Abstract
Trimethoprim, an antifolate commonly prescribed in combination with sulfamethoxazole, potently inhibits several prokaryotic species of dihydrofolate reductase (DHFR). However, several eukaryotic pathogenic organisms are resistant to trimethoprim, preventing its effective use as a therapeutic for those infections. We have been building a program to reengineer trimethoprim to more potently and selectively inhibit eukaryotic species of DHFR as a viable strategy for new drug discovery targeting several opportunistic pathogens. We have developed a series of compounds that exhibit potent and selective inhibition of DHFR from the parasitic protozoa Cryptosporidium and Toxoplasma as well as the fungus Candida glabrata. A comparison of the structures of DHFR from the fungal species Candida glabrata and Pneumocystis suggests that the compounds may also potently inhibit Pneumocystis DHFR.
Reduced folate cofactors such as tetrahydrofolate are required for critical cellular functions, such as the production of dTMP, several amino acids, and purines. Dihydrofolate reductase (DHFR), the sole source of tetrahydrofolate, is one of several enzymes in the folate biosynthetic pathway. DHFR has been a recognized and validated drug target since the 1960s, with the discovery of methotrexate (5). Fortunately, since pathogenic and human forms of DHFR exhibit several critical sequence differences, it has also been possible to develop species-selective antifolates for several infectious diseases, including malaria, toxoplasmosis, and urinary tract infections. Trimethoprim (TMP) (Fig. 1) is a commonly administered antifolate, primarily in combination with sulfamethoxazole (TMP-SMZ), which inhibits dihydropteroate synthase (DHPS), another enzyme in the folate pathway (14). TMP-SMZ is most effective against prokaryotic pathogens. However, it is also recognized as first-line therapy in treating and preventing the common eukaryotic opportunistic pathogen Pneumocystis jirovecii, which causes life-threatening pneumonia in immunocompromised patients (20).
FIG. 1.
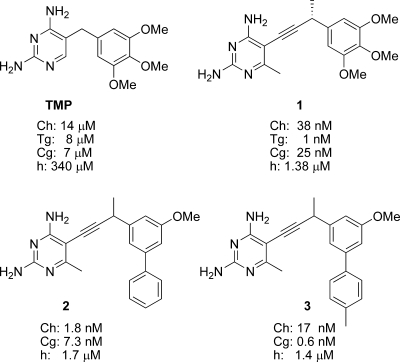
Antifolates and their corresponding IC50s against various eukaryotic species of DHFR. Ch, Cryptosporidium hominis; Tg, Toxoplasma gondii; Cg, Candida glabrata; h, human.
Interestingly, while TMP inhibits bacterial species of DHFR at concentrations in the low nanomolar range, it inhibits many eukaryotic species of the enzyme at concentrations in the micromolar range, resulting in 3 orders of magnitude lower potency. Even the use of TMP-SMZ as a prophylactic agent against pneumocystis relies heavily on the sulfa component (31) and only on relatively weak binding between TMP and P. jirovecii DHFR (6, 18). In fact, studies have reported that mutations conferring resistance to TMP-SMZ arise in DHPS, not in DHFR (17, 28). In contrast, when the DHFR inhibitor pyrimethamine, which is four times more potent than TMP (18), is used in combination with sulfadiazine against pneumocystis, mutations arise in DHFR as well as DHPS (20). A logical conclusion of these studies is that the low potency of TMP against P. jirovecii DHFR is preventing it from reaching its full potential as an effective therapy for this eukaryotic opportunistic pathogen. In contrast, high-affinity DHFR inhibitors such as trimetrexate and piritrexim potently inhibit the growth of cultures of Pneumocystis (2, 23) even when administered as single agents. These inhibitors have limited use, however, because of their toxicity to the human enzyme.
In an attempt to discover more potent analogs of TMP that would eliminate the need for the sulfonamide component and selectively inhibit DHFR from Pneumocystis and Toxoplasma, several DHFR inhibitors have been developed over the past 2 decades (10-12, 24-27). At the time, investigators were examining interactions with Pneumocystis carinii instead of Pneumocystis jirovecii, primarily because of the late recognition of P. jirovecii as the causative agent of human pneumocystis pneumonia during compound development and the ability to use the P. carinii DHFR crystal structures (7-9) in design. During this time, compounds were developed that exhibited nanomolar levels of inhibition for the Toxoplasma gondii and P. carinii enzymes.
We have been building a program to reengineer TMP to more potently inhibit eukaryotic species of DHFR as a viable strategy for new drug discovery targeting several eukaryotic opportunistic pathogens. These novel DHFR inhibitors, originally based on the TMP scaffold, exhibit potent inhibition of DHFR from the parasitic protozoa Cryptosporidium and Toxoplasma and the fungus Candida glabrata. We chose to study these opportunistic infections because for immunocompromised patients there are no approved agents for treatment of cryptosporidiosis (1, 30), the severity of toxoplasmosis can be devastating (3), and strains of Candida (such as C. glabrata) resistant to the commonly administered antifungals now comprise at least 20% of candidemia cases (13, 22, 29). Herein, we analyze the interactions of these inhibitors across a wide variety of eukaryotic opportunistic pathogens and extend this analysis to Pneumocystis jirovecii DHFR. A comparison of the structures of Candida glabrata and Pneumocystis carinii DHFR, along with a sequence comparison of P. jirovecii and P. carinii, suggests that these compounds may additionally inhibit Pneumocystis jirovecii DHFR.
MATERIALS AND METHODS
Cryptosporidium hominis and T. gondii DHFR-TS preparation.
DHFR-thymidylate synthase (DHFR-TS) from C. hominis (ChDHFR-TS) was expressed in Escherichia coli and purified using a methotrexate agarose column (Sigma). Briefly, crude cell lysate was loaded on the column, which was then washed with 4 column volumes of buffer A [0.2 M N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES), pH 7.0, 1 mm EDTA, 5 mM dithiothreitol] containing 0.2 M NaCl, followed by 8 column volumes of buffer A containing 0.5 M NaCl. The protein was eluted from the column with buffer A containing 2 mM dihydrofolate. The gene for DHFR from T. gondii was cloned and the protein expressed and purified as previously described (21).
C. glabrata DHFR preparation, crystallization, and structure solution.
The gene for C. glabrata DHFR (CgDHFR) was amplified from genomic DNA obtained from ATCC (36909D), inserted into a pET41 vector with a C-terminal His tag, and expressed in E. coli BL21(DE3) cells. The protein was purified using nickel affinity chromatography, desalted with a PD-10 column, and concentrated to 13 mg/ml. CgDHFR was incubated with 1.5 mM NADPH and 1 mM inhibitor and crystallized by hanging drop vapor diffusion using a mother liquor with 0.1 M Tris, pH 8.5, 30% polyethylene glycol 4000, and 0.2 M MgCl2. Diffraction data were collected at Brookhaven National Laboratory, beam line X25, and processed using the program HKL2000. The structure was determined by molecular replacement, using a structure of C. albicans DHFR (32), and refined using the program Refmac5 (19). Synthesis and characterization of compounds 1 to 3 and their analogs have been described previously (16, 21).
Enzyme assays.
Enzyme assays were performed at 25°C by monitoring the rate of NADPH oxidation at 340 nm over 5 min. Reactions were performed in the presence of 20 mM TES, pH 7.0, 50 mM KCl, 10 mM 2-mercaptoethanol, 0.5 mM EDTA, 1 mg/ml bovine serum albumin, 100 μM NADPH, and 1 mM dihydrofolate.
Protein structure accession number.
The coordinates for the CgDHFR protein have been deposited at the Protein Data Bank with identifier 3CSE.
RESULTS AND DISCUSSION
Unlike prokaryotic, fungal, and human species of DHFR, parasitic protozoa, including C. hominis and T. gondii, have a bifunctional form of the enzyme (DHFR-TS), wherein DHFR and TS are translated from the same polypeptide. The DHFR and TS domains are distinct, and the active sites are separated by approximately 46 Å, allowing inspection of each active site individually. Therefore, in order to design more effective DHFR inhibitors for eukaryotic pathogens, we first examined the crystal structure of ChDHFR (4), with TMP modeled in the active site. It was evident from this analysis that the trimethoxyphenyl ring of TMP did not form maximal contacts with the hydrophobic pocket that normally houses the para-aminobenzoic acid moiety of the substrate, dihydrofolate (residues Thr 58, Ser 61, Ile 62, and Leu 67). Specifically, there appeared to be at least a 1- to 2-Å gap between the trimethoxyphenyl ring and the ideal distance for van der Waals contacts with residues Ile 62, Thr 58, and Ser 61, thereby reducing the potential van der Waals energy. We hypothesized that new inhibitors with an extended bridge between the diaminopyrimidine and the trimethoxyphenyl rings of TMP may increase the favorable contacts between the phenyl ring and the residues in the hydrophobic pocket. After synthesizing a series of analogs to probe the proper conformational freedom and length of the extended compounds, we found that the propargyl-linked compound 1 (Fig. 1) exhibited 368-fold greater potency than that of TMP against the ChDHFR enzyme (21). Compound 1 then formed the basis of new lead discovery for potent antifolates effective against eukaryotic pathogens.
We then constructed homology models of DHFR from Toxoplasma gondii and Candida glabrata (TgDHFR and CgDHFR) and predicted that the extended TMP compounds may be effective inhibitors of these eukaryotic DHFR enzymes as well. Excitingly, in vitro enzyme inhibition assays with TgDHFR, CgDHFR, and compound 1 confirmed that this scaffold could serve as a base structure of an inhibitor for these species of DHFR, since 50% inhibitory concentrations (IC50s) were 1 and 25 nM, respectively (Fig. 1) (16, 21).
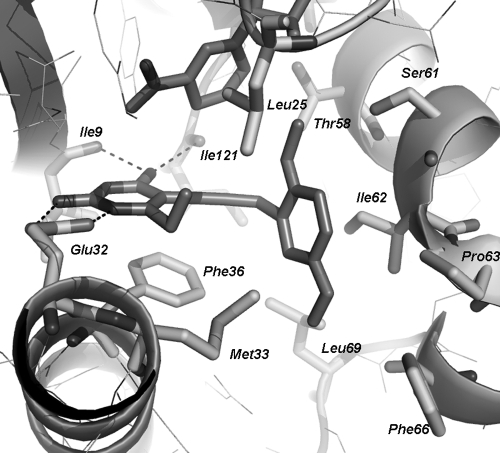
We crystallized the C. glabrata DHFR enzyme with an analog of compound 1 (a compound with a dimethoxyphenyl ring and an ethyl group at the C-6 position of the pyrimidine instead of a methyl) in order to validate the proposed binding mode and to gain new insights for future inhibitor design. The structure (PDB accession number 3CSE) was determined to 1.6-Å resolution and was refined to an R factor of 0.18 and an Rfree of 0.23. The structure directly confirms that the inhibitor binds in the active site, as predicted (Fig. 2) (16). With the extended bridge between the two rings, the dimethoxyphenyl ring reaches deeper into the eukaryotic pocket than a single methylene bridge and interacts with several eukaryotic hydrophobic residues, including Thr 58, Ser 61, Ile 62, and Pro 63 of CgDHFR. There appear to be ideal distances between the van der Waals radii of the residues and the inhibitor, explaining the large increase in potency.
FIG. 2.
The crystal structure of CgDHFR bound to an analog of compound 1 and NADPH reveals details of the interaction between the ligand and the eukaryotic hydrophobic pocket. Hydrogen bonds are shown with dashed lines.
Using this first crystal structure of CgDHFR bound to one of the novel antifolates and interpreting docked complexes of compound 1 bound to ChDHFR, we designed second-generation inhibitors that have increased potency for the pathogenic enzymes. Compound 2 has an IC50 of 1.8 nM against ChDHFR (a 21-fold improvement relative to compound 1), and compound 3 exhibits picomolar inhibition against CgDHFR (a 42-fold improvement relative to compound 1). These new compounds also exhibit potent inhibition of the growth of C. glabrata in culture, with a MIC of 1.5 μg/ml (16).
Subtle differences in eukaryotic species of DHFR also allow for selectivity against the human form of the enzyme. In fact, compounds 1 to 3 are poor inhibitors of the human enzyme, with IC50s in the micromolar range (Fig. 1). There are several residue substitutions between the opportunistic pathogens and human DHFR (hDHFR), two of which are critical in the active site. Near the pyrimidine ring, CgDHFR has Met 33 and ChDHFR has Leu 33, while hDHFR has the much larger and less flexible residue Phe 31. Near the opening to the active site, there is a loop in hDHFR, containing residues Pro 61 to Asn 64, which is absent in ChDHFR. While the loop is present in CgDHFR (Pro 63 to Phe 66), it is translated by 1.2 Å and contains a hydrophobic Phe residue instead of the polar Asn residue in hDHFR. The structural differences translate into potency differences for inhibition of the enzyme (Fig. 1) and growth of the cell culture. Compound 3 inhibits the growth of Candida glabrata 13.5-fold more potently than the growth of human cell lines (16). Cellular selectivity indices of >10 are considered to be within an acceptable range (15), although obviously further designs will focus on increasing this selectivity.
Interestingly, in comparing compounds 1 and 3, the improvement in potency (42-fold) at the enzyme level is in the same range as the improvement in potency of growth inhibition (13-fold). This correlation suggests that high enzyme potency is an important factor in developing an efficacious drug lead. In addition to correlations between potency at the level of the enzyme and fungal growth inhibition, there appear to be correlations between the MIC and the hydrophobic character of the compounds, suggesting that we can optimize cell permeation in future generations of compound design.
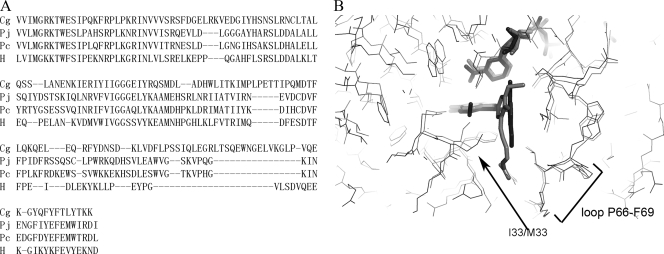
As discussed in the introduction, TMP-SMZ is used prophylactically for Pneumocystis pneumonia, despite the weak affinity of TMP for Pneumocystis DHFR (IC50 = 4.8 μM for P. jirovecii DHFR) (18). Since the propargyl-based antifolates are significantly more potent than TMP against the fungal C. glabrata DHFR, they may also be effective against the fungal Pneumocystis DHFR. In order to begin to explore whether these compounds may exhibit in vitro potency for Pneumocystis DHFR, we compared the sequences of P. carinii DHFR (PcDHFR), P. jirovecii DHFR, and C. glabrata DHFR (Fig. 3A). We also compared the available structures (Fig. 3B) of P. carinii DHFR (9) and C. glabrata DHFR (16).
FIG. 3.
Comparison of sequences of DHFR from C. glabrata (CgDHFR), P. jirovecii (PjDHFR), P. carinii (PcDHFR), and human (hDHFR) (A) and of the structures of CgDHFR (black) and PcDHFR (gray) bound to antifolates and NADPH (ligands are shown in stick form) (B).
Overall, the structures of the fungal C. glabrata and P. carinii enzymes are very similar and superimpose with a root mean square deviation of 3.6 Å over 161 C-α atoms. The active site residues exhibit even more structural similarity (Fig. 3B), reinforcing the similarity between these enzymes. The only residue difference between CgDHFR and PcDHFR is located at Met 33 (CgDHFR) and Ile 33 (PcDHFR). Interestingly, P. jirovecii DHFR maintains the methionine at position 33 (Fig. 3A). Specifically, it is likely that the propargyl-linked antifolates will be effective inhibitors of Pneumocystis DHFR based on the similarity of the active site size and the apparent fit of the docked lead to the Pneumocystis enzyme. The compounds are likely to maintain their specificity for the pathogenic species of DHFR over hDHFR, since the key loop at the active site, residues Pro 66 to Phe 69 in PcDHFR, maintains the same position and conformation as those observed in CgDHFR and includes the Phe residue, as opposed to the Asn residue in hDHFR. When the structure of P. jirovecii DHFR becomes available, it will be important to include it in a wider comparison with other fungal enzymes.
In conclusion, DHFR is a potentially effective drug target for opportunistic infectious diseases. We exploited the overall similarities in the structures of eukaryotic species of DHFR to design a series of antifolates that include an extended propargyl linker, intended to force the interaction of the substituted phenyl ring with the eukaryotic hydrophobic pocket. These novel antifolates are between 300- and 11,000-fold more potent than TMP against the eukaryotic pathogenic enzymes. A high-resolution crystal structure of CgDHFR bound to one of these extended compounds confirms that the inhibitor is bound to the active site, with the phenyl ring buried in a hydrophobic pocket, accounting for the increased affinity. Using the subtle differences between eukaryotic species of DHFR, we customized the lead compounds and synthesized very potent and selective second-generation inhibitors for ChDHFR and CgDHFR. A structural comparison with Pneumocystis DHFR suggests that the propargyl-linked antifolates may also be effective inhibitors of this important pathogen.
Acknowledgments
This work was supported by NIH grant GM067542 to A.C.A., NIH grant AI065143 to D.L.W., and grant R13AI078718 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 23 January 2009.
REFERENCES
- 1.Abubakar, I., S. Aliyu, C. Arumugam, N. Usman, and P. Hunter. 2007. Treatment of cryptosporidiosis in immunocompromised individuals: systemic review and meta-analysis. Br. J. Clin. Pharmacol. 63387-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allegra, C., B. Chabner, C. Tuazon, D. Ogata-Arakaki, B. Baird, J. Drake, J. Simmons, E. Lack, J. Shelhamer, F. Balis, R. Walker, J. Kovacs, H. Lane, and H. Masur. 1987. Trimetrexate for the treatment of Pneumocystis carinii pneumonia in patients with the acquired immunodeficiency syndrome. N. Engl. J. Med. 317978-985. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, A. 2005. Targeting DHFR in parasitic protozoa. Drug Discov. Today 10121-128. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, A. 2005. Two crystal structures of dihydrofolate reductase-thymidylate synthase from Cryptosporidium hominis reveal protein:ligand interactions including a structural basis for observed antifolate resistance. Acta Crystallogr. F 61258-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertino, J. 1993. Karnofsky Memorial Lecture: ode to methotrexate. J. Clin. Oncol. 115-14. [DOI] [PubMed] [Google Scholar]
- 6.Chan, D., and A. Anderson. 2006. Towards species-specific antifolates. Curr. Med. Chem. 13377-398. [DOI] [PubMed] [Google Scholar]
- 7.Cody, V. 2002. Structure-based modeling of reversed N9-C10 bridge antifols with human, Pc and Tg DHFR, p. 525-529. In Chemistry and biology of pteridines and folates. Proceedings of the 12th International Symposium on Pteridines and Folates. National Institutes of Health, Bethesda, MD.
- 8.Cody, V., N. Galitsky, D. Rak, J. Luft, W. Pangborn, and S. Queener. 1999. Ligand-induced conformational changes in the crystal structures of Pneumocystis carinii dihydrofolate reductase complexes with folate and Nadp+. Biochemistry 384303-4312. [DOI] [PubMed] [Google Scholar]
- 9.Cody, V., J. Pace, K. Chisum, and A. Rosowsky. 2006. New insights into DHFR interactions: analysis of Pneumocystis carinii and mouse DHFR complexes with NADPH and two highly potent trimethoprim derivatives. Proteins 65959-969. [DOI] [PubMed] [Google Scholar]
- 10.Gangjee, A., O. Adair, and S. Queener. 2003. Synthesis and biological evaluation of 2,4-diamino-6-(arylaminomethyl)pyrido[2,3-d]pyrimidines as inhibitors of Pneumocystis carinii and Toxoplasma gondii dihydrofolate reductase and as antiopportunistic infection and antitumor agents. J. Med. Chem. 465074-5082. [DOI] [PubMed] [Google Scholar]
- 11.Gangjee, A., and X. Lin. 2005. CoMFA and CoMSIA analyses of Pneumocystis carinii dihydrofolate reductase, Toxoplasma gondii dihydrofolate reductase and rat liver dihydrofolate reductase. J. Med. Chem. 481448-1469. [DOI] [PubMed] [Google Scholar]
- 12.Gangjee, A., X. Lin, and S. E. Queener. 2003. 7-Methyl trimethoprim analogs as inhibitors of the folate metabolizing enzymes. J. Heterocyclic Chem. 40507-512. [Google Scholar]
- 13.Hajjeh, R., A. Sofair, L. Harrison, G. M. Lyon, B. Arthington-Skaggs, S. Mirza, M. Phelan, J. Morgan, W. Lee-Yang, M. Ciblak, L. Benjamin, L. Sanza, S. Huie, S. Yeo, M. Brandt, and D. Warnock. 2004. Incidence of bloodstream infections due to Candida species and in vitro susceptibilities of isolates collected from 1998 to 2000 in a population-based active surveillance program. J. Clin. Microbiol. 421519-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawser, S., S. Luciuro, and K. Islam. 2006. Dihydrofolate reductase inhibitors as antibacterial agents. Biochem. Pharmacol. 71941-948. [DOI] [PubMed] [Google Scholar]
- 15.Kamchonwongpaisan, S., R. Quarrell, N. Chareonsethakul, R. Ponsinet, T. Vilaivan, J. Vanichtanankul, B. Tarnchompoo, W. Sirawaraporn, G. Lowe, and Y. Yuthavong. 2004. Inhibitors of multiple mutants of Plasmodium falciparum dihydrofolate reductase and their antimalarial activities. J. Med. Chem. 47673-680. [DOI] [PubMed] [Google Scholar]
- 16.Liu, J., D. Bolstad, A. Smith, N. Priestley, D. Wright, and A. Anderson. 2008. Structure-guided development of efficacious antifungal agents targeting Candida glabrata dihydrofolate reductase. Chem. Biol. 15990-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma, L., L. Borio, H. Masur, and J. Kovacs. 1999. Pneumocystis carinii dihydropteroate synthase but not dihydrofolate reductase gene mutations correlate with prior trimethoprim-sulfamethoxazole or dapsone use. J. Infect. Dis. 1801969-1978. [DOI] [PubMed] [Google Scholar]
- 18.Ma, L., and J. Kovacs. 2000. Expression and characterization of recombinant human-derived Pneumocystis carinii dihydrofolate reductase. Antimicrob. Agents Chemother. 443092-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murshudov, G., A. Vagin, and E. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53240-255. [DOI] [PubMed] [Google Scholar]
- 20.Nahimana, A., M. Rabodonirina, J. Bille, P. Francioli, and P. Hauser. 2004. Mutations of Pneumocystis jirovecii dihydrofolate reductase associated with failure of prophylaxis. Antimicrob. Agents Chemother. 484301-4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelphrey, P., V. Popov, T. Joska, J. Beierlein, E. Bolstad, Y. Fillingham, D. Wright, and A. Anderson. 2007. Highly efficient ligands for DHFR from Cryptosporidium hominis and Toxoplasma gondii inspired by structural analysis. J. Med. Chem. 50940-950. [DOI] [PubMed] [Google Scholar]
- 22.Pfaller, M., and D. Diekema. 2004. Rare and emerging opportunistic fungal pathogens: concern for resistance beyond Candida albicans and Aspergillus fumigatus. J. Clin. Microbiol. 424419-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Queener, S., M. Bartlett, M. Jay, M. Durkin, and J. Smith. 1987. Activity of lipid-soluble inhibitors of dihydrofolate reductase against Pneumocystis carinii in culture and in a rat model of infection. Antimicrob. Agents Chemother. 311323-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosowsky, A., H. Chen, H. Fu, and S. Queener. 2003. Synthesis of new 2,4-diaminopyrido[2,3-d]pyrimidine and 2,4-diaminopyrrolo[2,3-d]pyrimidine inhibitors of Pneumocystis carinii, Toxoplasma gondii and Mycobacterium avium dihydrofolate reductase. Bioorg. Med. Chem. 1159-67. [DOI] [PubMed] [Google Scholar]
- 25.Rosowsky, A., R. Forsch, and S. Queener. 2002. Inhibition of Pneumocystis carinii, Toxoplasma gondii and Mycobacterium avium dihydrofolate reductases by 2,4-diamino-5-[2-methoxy-5-(ω-carboxyalkoxy)benzyl]pyrimidines: marked improvement in potency relative to trimethoprim and species selectivity relative to piritrexim. J. Med. Chem. 45233-241. [DOI] [PubMed] [Google Scholar]
- 26.Rosowsky, A., R. Forsch, and S. Queener. 2003. Further studies on 2,4-diamino-5-(2′,5′-disubstituted benzyl)pyrimidines as potent and selective inhibitors of dihydrofolate reductases from three major opportunistic pathogens of AIDS. J. Med. Chem. 461726-1736. [DOI] [PubMed] [Google Scholar]
- 27.Rosowsky, A., R. Forsch, C. Sibley, C. Inderlied, and S. Queener. 2004. New 2,4-diamino-5-(2′,5′-substituted benzyl)pyrimidines as potential drugs against opportunistic infections of AIDS and other immune disorders. Synthesis and species-dependent antifolate activity. J. Med. Chem. 471475-1486. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi, T., T. Endo, T. Nakamura, H. Sakashitat, K. Kimurat, K. Ohnishit, Y. Kitamura, and A. Iwamoto. 2002. Dihydrofolate reductase gene polymorphisms in Pneumocystis carinii f. sp. hominis in Japan. J. Med. Microbiol. 51510-515. [DOI] [PubMed] [Google Scholar]
- 29.Trick, W., S. Fridkin, J. Edwards, R. Hajjeh, and R. Gaynes. 2002. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989-1999. Clin. Infect. Dis. 35627-630. [DOI] [PubMed] [Google Scholar]
- 30.Tzipori, S., and G. Widmer. 2008. A hundred-year retrospective on cryptosporidiosis. Trends Parasitol. 24184-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walzer, P., J. Foy, P. Steele, C. K. Kim, M. White, R. Klein, B. Otter, and C. Allegra. 1992. Activities of antifolate, antiviral and other drugs in an immunosuppressed rat model of Pneumocystis carinii pneumonia. Antimicrob. Agents Chemother. 361935-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitlow, M., A. Howard, D. Stewart, K. Hardman, L. Kuyper, D. Baccanari, M. Fling, and R. Tansik. 1997. X-ray crystallographic studies of Candida albicans dihydrofolate reductase. J. Biol. Chem. 27230289-30298. [DOI] [PubMed] [Google Scholar]