Abstract
PCSK9 (proprotein convertase subtilisin/kexin type 9) promotes degradation of the LDLR [LDL (low-density lipoprotein) receptor] through an as-yet-undefined mechanism, leading to a reduction in cellular LDLc (LDL-cholesterol) and a concomitant increase in serum LDLc. Central to the function of PCSK9 is a direct protein–protein interaction formed with the LDLR. In the present study, we investigated a strategy to modulate LDL uptake by blocking this interaction using specific antibodies directed against PCSK9. Studies using surface plasmon resonance demonstrated that direct binding of PCSK9 to the LDLR could be abolished with three different anti-PCSK9 antibodies. Two of these antibodies were raised against peptide epitopes in a region of the catalytic domain of PCSK9 that is involved in the interaction with the LDLR. Such antibodies restored LDL uptake in HepG2 cells treated with exogenous PCSK9 and in HepG2 cells engineered to overexpress recombinant PCSK9. This latter observation indicates that antibodies blocking the PCSK9–LDLR interaction can inhibit the action of PCSK9 produced endogenously in a cell-based system. These antibodies also disrupted the higher-affinity interaction between the natural gain-of-function mutant of PCSK9, D374Y, and the LDLR in both the cell-free and cell-based assays. These data indicate that antibodies targeting PCSK9 can reverse the PCSK9-mediated modulation of cell-surface LDLRs.
Keywords: antibody-mediated disruption, cholesterol, low-density lipoprotein receptor, proprotein convertase subtilisin/kexin type 9 (PCSK9), protein–protein interaction, surface plasmon resonance
Abbreviations: aa, amino acids; BODIPY®, boron dipyrromethene (4,4-difluoro-4-bora-3a,4a-diaza-s-indacene; CHRD, cysteine/histidine-rich domain; EGF-A, epidermal growth factor-like repeat A; FBS, fetal bovine serum; HEK-F cells, Freestyle™ suspension human embryonic kidney cells; IL1Ra, interleukin-1 receptor antagonist; LDL, low-density lipoprotein; LDLc, LDL-cholesterol; LDLR, LDL receptor; MOI, multiplicity of infection; PCSK9, proprotein convertase subtilisin/kexin type 9; RU, resonance units; SPR, surface plasmon resonance
INTRODUCTION
Cardiovascular disease is one of the great plagues of our affluent society, responsible for approx. 30% of all deaths each year. In addition to lifestyle risk factors, such as poor diet and smoking, genetic predisposition also contributes to the prevalence of the disease. Sufferers of familial hypercholesterolaemia exhibit highly elevated serum LDLc [LDL (low-density lipoprotein)-cholesterol] levels, leading to premature atherosclerosis [1]. Recently, a novel serine protease, PCSK9 (proprotein convertase subtilisin/kexin type 9) was identified as the third genetic locus for autosomal dominant hypercholesterolaemia [2]. PCSK9 is involved in the metabolism of LDLc through its action on the LDLR (LDL receptor), degrading the receptor via a mechanism which has yet to be fully elucidated [3,4].
Structurally, PCSK9 consists of a 30 aa (amino acid) signal peptide, followed by a prodomain (aa 31–152), a catalytic domain (aa 153–425) and a CHRD (C-terminal cysteine/histidine-rich domain) (aa 426–692) [3]. PCSK9 is synthesized as a ∼72 kDa precursor protein that undergoes autocatalytic cleavage in the endoplasmic reticulum [5]. After cleavage, the ∼14 kDa prodomain remains tightly associated with the active site, rendering the mature protein catalytically inactive [6,7]. PCSK9 undergoes a variety of post-translational modifications before being secreted efficiently from the cell [5,8].
In cell-culture systems, overexpression of PCSK9 resulted in a decrease in LDLR levels via a post-transcriptional mechanism [9]. However, PCSK9 does not affect the synthesis or trafficking of the LDLR [10]. The addition of media containing recombinant PCSK9 to HepG2 cells caused a reduction in the number of surface LDLRs [11–13]. It has now been established that PCSK9 forms a direct protein–protein interaction with the EGF-A (epidermal growth factor-like repeat A) domain of the LDLR that results in targeting of the LDLR to lysosomes for degradation [14,15]. This interaction appears to be necessary, and may be sufficient, for PCSK9-mediated LDLR degradation, a hypothesis supported by two studies showing that the addition of catalytically inactive PCSK9 to HepG2 cells decreased LDLR levels to the same extent as wild-type PCSK9 [16,17]. Further evidence supporting the importance of the PCSK9–LDLR interaction was the discovery that the natural PCSK9 gain-of-function mutant D374Y, which causes a particularly severe form of hypercholesterolaemia, has a significantly increased binding affinity for the LDLR [6,13].
In the present study, we investigated a strategy to disrupt the interaction between PCSK9 and the LDLR using specific antibodies directed against PCSK9, and examined the effects this has on LDL uptake in cellular models.
EXPERIMENTAL
cDNA and site-directed mutagenesis
The cDNA encoding human PCSK9 (GenBank® accession number AX207686) was amplified by PCR using gene-specific primers and incorporating a 3′ FLAG tag sequence (DYKDDDDK) to aid purification of the expressed product, and attB1 and attB2 sites at the 5′ and 3′ ends respectively. The resulting PCR product was inserted into the Gateway® donor vector pDONR221 (Invitrogen) using the BP clonase reaction (Invitrogen) and subsequently transferred to the Gateway® destination vector, pFastBacMam-2-rfa (a variant of the plasmid described in [18], containing additional polylinker sequences), via the LR clonase reaction (Invitrogen). To generate the PCSK9 mutants, site-directed mutagenesis was carried out on pDONR221-PCSK9-FLAG using the QuikChange® II XL kit (Stratagene). For the D374Y mutant, the mutagenic primers 5′-CATTGGTGCCTCCAGCTATTGCAGCACCTGCTTTG-3′ (forward) and 3′-GTAACCACGGAGGTCGATAACGTCGTGGACGAAAC-5′ (reverse) were used. For the RRRR218EL mutant, the mutagenic primers 5′-GGACGGGACCCGCCGCCGCAGAGAGCTAAGCAAGTGTGAC-3′ (forward) and 5′-GTCACACTTGCTTAGCTCTCTGCGGCGGCGGGTCCCGTCC-3′ (reverse) were used. The residues in bold indicate the site of mutation.
Antibodies
The affinity-purified anti-PCSK9 polyclonal antibody AF3888, raised against mature (aa 31–692) PCSK9, was from R&D Systems. The affinity-purified goat anti-PCSK9 polyclonal antibody (Ab28770), raised against peptide aa 679–692, was from Abcam. The anti-PCSK9 polyclonal antibodies, E6675 and E6660, were raised in rabbits against the peptides GEDIIGASSDCSTCFVSQSG (aa 365–384) and NVPEEDGTRFHRQASKC (aa 207–223) respectively and were affinity-purified using the respective peptides. A non-specific control polyclonal antibody [rabbit anti-IL1Ra (interleukin-1 receptor antagonist)] was from Abcam.
Cell culture
Spodoptera frugiperda (Sf9) cells (Invitrogen) were cultured in ExCell 420 insect cell medium (JRH Biosciences) at 27 °C with shaking at 125 rev./min. Cell densities were maintained between 0.3×106 and 5×106 cells/ml. HEK-F (Freestyle™ suspension human embryonic kidney) cells (Invitrogen) were cultured in Freestyle™ 293 serum-free medium (Gibco, Invitrogen) at 37 °C under 5% CO2 with shaking at 125 rev./min. HEK-F cells were routinely passaged to maintain a cell density between 0.5×106 and 3×106 cells/ml. HepG2 cells were cultured in advanced minimum essential medium (Gibco) containing 110 mg/l sodium pyruvate and non-essential amino acids, supplemented with 2 mM L-glutamine, 10% (v/v) FBS (fetal bovine serum) and 100 units/ml penicillin/streptomycin (all Gibco) at 37 °C under 5% CO2. The cells were routinely passaged with 50% (v/v) TrypLE™ Express in Versene (Gibco).
BacMam virus generation and transduction of mammalian cell lines
BacMam viruses were generated using standard procedures for the Bac-to-Bac system (Invitrogen) as described previously [18,19]. For transduction to enable protein purification, the HEK-F cells were cultured to a density of 2.5×106 cells/ml and to this, the BacMam virus inoculum at 108 pfu (plaque-forming units)/ml was added at 20% (v/v) to give a final cell density of 2×106 cells/ml and an MOI (multiplicity of infection) of 10. The transduced culture was then incubated for 3 days at 37 °C under 5% CO2 with shaking at 125 rev./min. For transduction to enable functional studies, the HepG2 cells were seeded in a 96-well tissue-culture plate at 30000 cells/well, and after 24 h, the medium was removed and replaced with 50 μl of recombinant BacMam virus inoculum, giving a MOI of ∼80. The cells were incubated with the virus for 1 h at 37 °C before replacing the inoculum with normal growth medium.
PCSK9 purification
The HEK-F cell culture medium from a 1 litre BacMam transduction of either wild-type PCSK9 or mutant D374Y was passed through a 0.22 μm pore size filter and mixed with 10 ml of anti-FLAG M2–agarose affinity chromatography resin (Sigma) overnight at 4 °C with rotation. The resin was collected in a 50 ml Econocolumn (Bio-Rad Laboratories) and washed twice with at least 10 column vol. of ice-cold PBS. The FLAG-tagged protein was eluted from the column using triple FLAG peptide (Sigma) diluted to 100 μg/ml in PBS. Peak fractions, determined by A280, were pooled and concentrated to a volume of 2.5 ml (approx. 5 mg/ml) using an Amicon Ultra-15 (30 kDa cut-off) concentrator (Millipore) before being loaded on to a HiLoad 16/60 Superdex 200 size-exclusion column (GE Healthcare) for separation in PBS at a flow rate of 1 ml/min (ÄKTA Explorer, GE Healthcare). Peak fractions were pooled and stored at −80 °C. To assess purity, 5 μg of the pooled protein was analysed on a NuPAGE Novex 4–12% acrylamide Bis-Tris gel (Invitrogen) under reducing conditions. Resolved proteins were visualized with GelCode Blue Stain Reagent (Pierce).
SPR (surface plasmon resonance)
Using the amine-coupling kit (Biacore) and the Biacore T100 immobilization wizard, purified LDLR ectodomain (His-tagged, R&D Systems) was immobilized to one of the four flow cells of a CM5 sensorchip to a level of approx. 150 RU (resonance units). Purified wild-type PCSK9 or the D374Y mutant were diluted in 10 mM Hepes (pH 7.4), 150 mM NaCl and 0.1 mM CaCl2 to a range of concentrations and passed over the sensorchip surface at a flow rate of 30 μl/min. Each cycle consisted of a 120 s analyte injection (the association phase), followed by a 300 s dissociation phase. Regeneration was achieved using a 60 s injection of 10 mM glycine/HCl (pH 2.0) with a 300 s stabilization period. The data were analysed using the Biacore T100 Evaluation software. Baselines were adjusted to zero for all curves and double-referenced by subtracting a sensorgram of buffer injected over the LDLR surface from the experimental sensograms to give curves representing specific binding. Curves were modelled assuming a simple 1:1 interaction to generate the kinetic data.
For antibody-blocking experiments, the ectodomain of the LDLR was amine-coupled to a CM5 sensorchip at an abundance of 2000 RU. Wild-type PCSK9 was diluted to 50 nM in Dulbecco's PBS plus Ca2+, Mg2+ and 0.005% Surfactant P20, and pre-incubated for 1 h at 4 °C with each antibody at 500 nM. The samples were then subject to SPR binding analysis using parameters identical with those described above. The Biacore T100 Evaluation software was used to calculate the maximum binding level achieved in the presence of each antibody.
BODIPY® [boron dipyrromethene (4,4-difluoro-4-bora-3a,4a-diazas-indacene)]–LDL uptake assay
HepG2 cells cultured in normal growth medium were seeded at 30000 cells/well in black walled 96-well tissue-culture plates. After 24 h, the medium was replaced with normal growth medium containing 1% FBS. After a further 18 h, the medium was replaced again, this time with normal growth medium lacking FBS, but containing 10 μg/ml BODIPY®–LDL (Invitrogen) as well as various concentrations (0–20 μg/ml) of purified wild-type PCSK9 or D374Y (1 μg/ml PCSK9=13.9 nM). The cells were then incubated for 6 h at 37 °C. After two washes with PBS, the fluorescence was measured at 485 nm excitation/535 nm emission using a Tecan ULTRA fluorescent plate reader. To assess cell viability, the Cell Titre-Glo® cell viability assay kit (Promega) was used following the manufacturer's instructions. Antibody-blocking experiments were carried out with the same protocol, this time using PCSK9 at 2.5 μg/ml and incorporating a pre-incubation step at 4 °C for 1 h before cell treatment with various concentrations (0–100 μg/ml) of antibody.
Alternatively, the cells were transduced 24 h after seeding with recombinant BacMam virus for wild-type, D374Y or RRRR218EL PCSK9. Cells were cultured in normal growth medium for 42 h, before exchange for serum-free growth medium containing 10 μg/ml BODIPY®–LDL. Plates were then incubated at 37 °C for 6 h and washed, and fluorescence was measured as described above. To conduct antibody-blocking experiments, the inoculum was replaced, immediately after BacMam transduction, with normal growth medium (+1% FBS) containing antibody at various concentrations (0, 10, 25 or 50 μg/ml). After incubation at 37 °C for 24 h, the medium was replaced with fresh antibody/medium mixture at the same concentration. After a further 18 h, the cells were replenished with antibody, this time in medium lacking FBS, but containing 10 μg/ml BODIPY®–LDL. After a further 6 h of incubation at 37 °C, the plates were washed with PBS and the fluorescence was measured as before.
RESULTS
Purification and functional validation of PCSK9
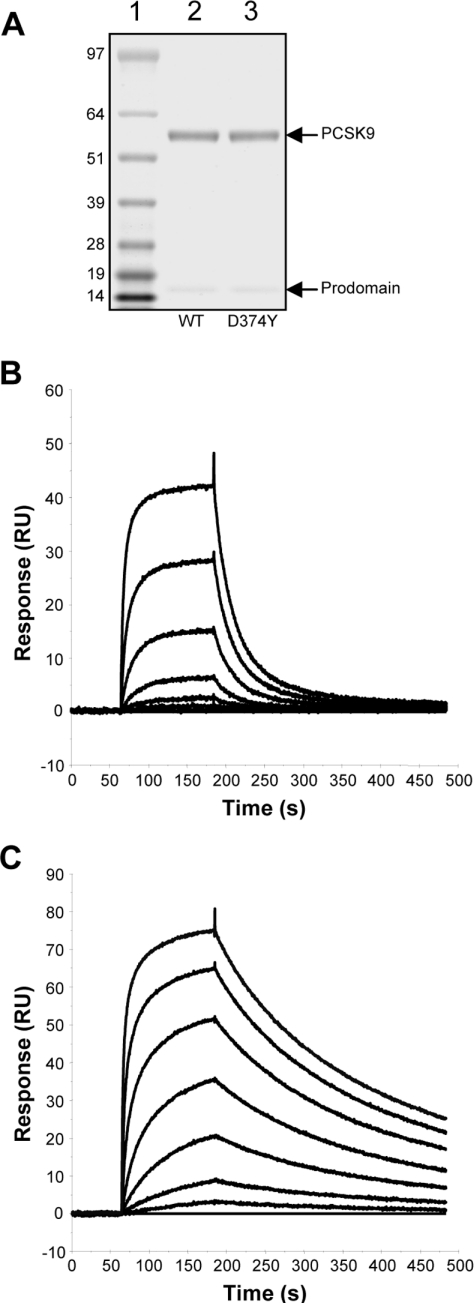
Wild-type PCSK9 and the D374Y gain-of-function mutant were expressed as C-terminal FLAG-tag-fusion proteins via BacMam-mediated transduction of HEK-F cells. The secreted mature form of PCSK9 was purified from the culture medium using anti-FLAG affinity chromatography followed by size-exclusion chromatography. The purified wild-type and D374Y forms of PCSK9 migrated with an apparent molecular mass of ∼60 kDa on SDS/PAGE (Figure 1A). Also visible, at ∼16 kDa, was the PCSK9 prodomain, which remained tightly associated throughout the purification, but dissociated under the denaturing conditions of SDS/PAGE. The identity of the preparation was confirmed by peptide mass fingerprinting and N-terminal sequencing (results not shown).
Figure 1. Binding affinity of wild-type PCSK9 and the D374Y mutant for the LDLR.
(A) FLAG-tagged versions of wild-type (WT) PCSK9 and the D374Y mutant were purified as described in the Experimental section, and 1 μg of each protein was subjected to electrophoresis on a 4–12% acrylamide NuPAGE Novex gel (Invitrogen) followed by staining with Coomassie Blue. Molecular-mass markers (sizes in kDa) are indicated on the left. The purified extracellular domain of the LDLR was immobilized on the surface of a CM5 sensorchip, and wild-type PCSK9 (B) and the D374Y mutant (C) at concentrations of 7000, 1750, 438, 109, 27, 7, 2 and 0 nM were passed over the surface as described in the Experimental section. The response from the reference surface was subtracted, and the baselines adjusted to zero to give response over time binding curves.
In order to functionally validate purified wild-type PCSK9 and the D374Y gain-of-function mutant, we characterized their binding to the LDLR using SPR. The purified extracellular domain of the LDLR was immobilized to the surface of a CM5 sensorchip via amine-coupling. Purified wild-type PCSK9 was passed over the surface of the chip at a range of concentrations, and the binding kinetics of the interaction were determined from the resulting sensograms (Figure 1B). By fitting a 1:1 binding model to these data, an affinity constant (Kd) of 810 nM at pH 7.4 was determined (Table 1), consistent with data published previously [7,13]. The D374Y mutant displayed an approx. 25-fold greater affinity for the LDLR than the wild-type protein (Figure 1C and Table 1), in agreement with previous reports [6,13]. The mean association (kon) and dissociation (koff) constants for each interaction are reported in Table 1. All χ2 values were below 5 RU2, indicating a good fit to the binding model.
Table 1. Binding affinities for wild-type PCSK9 and the D374Y mutant with the LDLR.
Results are means±S.E.M. (n=4).
| PCSK9 | kon (×10−4 M−1·s−1) | koff (×102 s−1) | Kd(nM) |
|---|---|---|---|
| Wild-type | 2.38 (±0.33) | 1.92 (±0.41) | 810 |
| D374Y | 12.8 (±2.09) | 0.42 (±0.05) | 33.0 |
Anti-PCSK9 antibodies block the PCSK9–LDLR interaction
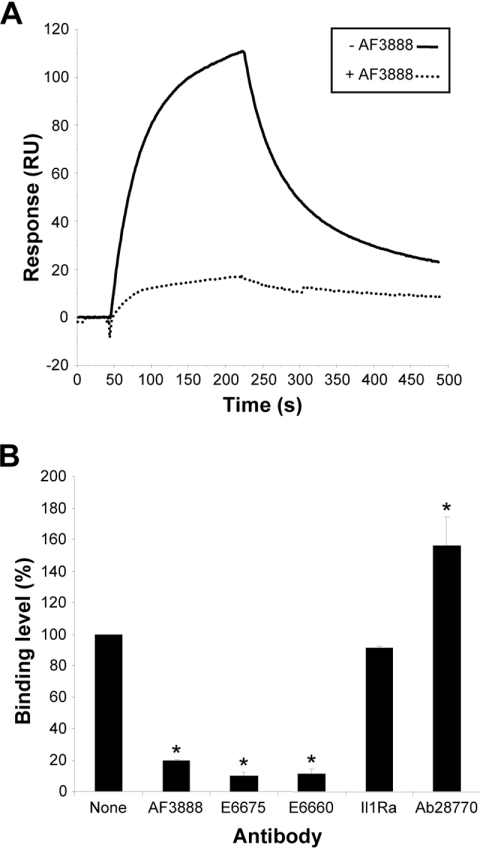
A potential strategy for blocking protein–protein interactions is to use an antibody directed against one of the binding partners. To investigate this approach, a variety of anti-PCSK9 antibodies were assessed for their ability to block the PCSK9–LDLR interaction in an SPR binding assay (Figure 2). Pre-incubation of PCSK9 with three affinity-purified anti-PCSK9 antibodies resulted in disruption of the interaction. These antibodies were AF3888 (a polyclonal antibody raised against mature PCSK9) and two polyclonal antibodies, E6675 and E6660, raised against PCSK9 peptide sequences (Figure 2). At a concentration of 500 nM, AF3888 reduced the binding of PCSK9 to the LDLR by 81%, whereas E6675 and E6660 reduced binding by 90 and 89% respectively. A fourth anti-PCSK9 antibody, Ab28770, raised against a C-terminal peptide did not block the binding of PCSK9 to the LDLR, but instead gave an apparent increased binding level (Figure 2B). This effect may be due to the inability of Ab28770 to prevent PCSK9 binding to the LDLR; the increased signal corresponds to the increased mass of the PCSK9–Ab28770 complex relative to PCSK9 alone. No effect was seen with the control antibody directed against IL1Ra (Figure 2B).
Figure 2. Antibody interference of the PCSK9–LDLR interaction.
LDLR was immobilized via amine coupling to a CM5 sensorchip. Wild-type PCSK9 at 50 nM was pre-incubated for 1 h at 4 °C with one of four anti-PCSK9 polyclonal antibodies (AF3888, E6675, E6660 or Ab28770) or a non-specific control antibody (anti-IL1Ra) and passed over the sensorchip surface as described in the Experimental section. (A) Representative sensorgram showing the significantly decreased binding level observed in the presence of 500 nM antibody AF3888. (B) Relative binding level observed in the presence of 500 nM of the indicated antibodies. Results are the means±S.E.M. for three independent experiments. *P<0.05 compared with no antibody.
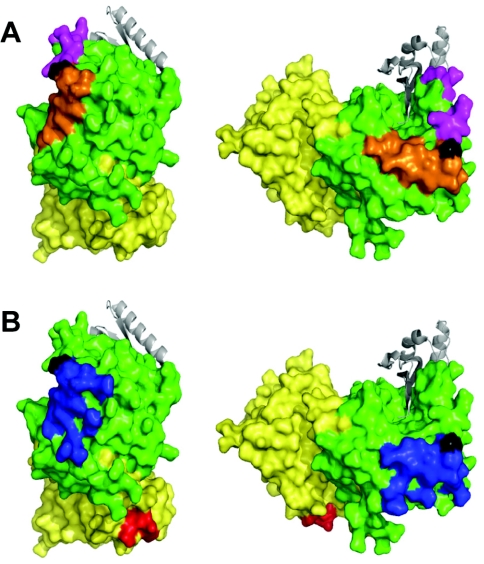
In order to locate the binding sites of the anti-PCSK9 antibodies, the immunizing peptide sequences were mapped on to a PCSK9 crystal structure. Consistent with our observations, the epitopes for the blocking antibodies E6675 (aa 365–384) and E6660 (aa 207–223) both map to a discrete area of the catalytic domain surrounding Asp374 (Figure 3A), a region which is critical in mediating the interaction with the LDLR EGF-A domain (Figure 3B) [14,15]. The epitope for Ab28770 (aa 679–692) lies within the CHRD, a region of PCSK9 that is not involved in binding to the LDLR (Figure 3B).
Figure 3. Predicted epitopes of the antibodies on the surface of PCSK9.
(A) The epitopes of the anti-peptide antibodies E6675 (orange) and E6660 (magenta) were mapped on to the surface of the crystal structure of PCSK9 (PDB code 2P4E). (B) The epitope for Ab28770 (red) and the region shown to interact with the EGF-A domain of the LDLR (blue) mapped on to the structure of PCSK9. The prodomain is shown in grey, the catalytic domain is shown in green, the CHRD is shown in yellow, and the site of the gain-of-function mutation D374Y is shown in black. Figure drawn using PyMOL (DeLano Scientific; http://pymol.sourceforge.net/).
Blocking the PCSK9–LDLR interaction inhibits PCSK9 function and restores cellular LDL uptake
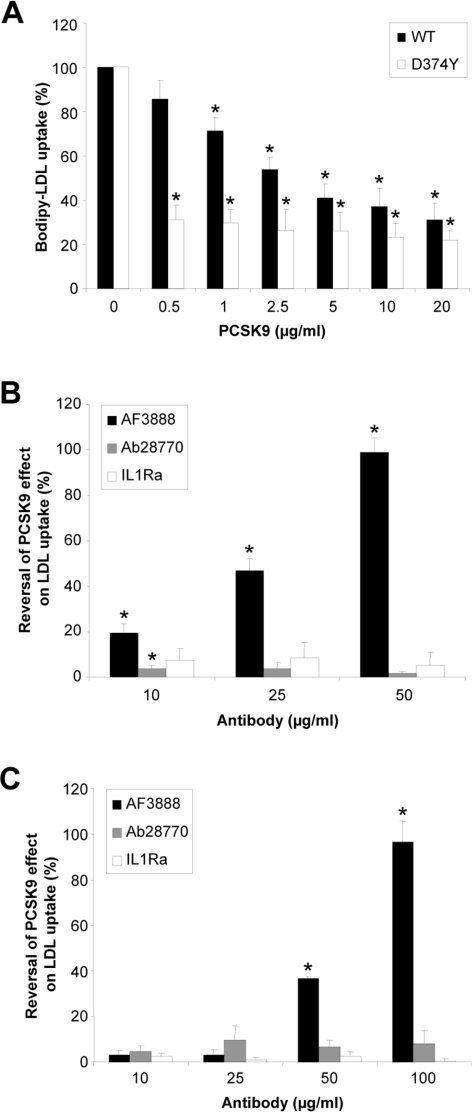
In order to monitor PCSK9 function, cell-surface LDLR levels were assessed indirectly by monitoring the uptake of fluorescently labelled LDL by HepG2 cells. Cells were treated for 6 h with various concentrations of purified recombinant PCSK9 (wild-type or D374Y) in the presence of BODIPY®–LDL particles. There was a dose-dependent reduction in BODIPY®–LDL uptake after treatment with purified PCSK9 (Figure 4A). The increased binding affinity of the gain-of-function mutant was clearly evident in this system. Following treatment with wild-type PCSK9 at a concentration of 1 μg/ml, there was a moderate decrease in LDL uptake of 28%, whereas a concentration of 20 μg/ml was required to get a near-maximal decrease in LDL uptake of 69%. In contrast, a near-maximal decrease in LDL uptake of 69% was obtained with D374Y at a concentration of 0.5 μg/ml.
Figure 4. Blocking the extracellular PCSK9–LDLR interaction inhibits PCSK9-mediated LDLR degradation and restores LDL uptake.
(A) HepG2 cells were incubated for 6 h with various concentrations of purified PCSK9 [wild-type (WT) or D374Y] in the presence of BODIPY®–LDL, the cells were then washed twice with PBS, and the fluorescence was measured at 485 nm excitation/535 nm emission. Results are presented as a percentage of the LDL taken up by the cells in the absence of PCSK9. Purified wild-type PCSK9 (B) or the D374Y mutant (C) at 2.5 μg/ml (34.7 nM) were pre-incubated for 1 h with increasing concentrations of antibody AF3888 or control antibodies IL1Ra and Ab28770 before addition to the HepG2 cells and determination of their relative LDL uptake. Results are the percentage reversal of the PCSK9 effect on LDL uptake, and are means±S.E.M. for three independent experiments each performed in triplicate. *P<0.05.
To assess whether blocking the PCSK9–LDLR interaction can prevent PCSK9-mediated LDLR degradation, LDL uptake was monitored following pre-incubation of purified PCSK9 using the blocking antibody AF3888, non-blocking antibody Ab28770 or control anti-IL1Ra antibody. The blocking antibody, but not the control antibodies, dose-dependently blocked the action of wild-type PCSK9 on the LDLR, thereby restoring uptake of LDL to normal levels (Figure 4B). An antibody concentration of 50 μg/ml (20-fold molar excess) was sufficient to give complete reversal of the PCSK9 effect on LDL uptake. In addition, we demonstrated that the action of the gain-of-function mutant, D374Y, could be similarly disrupted, obtaining complete reversal of its effect on LDL uptake with blocking antibody at 100 μg/ml (Figure 4C). The non-blocking and control antibodies did not reverse the effects of either the wild-type or mutant PCSK9 on BODIPY®–LDL uptake (Figures 4B and 4C).
Antibody-mediated inhibition of the effect of overexpressed PCSK9 in HepG2 cells
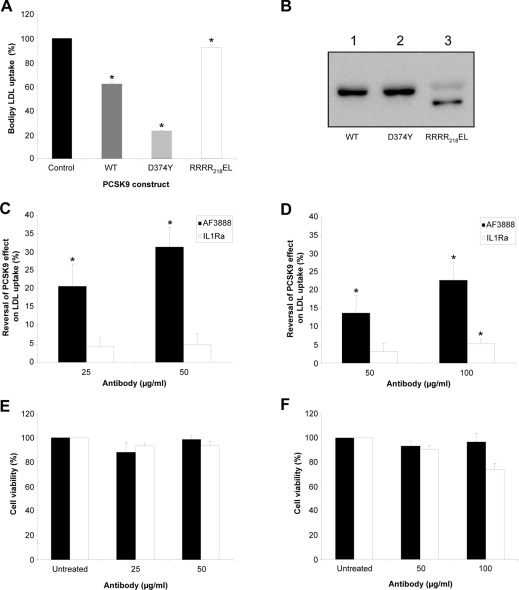
To assess further the function of the blocking antibody, a novel PCSK9-overexpression cellular system was employed. The relative amount of BODIPY®–LDL uptake was quantified after BacMam-mediated PCSK9 overexpression in HepG2 cells. LDL uptake by the cells was measured 48 h after treatment with recombinant BacMam virus expressing wild-type PCSK9, D374Y or RRRR218EL. The latter construct has been mutated from RFHR218QA to give a site within PCSK9 that is optimized for furin cleavage. This construct acts as a control for this experiment, since it is completely processed by furin, resulting in a truncated form of the protein (PCSK9-ΔN218) that is secreted, but is no longer sorted to endosomes and does not affect the levels of LDLR [8]. Overexpression of wild-type PCSK9 reduced LDL uptake by 38%, whereas the D374Y gain-of-function mutant construct reduced it by 77% (Figure 5A). As expected, the control construct, RRRR218EL, had little effect on LDL uptake by the HepG2 cells. All three constructs were expressed and secreted into the medium at similar levels (Figure 5B), indicating that the differences observed in LDL uptake between each construct were not due to variation in expression. The RRRR218EL mutant appeared as a band of approx. 53 kDa, representing PCSK9-ΔN218 (Figure 5B, lane 3), as described previously [8].
Figure 5. Antibody AF3888 counteracts the effect of PCSK9 expression on LDLR degradation in HepG2 cells.
(A) HepG2 cells were transduced with recombinant BacMam virus constructs encoding wild-type (WT) PCSK9, D374Y or RRRR218EL as described in the Experimental section. After 42 h, the medium was changed and replaced with serum-free growth medium containing 10 μg/ml BODIPY®–LDL. After 6 h, the cells were washed twice with PBS and the fluorescence was measured at 485 nm excitation/535 nm emission. Results are presented as a percentage of the LDL taken up by untransduced cells and are representative of three independent experiments, each carried out in triplicate. *P<0.05. (B) Western blot of secreted PCSK9 (wild-type, D374Y and RRRR218EL) in the medium of HepG2 cells after BacMam-mediated overexpression. PCSK9 was detected via its FLAG tag using an anti-FLAG M2 peroxidase-conjugated antibody. For antibody-blocking experiments, immediately after transduction with either wild-type (WT) PCSK9 (C) or D374Y (D) BacMam virus, the inoculum was replaced with normal growth medium containing various concentrations of antibody AF3888 or control antibody IL1Ra. After 24 h, the medium was replaced with fresh medium containing antibody at the same concentration. After a further 16 h, the medium/antibody mixture was replenished, this time with the addition of 10 μg/ml BODIPY®–LDL. After 6 h, the cells were washed twice with PBS and their relative fluorescence was determined. Results are presented as percentage reversal of the PCSK9 effect on LDL uptake. Cell viability (determined using the Cell Titre-Glo® kit from Promega) is presented as a percentage of control (BacMam-transduced, antibody-untreated cells) after antibody-blocking experiments for wild-type PCSK9 (E) and D374Y (F) transductions. For (C–F), black bars indicate AF3888, and white bars indicate IL1Ra. Results are the means±S.E.M. for three independent experiments each performed in triplicate, except for (D) which are the mean±S.E.M. for three independent experiments each performed in duplicate. *P<0.05.
For antibody-inhibition studies, blocking or control antibodies were incubated with the cells immediately after BacMam virus transduction, replenishing with antibody after 24 and 42 h, followed by quantification of BODIPY®–LDL uptake after a subsequent 6 h incubation period. Antibody AF3888 restored LDL uptake in a dose-dependent manner by 31% at a concentration of 50 μg/ml for wild-type PCSK9 (Figure 5C) and by 23% at a concentration of 100 μg/ml for D374Y (Figure 5D). There was no comparable increase in LDL uptake when cells were treated in the same way with the control antibody against IL1Ra. The viability of cells expressing wild-type PCSK9 or the D374Y mutant was unaffected by the presence of AF3888 (Figures 5E and 5F).
DISCUSSION
The present study represents the first demonstration that the interaction between PCSK9 and the LDLR can be successfully disrupted using antibodies directed against PCSK9. Furthermore, we have shown that blocking the interaction in this way reduces PCSK9-mediated LDLR degradation in a hepatocyte cell-based system. Three antibodies capable of mediating this effect were identified: a commercial polyclonal antibody raised against full-length PCSK9 (AF3888) and two polyclonal antibodies raised against peptides corresponding to regions of PCSK9 known to be crucial for LDLR binding [14,15]. A fourth anti-PCSK9 antibody to an epitope in the CHRD that is not involved in the interaction with the LDLR was used to demonstrate that antibody binding alone was not sufficient to disrupt the interaction.
In a cell-based functional assay monitoring uptake of fluorescently labelled LDL, pre-incubation of purified recombinant PCSK9 with antibody AF3888 led to a reduction in PCSK9-mediated LDLR degradation, allowing levels of cellular LDL uptake to return to normal. In order to assess further the effect of the antibody, the BODIPY®–LDL uptake assay was modified to incorporate BacMam virus-mediated overexpression of PCSK9 in place of treatment with purified protein. When transduced cells overexpressing PCSK9 (wild-type or the gain-of-function mutant D374Y) were incubated with the blocking antibody, AF3888, there was reversal of the PCSK9 effect on LDL uptake relative to transduced cells in the absence of blocking antibody. Additionally, the results of the present study demonstrate that the higher-affinity interaction between the D374Y mutant and LDLR could also be disrupted, albeit with higher concentrations of antibody than were required to disrupt the wild-type PCSK9–LDLR interaction.
The antibody-mediated reversal of the PCSK9 effect on LDL uptake was lower in the cell-based model when PCSK9 was overexpressed in the cells rather than being added exogenously as a purified protein. This may reflect that insufficient antibody was present to neutralize the continual overexpression of PCSK9. However, we did seek to overcome this by replenishing the medium during the time course of the experiment with the maximum amount of fresh antibody that the experimental system would allow. An alternative possibility is that there may be an additional cellular site of interaction between PCSK9 and LDLR that is inaccessible to the antibody. Indeed, there is evidence to suggest that PCSK9 may act upon the LDLR in a post-endoplasmic reticulum compartment before its secretion [10,20], in addition to the well-characterized effect of secreted PCSK9 on cell-surface LDLR. Nevertheless, our data clearly show that antibodies blocking the PCSK9–LDLR interaction can inhibit the action of PCSK9 endogenously produced in a cell-based system.
During the course of the present study, it was reported that the isolated EGF-A domain of the LDLR was sufficient to inhibit the LDLR-lowering effect of purified PCSK9 added exogenously to HEK-293 cells [21]. Epitope excision and MS using PCSK9 and the PCSK9–EGF-A complex revealed that aa 175–210 of PCSK9 remained bound to EGF-A after protease treatment and washing. Interestingly, this region overlaps with the peptide (aa 207–223) against which antibody E6660 was raised and which we have shown blocks the interaction of PCSK9 with the LDLR.
Despite the success of currently available lipid-lowering therapeutics, there are still >267 million people worldwide that have a plasma total cholesterol concentration of >200 mg/dl (5.17 mmol/l) [22], significantly increasing their risk of coronary heart disease, a condition that accounts for >30% of all deaths in North America [22]. In addition to their inhibition of cholesterol synthesis and the resultant up-regulation of the LDLR, the most widely prescribed class of drugs, the statins, also have the effect of up-regulating PCSK9 [23,24]. This potentially limits their efficacy, and suggests that supplementing this therapy with an agent inhibiting the action of PCSK9 may have significant benefit. This approach is supported by studies of Pcsk9-knockout mice, as well as humans with missense mutations in PCSK9, which both exhibit increased sensitivity to statins [23,25]. Furthermore, reducing PCSK9 expression, either via antisense or RNAi (RNA interference), effectively reduces plasma LDLc levels in rodents and non-human primates [26,27]. An alternative approach to prevent the biological effect of PCSK9 is to abrogate binding of PCSK9 to the LDLR using antibodies or small molecules [28], rather than reducing expression of endogenous PCSK9.
In summary, the present paper reports the first demonstration that antibodies directed against PCSK9 are able to specifically block its interaction with the LDLR. in vitro, such antibodies were able to inhibit the interaction between purified recombinant PCSK9 and the extracellular domain of the LDLR. Those antibodies capable of blocking the interaction mapped to the region on PCSK9 were reported recently to interact directly with the LDLR EGF-A domain [14,15]. We have demonstrated further using cell-based LDL-uptake assays that one of these blocking antibodies could inhibit the effect of both wild-type PCSK9 and the D374Y natural gain-of-function mutant. The results suggest that targeting PCSK9 using a specific antibody may represent a strategy for the treatment of dyslipidaemia by reversing PCSK9-mediated modulation of cell-surface LDLRs.
FUNDING
C. J. D. was in receipt of a studentship from the Biotechnology and Biological Sciences Research Council.
References
- 1.Rader D. J., Cohen J., Hobbs H. H. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. J. Clin. Invest. 2003;111:1795–1803. doi: 10.1172/JCI18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abifadel M., Varret M., Rabes J. P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 3.Horton J. D., Cohen J. C., Hobbs H. H. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem. Sci. 2007;32:71–77. doi: 10.1016/j.tibs.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez D. PCSK9: an enigmatic protease. Biochim. Biophys. Acta. 2008;1781:184–191. doi: 10.1016/j.bbalip.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Benjannet S., Rhainds D., Essalmani R., Mayne J., Wickham L., Jin W., Asselin M. C., Hamelin J., Varret M., Allard D., et al. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Biol. Chem. 2004;279:48865–48875. doi: 10.1074/jbc.M409699200. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham D., Danley D. E., Geoghegan K. F., Griffor M. C., Hawkins J. L., Subashi T. A., Varghese A. H., Ammirati M. J., Culp J. S., Hoth L. R., et al. Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia. Nat. Struct. Mol. Biol. 2007;14:413–419. doi: 10.1038/nsmb1235. [DOI] [PubMed] [Google Scholar]
- 7.Piper D. E., Jackson S., Liu Q., Romanow W. G., Shetterly S., Thibault S. T., Shan B., Walker N. P. The crystal structure of PCSK9: a regulator of plasma LDL-cholesterol. Structure. 2007;15:545–552. doi: 10.1016/j.str.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Benjannet S., Rhainds D., Hamelin J., Nassoury N., Seidah N. G. The proprotein convertase (PC) PCSK9 is inactivated by furin and/or PC5/6A: functional consequences of natural mutations and post-translational modifications. J. Biol. Chem. 2006;281:30561–30572. doi: 10.1074/jbc.M606495200. [DOI] [PubMed] [Google Scholar]
- 9.Park S. W., Moon Y. A., Horton J. D. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver. J. Biol. Chem. 2004;279:50630–50638. doi: 10.1074/jbc.M410077200. [DOI] [PubMed] [Google Scholar]
- 10.Maxwell K. N., Fisher E. A., Breslow J. L. Overexpression of PCSK9 accelerates the degradation of the LDLR in a post-endoplasmic reticulum compartment. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2069–2074. doi: 10.1073/pnas.0409736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cameron J., Holla O. L., Ranheim T., Kulseth M. A., Berge K. E., Leren T. P. Effect of mutations in the PCSK9 gene on the cell surface LDL receptors. Hum. Mol. Genet. 2006;15:1551–1558. doi: 10.1093/hmg/ddl077. [DOI] [PubMed] [Google Scholar]
- 12.Lagace T. A., Curtis D. E., Garuti R., McNutt M. C., Park S. W., Prather H. B., Anderson N. N., Ho Y. K., Hammer R. E., Horton J. D. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J. Clin. Invest. 2006;116:2995–3005. doi: 10.1172/JCI29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher T. S., Lo Surdo P., Pandit S., Mattu M., Santoro J. C., Wisniewski D., Cummings R. T., Calzetta A., Cubbon R. M., Fischer P. A., et al. Effects of pH and low density lipoprotein (LDL) on PCSK9-dependent LDL receptor regulation. J. Biol. Chem. 2007;282:20502–20512. doi: 10.1074/jbc.M701634200. [DOI] [PubMed] [Google Scholar]
- 14.Kwon H. J., Lagace T. A., McNutt M. C., Horton J. D., Deisenhofer J. Molecular basis for LDL receptor recognition by PCSK9. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1820–1825. doi: 10.1073/pnas.0712064105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D. W., Lagace T. A., Garuti R., Zhao Z., McDonald M., Horton J. D., Cohen J. C., Hobbs H. H. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 2007;282:18602–18612. doi: 10.1074/jbc.M702027200. [DOI] [PubMed] [Google Scholar]
- 16.Li J., Tumanut C., Gavigan J. A., Huang W. J., Hampton E. N., Tumanut R., Suen K. F., Trauger J. W., Spraggon G., Lesley S. A., et al. Secreted PCSK9 promotes LDL receptor degradation independently of proteolytic activity. Biochem. J. 2007;406:203–207. doi: 10.1042/BJ20070664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNutt M. C., Lagace T. A., Horton J. D. Catalytic activity is not required for secreted PCSK9 to reduce low density lipoprotein receptors in HepG2 cells. J. Biol. Chem. 2007;282:20799–20803. doi: 10.1074/jbc.C700095200. [DOI] [PubMed] [Google Scholar]
- 18.Condreay J. P., Witherspoon S. M., Clay W. C., Kost T. A. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. Proc. Natl. Acad. Sci. U.S.A. 1999;96:127–132. doi: 10.1073/pnas.96.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott M. J., Modha S. S., Rhodes A. D., Broadway N. M., Hardwicke P. I., Zhao H. J., Kennedy-Wilson K. M., Sweitzer S. M., Martin S. L. Efficient expression of secreted proteases via recombinant BacMam virus. Protein Expression Purif. 2007;52:104–116. doi: 10.1016/j.pep.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 20.Nassoury N., Blasiole D. A., Tebon Oler A., Benjannet S., Hamelin J., Poupon V., McPherson P. S., Attie A. D., Prat A., Seidah N. G. The cellular trafficking of the secretory proprotein convertase PCSK9 and its dependence on the LDLR. Traffic. 2007;8:718–732. doi: 10.1111/j.1600-0854.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 21.Bottomley M. J., Cirillo A., Orsatti L., Ruggeri L., Fisher T. S., Santoro J. C., Cummings R. T., Cubbon R. M., Lo Surdo P., Calzetta A., et al. Structural and biochemical characterization of the wild type PCSK9/EGF-AB complex and natural FH mutants. J. Biol. Chem. 2009;284:1313–1323. doi: 10.1074/jbc.M808363200. [DOI] [PubMed] [Google Scholar]
- 22.Pollex R. L., Joy T. R., Hegele R. A. Emerging antidyslipidemic drugs. Expert Opin. Emerg. Drugs. 2008;13:363–381. doi: 10.1517/14728214.13.2.363. [DOI] [PubMed] [Google Scholar]
- 23.Rashid S., Curtis D. E., Garuti R., Anderson N. N., Bashmakov Y., Ho Y. K., Hammer R. E., Moon Y. A., Horton J. D. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5374–5379. doi: 10.1073/pnas.0501652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubuc G., Chamberland A., Wassef H., Davignon J., Seidah N. G., Bernier L., Prat A. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 2004;24:1454–1459. doi: 10.1161/01.ATV.0000134621.14315.43. [DOI] [PubMed] [Google Scholar]
- 25.Berge K. E., Ose L., Leren T. P. Missense mutations in the PCSK9 gene are associated with hypocholesterolemia and possibly increased response to statin therapy. Arterioscler. Thromb. Vasc. Biol. 2006;26:1094–1100. doi: 10.1161/01.ATV.0000204337.81286.1c. [DOI] [PubMed] [Google Scholar]
- 26.Frank-Kamenetsky M., Grefhorst A., Anderson N. N., Racie T. S., Bramlage B., Akinc A., Butler D., Charisse K., Dorkin R., Fan Y., et al. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham M. J., Lemonidis K. M., Whipple C. P., Subramaniam A., Monia B. P., Crooke S. T., Crooke R. M. Antisense inhibition of proprotein convertase subtilisin/kexin type 9 reduces serum LDL in hyperlipidemic mice. J. Lipid Res. 2007;48:763–767. doi: 10.1194/jlr.C600025-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Shan L., Pang L., Zhang R., Murgolo N. J., Lan H., Hedrick J. A. PCSK9 binds to multiple receptors and can be functionally inhibited by an EGF-A peptide. Biochem. Biophys. Res. Commun. 2008;375:69–73. doi: 10.1016/j.bbrc.2008.07.106. [DOI] [PubMed] [Google Scholar]