Abstract
Schistosomiasis is a major neglected tropical disease that currently affects over 200 million people and leads to over 200,000 annual deaths. Schistosoma mansoni parasites survive in humans in part because of a set of antioxidant enzymes that continuously degrade reactive oxygen species produced by the host. A principal component of this defense system has been recently identified as thioredoxin glutathione reductase (TGR), a parasite-specific enzyme that combines the functions of two human counterparts, glutathione reductase and thioredoxin reductase, and as such this enzyme presents an attractive new target for anti-schistosomiasis drug development. Herein, we present the development of a highly miniaturized and robust screening assay for TGR. The 5-μl final volume assay is based on the Ellman reagent [5,5′-dithiobis(2-nitrobenzoic acid) (DTNB)] and utilizes a high-speed absorbance kinetic read to minimize the effect of dust, absorbance interference, and meniscus variation. This assay is further applicable to the testing of other redox enzymes that utilize DTNB as a model substrate.
Introduction
Schistosomiasis, a parasitic disease also known as bilharzia and snail fever, is caused by different species of flatworms, such as Schistosoma mansoni. It currently affects over 200 million people, mostly in developing countries,1 while an estimated 280,000 people die annually from the disease in sub-Saharan Africa alone.2 Praziquantel has remained the single drug of choice for the past decades,3,4 and preliminary reports of praziquantel-resistant cases, as well as the generation of praziquantel-resistant parasites in the laboratory,5–7 highlight the need for new drugs to treat the disease. Once inside humans, the schistosome parasite can survive decades8 without being destroyed by the immune system and the associated assault by various reactive oxygen species (ROS). One uniquely positioned S. mansoni enzyme, thioredoxin glutathione reductase (TGR), has been recently identified as a major component of this protective antioxidant “firewall.”9 In contrast to humans, who possess two distinct enzymes to maintain high cellular levels of reduced glutathione (GSH) and thioredoxin, glutathione reductase and thioredoxin reductase, which specifically recognize GSH and thioredoxin as substrates, respectively, TGR is a multifunctional enzyme that catalyzes the interconversion between reduced and oxidized forms of both GSH and thioredoxin.9–11 The existence of one worm enzyme in place of two different human counterparts represents a bottleneck in ROS processing and maintenance of redox balance in S. mansoni and has in turn made TGR an attractive new antiparasitic target.9,12 Indeed, RNA interference experiments and our recent identification of a novel, highly active TGR inhibitor indicate that inactivation of TGR has profound effects on S. mansoni survival rates both in culture and in worm-infected mice.9,13,14
We recently reported on the identification of furoxan (4-phenyl-3-furoxancarbonitrile, PubChem compound ID 1756; structure shown in Fig. 1C) as a powerful anti-schistosomiasis agent that acts as an inhibitor of TGR.12,13 Furoxan and related oxadiazole 2-oxides were discovered via a quantitative high-throughput screen (qHTS) of a reconstituted S. mansoni redox cascade consisting of TGR, glutathione, and peroxiredoxin 2 (an H2O2-reducing enzymatic component of the S. mansoni redox “firewall”15) by following the decrease of NADPH fluorescence.12 By performing HTS against both enzymes, we were able to address both targets simultaneously. However, weaknesses of this approach included post-screen target deconvolution required in order to further characterize the actives and the susceptibility of the assay to fluorescence interference from compound library members.16 While the screening assay used TGR and NADPH to feed a downstream enzyme, peroxiredoxin 2, which cannot be assayed independently, and the outcome of the cascade reaction was monitored by fluorescence intensity measurement, the assay described in the present work addresses TGR as a sole target. In order to further explore compound libraries for inhibitors of this recently validated anti-schistosomiasis target, we sought to develop and miniaturize a TGR-specific assay amenable to automated HTS. TGR can be assayed in a simple colorimetric assay format by following the catalytic reduction of 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) (Ellman's reagent) by NADPH,9 but an HTS-compatible assay17 is yet to be established. Here, we describe the development of a 1,536-well based assay for TGR that utilizes as a quantitative measure the increase in absorbance at 412 nm as a result of the reduction of DTNB. This assay and detection scheme can be modified to enable the qHTS of other important biochemical pathways and enzymes targeted in drug development for other diseases.
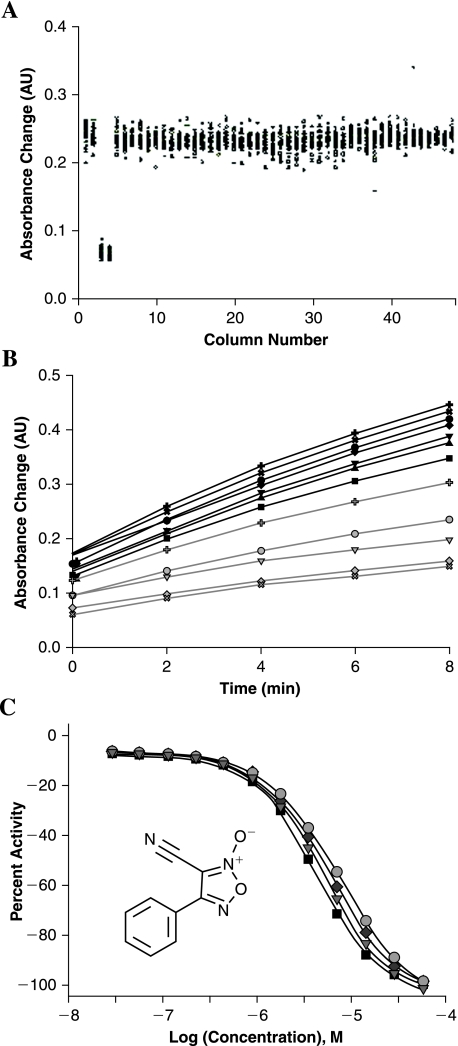
FIG. 1.
(A) Whole-plate scatter plot: columns 1, 2, and 5–48 represent complete reaction samples; columns 3 and 4 contain no-enzyme controls. Z′ factor of 0.70 and signal-to-background ratio of 3.5 were calculated based on the absorbance increase over 8 min (n = 64 wells each for the positive and negative controls). AU, absorbance units. (B) Single-well reaction time courses in the presence of furoxan (12 concentrations ranging from 57 μM [bottom line] to 28 nM [top line] in twofold dilution steps). The zero-time point refers to the first plate read, while the actual reaction initiation is estimated to take place approximately 1.5 min earlier. Reaction time-course data from a no-enzyme control, overlapping with those from the top furoxan concentrations, are provided as larger-size open circles. (C) Concentration-response curves of four furoxan samples delivered as 12-point dilution series with final concentrations ranging from 28 nM to 57 μM. (Inset) Structure of furoxan.
Materials and Methods
Reagents
NADPH and DTNB (Ellman's reagent) were obtained from Sigma-Aldrich (St. Louis, MO). Dimethyl sulfoxide (DMSO) certified ACS grade was from Fisher (Fair Lawn, NJ). The screening assay was performed in 0.1 M potassium phosphate, pH 7.0, containing 0.01% Tween-20.
Preparation of recombinant S. mansoni TGR
Recombinant TGR with a fused bacterial-type SECIS element was expressed in Escherichia coli strain BL21(DE3) (Invitrogen, Carlsbad, CA) and purified according to previously published methods.9 TGR concentration was determined from the flavin adenine dinucleotide absorption (ɛ463 = 11.3 mM−1 cm−1).
Compound plate
Furoxan powders were dissolved in DMSO to produce 10 mM initial stock solutions. The samples were then serially diluted row-wise in 384-well plate in twofold steps for a total of 12 concentrations. Upon completion of the serial dilution protocol, solutions from up to two 384-well plates were transferred to duplicate wells of a 1,536-well compound plate at 7 μl per well. The last two rows of the 1,536-well plate did not contain any test compound and were reserved for placement of positive and negative controls. A Flying Reagent Dispenser (FRD) (formerly Aurora Discovery, presently Beckman-Coulter, San Diego, CA) was used to dispense reagents into the assay plates.
qHTS protocol
Three microliters of reagents (100 μM NADPH in row 32 as no-enzyme control and 100 μM NADPH/15 nM TGR, in rows 1–31) were dispensed into 1,536-well Greiner (Monroe, NC) black clear-bottom assay plates. Compounds (23 nl) were transferred via a Kalypsys Pin-Tool equipped with a 1,536-pin array (10 nL slotted pins, V&P Scientific, Palo Alto, CA). The plate was incubated for 15 min at room temperature, and then a 1-μl aliquot of 500 μM NADPH was added, immediately followed by a 1-μl aliquot of 15 mM DTNB to start the reaction. The second addition of NADPH is done to minimize the effect of non-inhibitory redox cyclers, that is, compounds that might exhaust NADPH during the 15-min incubation and thus give the erroneous appearance of inhibition. The plate was transferred to a ViewLux™ high-throughput charge-coupled device (CCD) imager (PerkinElmer, Waltham, MA) where kinetic measurements (five reads, one read every 2 min) of the 5-thio-2-nitrobenzoic acid (TNB) absorbance were acquired using a 405 nm excitation filter.
Analysis of qHTS data
For activity calculations, delta values, computed as the difference in absorbance between last and first time points, were used, while the calculated slope, intercept, and the raw time-course data were stored in the database. Screening data were corrected and normalized, and concentration-effect relationships were derived by using in-house software (http://ncgc.nih.gov/pub/openhts/). Percentage activity was computed from the median values of the uninhibited, or neutral, control and the no-enzyme, or 100% inhibited, control, respectively.
Results and Discussion
In this report, we present the miniaturization of an enzymatic assay for the recently identified TGR using DTNB as a model substrate and its further optimization for HTS by the implementation of kinetic mode of data acquisition. DTNB was initially established as a reagent to quantitate thiol groups in native and denatured proteins.18 It reacts with thiols in a 1:1 ratio and produces TNB, which has an absorbance maximum of 412 nm and ɛ412 of 14,150 M−1 cm−1.19 Here, we utilized the catalytic reduction of DTNB by NADPH in the presence of TGR.9 The assay was miniaturized to 1,536-well format by volume reduction and appropriate adjustment of stock concentrations of enzyme and substrates to reflect the volumes being combined. The final enzyme concentration was elevated from 10 nM (used in the cuvette and 96-well plate experiments) to 15 nM in order to obtain a larger signal window, and 0.01% Tween-20 was included in order to minimize the effect of promiscuous inhibitors acting by colloidal aggregation.20 During optimization, extended time-course experiments were performed to evaluate the linearity of absorbance increase and to establish a time frame for monitoring the assay reaction during an actual screen. We observed slight loss of linearity after extended incubation when well absorbance values crossed above 0.7 absorbance units (data not shown). This appeared to be primarily due to loss of linear response of the plate reader and/or gradual deviation from the Beer's law than to substrate exhaustion because more than 70% of the limiting reagent (NADPH) was estimated to still remain at that advanced point. The kinetic window for the assay was thus set to 8 min, at which point the well absorbance reached by the enzyme-containing reaction was approximately 0.4 absorbance units and the substrate conversion was estimated to be below 20%. In order to evaluate the assay for stability and potential positional effects, an entire 1,536-well plate was tested in the following manner: columns 3 and 4 contained no-enzyme controls, whereas the rest of the plate was filled with complete enzyme mix. An example of such whole-plate scatter plot is shown in Fig. 1A. The use of kinetic read and the computation of reaction progress based on the increase of absorbance instead of end point values resulted in a signal-to-background ratio of 3.5 and Z′ factor of 0.721 (see discussion below).
After initial miniaturization and optimization, we proceeded to validate a screening protocol for this assay. Inhibitor samples were arrayed as a 12-concentration dose-response series across the 1,536-well source plate and were added (23 nl) to the assay mixture (3 μl in black clear-bottom assay plates). The assay plate was incubated for 15 min at room temperature, and then substrate was added to start the reaction. The plate was transferred to a ViewLux high-throughput CCD imager where kinetic measurements of the TNB absorbance were acquired. The assay protocol is detailed in Table 1. The enzymatic activity associated with each well was computed from the change in absorbance over the time course measured, normalized against control wells.
Table 1.
TGR Assay Protocol
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Reagent | 3 μl | Enzyme and no-enzyme solutions |
| 2 | Library compounds | 23 nl | 57.2 μM–28 nM titration series |
| 3 | Incubation time | 15 min | Compound interaction with targets |
| 4 | Reagent | 1 μl | Second equivalent of NADPH |
| 5 | Reagent | 1 μl | Substrate |
| 6 | Assay readout | 405 nm | Absorbance kinetic read |
| Step notes | |
|---|---|
| 1 | Black clear-bottom plates, 15 nM TGR and 100 μM NADPH final concentrations. |
| 2 | Pintool transfer of DMSO solutions of test compounds. |
| 3 | Room temperature incubation. |
| 4 | Solution of 500 μM NADPH; necessary in order to minimize the effect of noninhibitory redox cyclers that might exhaust NADPH during step 3 and thus give the erroneous appearance of inhibition. |
| 5 | DTNB at 3 mM final concentration. |
| 6 | ViewLux CCD imager (PerkinElmer), 405/10 nm excitation filter, excitation energy 4,000, exposure time 2 s, clear emission filter, one read per 120 s for 8 min |
To demonstrate the assay sensitivity and the overall screening process robustness, the previously identified TGR inhibitor furoxan was tested using the above protocol in a concentration-response format, at 12 twofold dilution points prepared as duplicates and covering the 28 nM–57 μM range. Two different sources of furoxan sample were used: one was purchased from Sigma-Aldrich, and the other was synthesized in-house using published methods.22,23 As illustrated by the well absorbance data shown in Fig. 1B, furoxan exhibited concentration-dependent inhibition towards TGR, recapitulating its property observed in the screening assay and upon testing in the cuvette-based assay.12 Furthermore, the four separate concentration-response curves were almost identical, with 50% inhibitory concentration values for the four samples falling within the 4.0–7.9 μM range (Fig. 1C). The Z′ factor and signal-to-background ratio associated with these experiments were 0.84 and 3.2, respectively.
Absorbance assays have remained very difficult to miniaturize and screen, yet they exist in great numbers in many academic labs, because they are easily run in low-throughput mode in 96-well plates or cuvettes. With the recent adoption of HTS by a number of academic institutions (at least 50 large- and mid-size HTS facilities exist in academic institutions in the United States alone), there has been a growing need by academic researchers, who have been historically unfamiliar with HTS, to develop robust screenable assays to run in these facilities. While the absorbance assay format has been widely used in lower-density microtiter plates, measuring absorbance in the 1,536-well density is rarely practical. At least two factors prevent the wide use of absorbance measurements in 1,536-well-based HTS. First, a significant fraction of organic molecules, as well as dust and buffer components, absorb light, thereby introducing unacceptably high levels of interference. Second, the short optical pathlength of the fluid in the well (generally between 2 and 4 mm) reduces the signal available for detection while making the measurements extremely sensitive to meniscus defects, bubbles, and volume variations. Generally, these problems have been addressed by the use of relatively large reaction volumes to increase the path length or by the application of assays that utilize intensely colored red-shifted substances.20 In the present system, we strived to maintain low assay volume while being additionally constrained by the absorption maximum and extinction coefficient of TNB. In order to improve the signal strength and to avoid the effect of the above interferences, we modified the detection format of the assay to measure the reaction progress in kinetic mode as opposed to collecting a single end-point read. While kinetic measurements are routinely used in assay development, their implementation in automated HTS remains challenging as they require either a fast detection instrument or involve multiple trips of each assay plate into and out of the reader. For relatively fast reactions only a fast-scanning reader or whole-plate imager (such as the ViewLux) can allow positionally unbiased and rapid repeated measurements of 1,536-well plates without slowing down the overall plate processing speed. Nevertheless, the benefits of performing HTS in kinetic mode are compelling. Effects of dust and absorbing but otherwise inert library members are subtracted out to reveal the true reaction course. The first time-point values associated with each assay well typically remain in the database and enable a further analysis to reveal quenching substances.24 Lastly, the signal-to-background ratios obtained from kinetic measurements are higher, and this may permit screening under conditions of lower substrate conversion.
In summary, we have described a 1,536-well based kinetic absorbance assay for the recently identified antiparasitic target TGR. The assay uses the catalytic reduction of DTNB by NADPH in the presence of TGR, and the application of kinetic read minimizes the interference related to fluidics, dust, or compound quenching. The excellent plate statistics observed upon repeated testing, as well as the concordance of 50% inhibitory concentration values obtained from different inhibitor samples, indicate that the present assay is sufficiently robust and sensitive to be utilized in HTS and could be applied to a wide variety of other redox enzyme targets.
Acknowledgments
This research was supported by the Molecular Libraries Initiative of the National Institutes of Health Roadmap for Medical Research and the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health, and in part by National Institutes of Health/National Institute of Mental Health grant 1R03MH076449 and National Institutes of Health/National Institute of Allergy and Infectious Diseases grant 5R01AI065622 (to D.L.W.).
Abbreviations
- CCD
charge-coupled device
- DMSO
dimethyl sulfoxide
- DTNB
5,5′-dithiobis(2-nitrobenzoic acid)
- qHTS
quantitative high-throughput screen
- ROS
reactive oxygen species
- TGR
thioredoxin glutathione reductase
- TNB
5-thio-2-nitrobenzoic acid.
References
- 1.Special Programme for Research and Training in Tropical Diseases. Report of the Scientific Working Group on Schistosomiasis 2005.TDR/SWG/07. Geneva: World Health Organization; 2006. [Google Scholar]
- 2.van der Werf MJ. de Vlas SJ. Brooker S. Looman CW. Nagelkerke NJ. Habbema JD, et al. Quantification of clinical morbidity associated with schistosome infection in sub-Saharan Africa. Acta Trop. 2003;86:125–139. doi: 10.1016/s0001-706x(03)00029-9. [DOI] [PubMed] [Google Scholar]
- 3.Cioli D. Pica-Mattoccia L. Praziquantel. Parasitol Res. 2003;90(Suppl 1):S3–S9. doi: 10.1007/s00436-002-0751-z. [DOI] [PubMed] [Google Scholar]
- 4.Doenhoff M. Kimani G. Cioli D. Praziquantel and the control of schistosomiasis. Parasitol Today. 2000;16:364–366. doi: 10.1016/s0169-4758(00)01749-x. [DOI] [PubMed] [Google Scholar]
- 5.Fallon PG. Doenhoff MJ. Drug-resistant schistosomiasis: resistance to praziquantel and oxamniquine induced in Schistosoma mansoni in mice is drug specific. Am J Trop Med Hyg. 1994;51:83–88. doi: 10.4269/ajtmh.1994.51.83. [DOI] [PubMed] [Google Scholar]
- 6.Cioli D. Pica-Mattoccia L. Archer S. Antischistosomal drugs: past, present … and future? Pharmacol Ther. 1995;68:35–85. doi: 10.1016/0163-7258(95)00026-7. [DOI] [PubMed] [Google Scholar]
- 7.Fallon PG. Sturrock RF. Niang AC. Doenhoff MJ. Short report: diminished susceptibility to praziquantel in a Senegal isolate of Schistosoma mansoni. Am J Trop Med Hyg. 1995;53:61–62. [PubMed] [Google Scholar]
- 8.Vermund SH. Bradley DJ. Ruiz-Tiben E. Survival of Schistosoma mansoni in the human host: estimates from a community-based prospective study in Puerto Rico. Am J Trop Med Hyg. 1983;32:1040–1048. doi: 10.4269/ajtmh.1983.32.1040. [DOI] [PubMed] [Google Scholar]
- 9.Kuntz AN. Davioud-Charvet E. Sayed AA. Califf LL. Dessolin J. Arnér ESJ, et al. Thioredoxin glutathione reductase from Schistosoma mansoni: an essential parasite enzyme, a key drug target. PLoS Med. 2007;4:e206. doi: 10.1371/journal.pmed.0040206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmgren A. Bjornstedt M. Thioredoxin and thioredoxin reductase. Methods Enzymol. 1995;252:199–208. doi: 10.1016/0076-6879(95)52023-6. [DOI] [PubMed] [Google Scholar]
- 11.Salinas G. Selkirk ME. Chalar C. Maizels RM. Fernandez C. Linked thioredoxin-glutathione systems in platyhelminths. Trends Parasitol. 2004;20:340–346. doi: 10.1016/j.pt.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Simeonov A. Jadhav A. Sayed AA. Wang Y. Nelson ME. Thomas CJ, et al. Quantitative high-throughput screen identifies inhibitors of the Schistosoma mansoni redox cascade. PLoS Negl Trop Dis. 2008;2(1):e127. doi: 10.1371/journal.pntd.0000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sayed AA. Simeonov A. Thomas CJ. Inglese J. Austin CP. Williams DL. Identification of oxadiazoles as new drug leads for the control of schistosomiasis. Nat Med. 2008;14:407–412. doi: 10.1038/nm1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simeonov A. Yasgar A. Jadhav A. Lokesh GL. Klumpp C. Michael S, et al. Dual-fluorophore quantitative high-throughput screen for inhibitors of BRCT-phosphoprotein interaction. Anal Biochem. 2008;375:60–70. doi: 10.1016/j.ab.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sayed AA. Williams DL. Biochemical characterization of 2-Cys peroxiredoxins from Schistosoma mansoni. J Biol Chem. 2004;279:26159–26166. doi: 10.1074/jbc.M401748200. [DOI] [PubMed] [Google Scholar]
- 16.Simeonov A. Jadhav A. Thomas CJ. Wang Y. Huang R. Southall NT, et al. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008;51:2363–2371. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 17.Inglese J. Johnson RL. Simeonov A. Xia M. Zheng W. Austin CP, et al. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 18.Ellman GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 19.Riddles PW. Blakeley RL. Zerner B. Ellman's reagent: 5,5′-dithiobis(2-nitrobenzoic acid)—a reexamination. Anal Biochem. 1979;94:75–81. doi: 10.1016/0003-2697(79)90792-9. [DOI] [PubMed] [Google Scholar]
- 20.Feng BY. Simeonov A. Jadhav A. Babaoglu K. Inglese J. Shoichet BK, et al. A high-throughput screen for aggregation-based inhibition in a large compound library. J Med Chem. 2007;50:2385–2390. doi: 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JH. Chung TD. Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 22.Boiani M. Cerecetto H. Gonzalez M. Cytotoxicity of furoxans: quantitative structure-activity relationships study. Farmaco. 2004;59:405–412. doi: 10.1016/j.farmac.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 23.Medana C. Ermondi G. Fruttero R. Di Stilo A. Ferretti C. Gasco A. Furoxans as nitric oxide donors. 4-Phenyl-3-furoxancarbonitrile: thiol-mediated nitric oxide release and biological evaluation. J Med Chem. 1994;37:4412–4416. doi: 10.1021/jm00051a020. [DOI] [PubMed] [Google Scholar]
- 24.Imbert P-E. Unterreiner V. Siebert D. Gubler H. Parker C. Gabriel D. Recommendations for the reduction of compound artifacts in time-resolved fluorescence resonance energy transfer assays. Assay Drug Dev Technol. 2007;5:363–372. doi: 10.1089/adt.2007.073. [DOI] [PubMed] [Google Scholar]