidylinositol kinase pathway in the regulation of vacuole morphology. Curr. Opin. Cell Biol. 17402-408. [DOI] [PubMed] [Google Scholar]
- 22.Efe, J. A., R. J. Botelho, and S. D. Emr. 2007. Atg18 regulates organelle morphology and Fab1 kinase activity independent of its membrane recruitment by phosphatidylinositol 3,5-bisphosphate. Mol. Biol. Cell 184232-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eitzen, G. 2003. Actin remodeling to facilitate membrane fusion. Biochim. Biophys. Acta 1641175-181. [DOI] [PubMed] [Google Scholar]
- 24.Eitzen, G., N. Thorngren, and W. Wickner. 2001. Rho1p and Cdc42p act after Ypt7p to regulate vacuole docking. EMBO J. 205650-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eitzen, G., L. Wang, N. Thorngren, and W. Wickner. 2002. Remodeling of organelle-bound actin is required for yeast vacuole fusion. J. Cell Biol. 158669-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evangelista, M., B. M. Klebl, A. H. Tong, B. A. Webb, T. Leeuw, E. Leberer, M. Whiteway, D. Y. Thomas, and C. Boone. 2000. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J. Cell Biol. 148353-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fagarasanu, M., A. Fagarasanu, and R. A. Rachubinski. 2006. Sharing the wealth: peroxisome inheritance in budding yeast. Biochim. Biophys. Acta 17631669-1677. [DOI] [PubMed] [Google Scholar]
- 28.Gary, J. D., T. K. Sato, C. J. Stefan, C. J. Bonangelino, L. S. Weisman, and S. D. Emr. 2002. Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol. Biol. Cell 131238-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gary, J. D., A. E. Wurmser, C. J. Bonangelino, L. S. Weisman, and S. D. Emr. 1998. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 14365-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Georgakopoulos, T., G. Koutroubas, I. Vakonakis, M. Tzermia, V. Prokova, A. Voutsina, and D. Alexandraki. 2001. Functional analysis of the Saccharomyces cerevisiae YFR021w/YGR223c/YPL100w ORF family suggests relations to mitochondrial/peroxisomal functions and amino acid signalling pathways. Yeast 181155-1171. [DOI] [PubMed] [Google Scholar]
- 31.Gulli, M. P., M. Jaquenoud, Y. Shimada, G. Niederhauser, P. Wiget, and M. Peter. 2000. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell 61155-1167. [DOI] [PubMed] [Google Scholar]
- 32.Haas, A., B. Conradt, and W. Wickner. 1994. G-protein ligands inhibit in vitro reactions of vacuole inheritance. J. Cell Biol. 12687-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haas, A., and W. Wickner. 1996. Organelle inheritance in a test tube: the yeast vacuole. Semin. Cell Dev. Biol. 7517-524. [Google Scholar]
- 34.Han, B. K., L. M. Bogomolnaya, J. M. Totten, H. M. Blank, L. J. Dangott, and M. Polymenis. 2005. Bem1p, a scaffold signaling protein, mediates cyclin-dependent control of vacuolar homeostasis in Saccharomyces cerevisiae. Genes Dev. 192606-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinrich, M., T. Kohler, and H. U. Mosch. 2007. Role of Cdc42-Cla4 interaction in the pheromone response of Saccharomyces cerevisiae. Eukaryot. Cell 6317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill, K. L., N. L. Catlett, and L. S. Weisman. 1996. Actin and myosin function in directed vacuole movement during cell division in Saccharomyces cerevisiae. J. Cell Biol. 1351535-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofken, T., and E. Schiebel. 2002. A role for cell polarity proteins in mitotic exit. EMBO J. 214851-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofken, T., and E. Schiebel. 2004. Novel regulation of mitotic exit by the Cdc42 effectors Gic1 and Gic2. J. Cell Biol. 164219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holly, S. P., and K. J. Blumer. 1999. PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 147845-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425686-691. [DOI] [PubMed] [Google Scholar]
- 41.Ishikawa, K., N. L. Catlett, J. L. Novak, F. Tang, J. J. Nau, and L. S. Weisman. 2003. Identification of an organelle-specific myosin V receptor. J. Cell Biol. 160887-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 984569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito, T., Y. Matsui, T. Ago, K. Ota, and H. Sumimoto. 2001. Novel modular domain PB1 recognizes PC motif to mediate functional protein-protein interactions. EMBO J. 203938-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnston, L. H., and A. L. Johnson. 1997. Elutriation of budding yeast. Methods Enzymol. 283342-350. [DOI] [PubMed] [Google Scholar]
- 45.Keniry, M. E., and G. F. Sprague, Jr. 2003. Identification of p21-activated kinase specificity determinants in budding yeast: a single amino acid substitution imparts Ste20 specificity to Cla4. Mol. Cell. Biol. 231569-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klionsky, D. J., P. K. Herman, and S. D. Emr. 1990. The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev. 54266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.LaGrassa, T. J., and C. Ungermann. 2005. The vacuolar kinase Yck3 maintains organelle fragmentation by regulating the HOPS tethering complex. J. Cell Biol. 168401-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamson, R. E., M. J. Winters, and P. M. Pryciak. 2002. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell. Biol. 222939-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lechler, T., A. Shevchenko, and R. Li. 2000. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J. Cell Biol. 148363-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leeuw, T., A. Fourest-Lieuvin, C. Wu, J. Chenevert, K. Clark, M. Whiteway, D. Y. Thomas, and E. Leberer. 1995. Pheromone response in yeast: association of Bem1p with proteins of the MAP kinase cascade and actin. Science 2701210-1213. [DOI] [PubMed] [Google Scholar]
- 51.Leeuw, T., C. Wu, J. D. Schrag, M. Whiteway, D. Y. Thomas, and E. Leberer. 1998. Interaction of a G-protein beta-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature 391191-195. [DOI] [PubMed] [Google Scholar]
- 52.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 53.Lowe, M., and F. A. Barr. 2007. Inheritance and biogenesis of organelles in the secretory pathway. Nat. Rev. Mol. Cell Biol. 8429-439. [DOI] [PubMed] [Google Scholar]
- 54.McCollum, D., and K. L. Gould. 2001. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 1189-95. [DOI] [PubMed] [Google Scholar]
- 55.Muller, O., D. I. Johnson, and A. Mayer. 2001. Cdc42p functions at the docking stage of yeast vacuole membrane fusion. EMBO J. 205657-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan, X., and D. S. Goldfarb. 1998. YEB3/VAC8 encodes a myristylated armadillo protein of the Saccharomyces cerevisiae vacuolar membrane that functions in vacuole fusion and inheritance. J. Cell Sci. 1112137-2147. [DOI] [PubMed] [Google Scholar]
- 57.Peter, M., A. M. Neiman, H. O. Park, M. van Lohuizen, and I. Herskowitz. 1996. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 157046-7059. [PMC free article] [PubMed] [Google Scholar]
- 58.Peterson, J., Y. Zheng, L. Bender, A. Myers, R. Cerione, and A. Bender. 1994. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J. Cell Biol. 1271395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raymond, C. K., P. J. O'Hara, G. Eichinger, J. H. Rothman, and T. H. Stevens. 1990. Molecular analysis of the yeast VPS3 gene and the role of its product in vacuolar protein sorting and vacuolar segregation during the cell cycle. J. Cell Biol. 111877-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richman, T. J., and D. I. Johnson. 2000. Saccharomyces cerevisiae cdc42p GTPase is involved in preventing the recurrence of bud emergence during the cell cycle. Mol. Cell. Biol. 208548-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richman, T. J., M. M. Sawyer, and D. I. Johnson. 2002. Saccharomyces cerevisiae Cdc42p localizes to cellular membranes and clusters at sites of polarized growth. Eukaryot. Cell 1458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rudge, S. A., D. M. Anderson, and S. D. Emr. 2004. Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol. Biol. Cell 1524-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saito, T. L., M. Ohtani, H. Sawai, F. Sano, A. Saka, D. Watanabe, M. Yukawa, Y. Ohya, and S. Morishita. 2004. SCMD: Saccharomyces cerevisiae Morphological Database. Nucleic Acids Res. 32D319-D322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 65.Seeley, E. S., M. Kato, N. Margolis, W. Wickner, and G. Eitzen. 2002. Genomic analysis of homotypic vacuole fusion. Mol. Biol. Cell 13782-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seshan, A., and A. Amon. 2005. Ras and the Rho effector Cla4 collaborate to target and anchor Lte1 at the bud cortex. Cell Cycle 4940-946. [DOI] [PubMed] [Google Scholar]
- 67.Seshan, A., A. J. Bardin, and A. Amon. 2002. Control of Lte1 localization by cell polarity determinants and Cdc14. Curr. Biol. 122098-2110. [DOI] [PubMed] [Google Scholar]
- 68.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simon, M. N., C. De Virgilio, B. Souza, J. R. Pringle, A. Abo, and S. I. Reed. 1995. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature 376702-705. [DOI] [PubMed] [Google Scholar]
- 70.Tang, F., E. J. Kauffman, J. L. Novak, J. J. Nau, N. L. Catlett, and L. S. Weisman. 2003. Regulated degradation of a class V myosin receptor directs movement of the yeast vacuole. Nature 42287-92. [DOI] [PubMed] [Google Scholar]
- 71.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56619-630. [DOI] [PubMed] [Google Scholar]
- 72.Toenjes, K. A., M. M. Sawyer, and D. I. Johnson. 1999. The guanine-nucleotide-exchange factor Cdc24p is targeted to the nucleus and polarized growth sites. Curr. Biol. 91183-1186. [DOI] [PubMed] [Google Scholar]
- 73.Vida, T. A., and S. D. Emr. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waddle, J. A., T. S. Karpova, R. H. Waterston, and J. A. Cooper. 1996. Movement of cortical actin patches in yeast. J. Cell Biol. 132861-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang, Y. X., N. L. Catlett, and L. S. Weisman. 1998. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J. Cell Biol. 1401063-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Warren, G., and W. Wickner. 1996. Organelle inheritance. Cell 84395-400. [DOI] [PubMed] [Google Scholar]
- 77.Weisman, L. S. 2003. Yeast vacuole inheritance and dynamics. Annu. Rev. Genet. 37435-460. [DOI] [PubMed] [Google Scholar]
- 78.Weisman, L. S. 2006. Organelles on the move: insights from yeast vacuole inheritance. Nat. Rev. Mol. Cell Biol. 7243-252. [DOI] [PubMed] [Google Scholar]
- 79.Weisman, L. S., R. Bacallao, and W. Wickner. 1987. Multiple methods of visualizing the yeast vacuole permit evaluation of its morphology and inheritance during the cell cycle. J. Cell Biol. 1051539-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weisman, L. S., and W. Wickner. 1988. Intervacuole exchange in the yeast zygote: a new pathway in organelle communication. Science 241589-591. [DOI] [PubMed] [Google Scholar]
- 81.Wild, A. C., J. W. Yu, M. A. Lemmon, and K. J. Blumer. 2004. The p21-activated protein kinase-related kinase Cla4 is a coincidence detector of signaling by Cdc42 and phosphatidylinositol 4-phosphate. J. Biol. Chem. 27917101-17110. [DOI] [PubMed] [Google Scholar]
- 82.Winters, M. J., and P. M. Pryciak. 2005. Interaction with the SH3 domain protein Bem1 regulates signaling by the Saccharomyces cerevisiae p21-activated kinase Ste20. Mol. Cell. Biol. 252177-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu, C., S. F. Lee, E. Furmaniak-Kazmierczak, G. P. Cote, D. Y. Thomas, and E. Leberer. 1996. Activation of myosin-I by members of the Ste20p protein kinase family. J. Biol. Chem. 27131787-31790. [DOI] [PubMed] [Google Scholar]
- 84.Wu, C., V. Lytvyn, D. Y. Thomas, and E. Leberer. 1997. The phosphorylation site for Ste20p-like protein kinases is essential for the function of myosin-I in yeast. J. Biol. Chem. 27230623-30626. [DOI] [PubMed] [Google Scholar]
- 85.Xu, H., and W. Wickner. 2006. Bem1p is a positive regulator of the homotypic fusion of yeast vacuoles. J. Biol. Chem. 28127158-27166. [DOI] [PubMed] [Google Scholar]
- 86.Xu, Z., and W. Wickner. 1996. Thioredoxin is required for vacuole inheritance in Saccharomyces cerevisiae. J. Cell Biol. 132787-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamaguchi, Y., K. Ota, and T. Ito. 2007. A novel Cdc42-interacting domain of the yeast polarity establishment protein Bem1. Implications for modulation of mating pheromone signaling. J. Biol. Chem. 28229-38. [DOI] [PubMed] [Google Scholar]
- 88.Yoshida, S., R. Ichihashi, and A. Toh-e. 2003. Ras recruits mitotic exit regulator Lte1 to the bud cortex in budding yeast. J. Cell Biol. 161889-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zheng, L., U. Baumann, and J. L. Reymond. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zheng, Y., A. Bender, and R. A. Cerione. 1995. Interactions among proteins involved in bud-site selection and bud-site assembly in Saccharomyces cerevisiae. J. Biol. Chem. 270626-630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.