Abstract
Bacteria communicate and cooperate to perform a wide range of social behaviors including production of extracellular products (public goods) that are crucial for growth and virulence. Their expression may be switched on by the detection of threshold densities of diffusible signals [Quorum-Sensing (QS)]. Studies using the opportunistic pathogen Pseudomonas aeruginosa suggest that QS “cheats”—individuals that don't respond to the QS signal, but are still able to use public goods produced by others—have a selective advantage in the presence of QS cooperators. It is, however, unclear whether this type of social exploitation is relevant in clinical contexts. Here, we report the evolutionary dynamics and virulence of P. aeruginosa populations during lung colonization of mechanically ventilated patients in the absence of antimicrobial treatments. We observed a large diversity of QS phenotypes among initial colonizing isolates. This diversity decreased over a matter of days, concomitant with a gradual increase in the proportion of QS cheating mutants (lasR mutants), which were found in 80% of the patients after 9 days of colonization. These mutants often evolved from initial wild-type genotypes. The fitness advantage of the lasR mutants is almost certainly due to social exploitation, because this advantage was only apparent in the presence of QS wild-type cells. Crucially, ventilator-associated pneumonia occurred significantly earlier in patients predominantly colonized by QS wild-type populations, highlighting the importance of QS in this clinical situation. These results demonstrate that social interactions can shape the short-term evolution and virulence of bacterial pathogens in humans, providing novel opportunities for therapy.
Keywords: infection, quorum sensing, social interactions
Pseudomonas aeruginosa is an opportunistic pathogen responsible for acute infections in immunocompromised hosts and for chronic diseases in cystic fibrosis (CF) patients. In this organism, Quorum Sensing (QS) regulates expression of approximately 5% of the total genome, including many factors associated with virulence (1–3). QS signaling is mediated essentially by 2 secreted acyl-homoserine lactones (HSL), N-(3-oxododecanoyl)-HSL (3-oxo-C12-HSL) and N-butyryl-HSL (C4-HSL) synthesized by the LasI and RhlI enzymes, respectively. In vitro, these signaling molecules accumulate in the medium and, on reaching a threshold concentration, bind to their cognate transcriptional activators LasR and RhlR respectively to induce expression of target genes (4).
The importance of QS for the virulence of P. aeruginosa has been clearly established in various animal models (5–7). In patients, the detection of QS signaling molecules (8, 9) suggests QS does occur; however, recent reports on QS deficient strains isolated from both acute infections (10–13) and from chronically colonized CF patients (14) have cast doubts on the role of QS in clinical contexts. The evolution of QS mutants after several years of chronic colonization (14) could be explained by the physiological cost of QS gene expression (15, 16) or increased immunogenicity (14), but neither explanation suggests a link between QS and virulence. However, recent in vitro data (17) suggest that QS is a social phenomenon which provides a benefit to a bacterial population because it allows extracellular “public goods,” such as catabolic enzymes, to be produced when bacteria are at a density when these public goods will be most beneficial for bacterial growth and survival (1). QS deficient lasR mutants were found to have a selective advantage in the presence of QS wild-type bacteria in vitro (17, 18) because they benefit from the extracellular public goods produced by QS cooperators in the vicinity without paying the metabolic costs themselves. If this explanation for the evolution of QS mutants is correct in vivo, then QS should be associated with effective host exploitation, and hence increased virulence, in a clinical context. Whether QS cheaters exploit QS wild-type strains and affect virulence during acute clinical situations remains to be established.
Here, we analyze the evolution of QS and associated virulence in acute clinical situations. Sampling methods in previous acute clinical studies (10–13) have not been sufficient to determine whether or not QS mutants have a selective advantage; what might be responsible for any realized advantage of the mutants; and whether or not mutants affect virulence. We followed the QS phenotype and the population dynamics of P. aeruginosa during the initial colonization (up to 20 days) and progression toward infection of intubated patients. We found that QS deficient lasR mutants rapidly increased in frequency, often evolving from QS wild-type genotypes in a matter of days. This growth rate advantage of QS mutants was only apparent in the presence of cocolonizing QS cooperators, suggesting that social exploitation provides a fitness advantage to the mutants. Furthermore we observed that virulence, as measured by the onset of Pseudomonas ventilator-associated pneumonia (VAP), was greatest in patients harboring only QS wild-type isolates.
Results and Discussion
Phenotypic Diversity of QS Phenotypes Among Colonizing Lung Isolates.
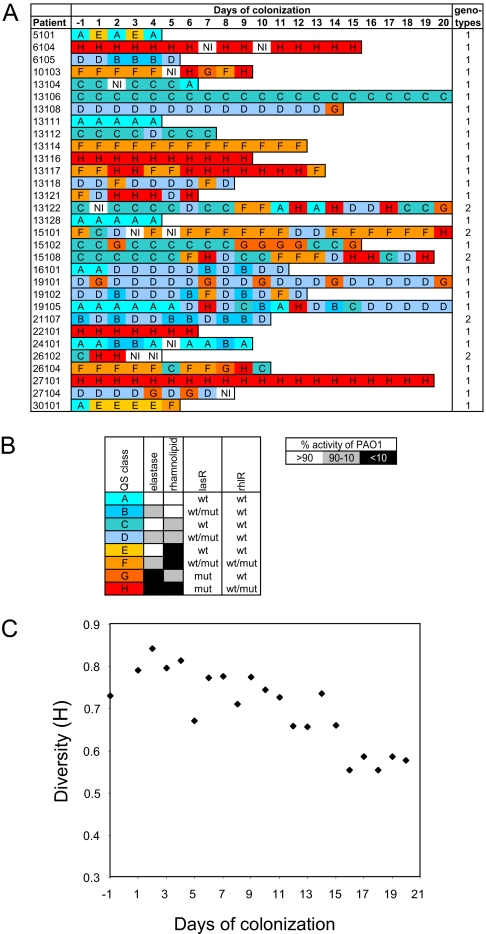
We followed prospectively 31 intubated patients (intubation times of 3–20 days), hospitalized in intensive care units of 13 different European hospitals and colonized by P. aeruginosa. Importantly, none of the patients received antipseudomonal drugs during the observation period. This observation allowed us to follow the adaptive behavior of P. aeruginosa in the absence of any external selective pressure. Every day we collected from each patient one tracheal aspirate, from which we obtained one P. aeruginosa isolate. To gain a global picture of the QS phenotypes, we determined the production of elastase and rhamnolipids in the 364 collected isolates, which are mainly under the control of the lasR and rhlR QS systems, respectively (Fig. 1). We observed a remarkable diversity in QS phenotypes, which allowed us to allocate the 364 isolates to 8 distinct classes (A through H) (Fig. 1 A and B). The majority of the patients (23 of 31) harbored at least 1 QS deficient isolate (classes E to H) at some point. We then calculated the diversity (H) of the QS phenotypes across the 31 patients through time by using the Shannon–Weaver index (19). Diversity among the initial colonizing isolates was high but decreased steadily during the colonization period (Fig. 1C). This loss in diversity was mainly due to a decrease in isolates of QS classes A and B, which were found in 30% of the patients on day −1, but had disappeared by day 15. The initial diversity probably reflects the different sources from which the isolates were acquired (endogenous, patient transmission, environment). Importantly this progressive loss of diversity was not due to selective pressure of antibiotics in our patient population and only resulted from the conditions that prevail in the lung environment.
Fig. 1.
QS phenotype diversity of isolates from intubated patients. (A) Elastase and rhamnolipid production of all 364 isolates was measured by ECR assay and halo diameter on modified SW-blue plates, respectively, and scored with respect to PAO1 activity (set to 100%), as shown in B. Genotypes of isolates from each individual patient were compared by RAPD. (B) The lasR and rhlR genes were sequenced in at least one isolate of each QS class from every patient. The presence of mutated lasR and rhlR alleles in the remaining isolates was assessed by high resolution melting (HRM) analysis of the corresponding PCR amplicons. Isolates of this class may carry either a wild-type (wt) or a mutated (mut) allele of the gene. (C) Diversity (H) of colonizing isolates across the 31 patients as calculated by the Shannon–Weaver index (19).
Rapid Emergence of lasR Mutants Among Isolates from Intubated Patients.
To understand how selection acts on QS during colonization of intubated patients, we sequenced both the lasR and rhlR genes in the first isolate obtained from each patient and then in subsequent isolates belonging to a different QS class (Fig. 1B, Table 1). As expected, QS proficient isolates (class A) all carried wild-type alleles of these genes, whereas QS deficient isolates (classes F to H) carried either a mutant lasR and/or rhlR allele(s). Surprisingly, isolates of classes C and E characterized by decreased rhamnolipid production all carried a wild-type rhlR gene, suggesting the presence of mutations in other regulatory loci. We identified 30 different genetic alterations in lasR, including 19 missense mutations, 7 base pair deletions and insertions, as well as 4 integrations of IS elements (Table 1). Most of the amino acid substitutions were predicted to be nontolerant by the SIFT algorithm (Table 1) (20) and were located either in the autoinducer binding-multimerization domain (amino acid 15–164) or in the DNA-binding domain (amino acid 174–231) of LasR (Table 1), suggesting loss of LasR function. We randomly chose 4 distinct lasR mutations (199IS, T115I, E196D, C79R) and complemented the corresponding isolates by inserting a Tn7::lasR cassette (10) at a neutral site on the chromosome. In all cases, wild-type elastase levels were restored, confirming that the lasR mutations were solely responsible for decreased elastase activity in these isolates.
Table 1.
Genotypes and LasR mutations of P. aeruginosa isolates
| Patient | SNP-type* | LasR, D −1 | LasR |
|---|---|---|---|
| 05101 | 6C22 | wt | |
| 06104 | AF9A | Y47-IS5 | |
| 06105 | D421 | L236P† | |
| 10103 | D421 | V199-IS30 | |
| 13104 | 0C1A | wt | |
| 13106 | 4F8A | wt | |
| 13108 | 1BAE | wt | E196D (14) |
| 13111 | 85AA | wt | |
| 13112 | C40A | wt | |
| 13114 | F469 | L148P | |
| 13116 | F469 | T222I | |
| 13117 | F469 | L148P | L148P + D156G (2), L148P + R224H† (8) |
| 13118 | 239A | wt | G191C† |
| 13121 | 239A | wt | V184A (2), C79R (3) |
| 13122 | F661 | wt | S204F† (6), T115I† (12) |
| E429 | NA | A231V† (20) | |
| 13128 | 062A | wt | |
| 15101 | 0C2E | wt | R180G† (2) |
| 239A | NA | P85 + 1bp (2), T75 + 1bp (10), Q94End (20) | |
| 15102 | E429 | wt | D43 81bp (2), Y93 81bp (2), Y93 408bp (9), C203R† (15) 408bp (9), C203R† (15) |
| 15108 | E429 | wt | |
| 239A | NA | L17 1bp (6), A134 1bp (6), A134 6bp (15), I197-IS4 (16) 6bp (15), I197-IS4 (16) |
|
| 16101 | 0C2E | wt |
 (3) (3) |
| 19101 | F469 | ||
| 19102 | 6D92 | A231V† | |
| 19105 | 7C2E | wt | F210L† (8) |
| 21107 | 6D92 | A231V† | |
| F469 | NA | wt (10) | |
| 22101 | F469 | P74L† | |
| 24101 | 2C25 | wt | T193I† (2) |
| 26102 | F469 | wt | D156Δ1bp (1) |
| 6D92 | NA | A231V† (2) | |
| 26104 | F469 | wt | D156 1bp (9) 1bp (9) |
| 27101 | EC4A | IS | |
| 27104 | 0812 | L110Q† | |
| 30101 | D421 | wt | T115I† (5) |
Numbers in parentheses indicate day when mutation first occurred;  indicates undefined deletion; NA, isolate not available. Amino acids 15–164, autoinducer binding, multimerization domain; amino acids 174–231, DNA binding domain. IS sequences were identified with IS-Finder (http://www-is.biotoul.fr/is.html).
indicates undefined deletion; NA, isolate not available. Amino acids 15–164, autoinducer binding, multimerization domain; amino acids 174–231, DNA binding domain. IS sequences were identified with IS-Finder (http://www-is.biotoul.fr/is.html).
*SNP type was established using the Clondiag array as described in ref. 22.
†Amino acid substitutions predicted by the SIFT algorithm to be nontolerated.
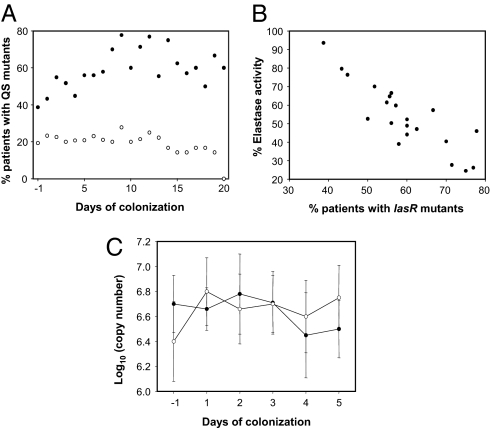
To test whether the proportion of QS mutants fluctuates during colonization, we calculated the daily average number of lasR and rhlR mutant alleles present in the isolates from all 31 patients. Strikingly, we found a significant increase (logistic regression: F1, 19 = 13.95, P < 0.01) in the proportion of patients harboring lasR mutants through time (filled circles in Fig. 2A), with peak frequency occurring on days 9 and 12. At this time, 80% of the patients were colonized by a lasR mutant, compared to 40% at the beginning of colonization. As expected, there was a highly significant relationship between the proportion of lasR mutants and mean elastase activity of the colonizing isolates (across all patients) through time (F1,19 = 54.17, P < 0.001) (Fig. 2B). In contrast, there was no significant change (logistic regression: F1,19 = 3.57, P < 0.1) in the proportion of patients harboring rhlR mutants through time (open circles in Fig. 2A). These data strongly suggest a short-term selective advantage of lasR, but not rhlR, mutants through time.
Fig. 2.
Emergence of QS mutants and population densities. (A) The proportion of patients harboring isolates with mutations in lasR (●) and rhlR (○) genes through time. Day −1 denotes the first day of detectable P. aeruginosa colonization. Note that sample sizes decreased from 31 patients to 5 patients by day 20. (B) The relationship between proportion of patients harboring isolates with lasR mutations and mean elastase production of the given isolates. Each data point represents a single day. (C) Comparison of bacterial loads between patients. Data shows mean (SEM) genomic copy number in patients harboring isolates with only wild-type (●) or mutant (○) lasR alleles. P. aeruginosa genomic copy numbers were determined on total DNA extractions from daily tracheal aspirates and quantified by qRT-PCR. Data only shows the initial periods of colonization because of small sample sizes in the wild-type group after day 5.
Why should there be a selective advantage of lasR but not rhlR mutants? The classical QS circuit assumes a hierarchy in which lasR partially controls rhlR expression (21), hence inhibition of genome-wide expression would be greater in lasR deficient mutants. The lasR deficient mutants are therefore likely to outcompete rhlR deficient mutants, and there would be little selection acting on rhlR in the presence of a lasR deficient mutation. It is also possible that some products which are under specific rhlR, but not lasR, control may in fact confer a direct fitness advantage.
We next addressed whether changes in the frequency of lasR mutants resulted only from selection acting on existing genetic variation or whether lasR mutants also evolved from wild-type genotypes during the course of colonization. We determined the genetic background of all isolates by RAPD [supporting information (SI) Fig. S1 and Fig. S2] and determined the clone type of 1 representative isolate of each phenotype by using the Clondiag P. aeruginosa microarray (22). We identified 19 different clones colonizing the 31 patients (Table 1). Crucially, the majority of the patients (26/31) were colonized by a single genotype, and the remaining 5 patients harbored isolates with 2 genotypes (Table 1). In 7 patients colonized by a single genotype with an initially wild-type lasR allele, lasR mutants were subsequently detected and elastase production showed a significant decrease (paired t = 3.17, P = 0.025), relative to when the patients were initially colonized, at this time. Given the high diversity of SNP types across the whole patient cohort, it is highly unlikely that 2 colonizations by genotypes with the same SNP type would occur. Furthermore, we observed lasR mutants following wild-type lasR alleles, but not vice versa: If multiple colonizations were occurring, we would expect to see both. These findings strongly suggest that lasR mutants evolved from the colonizing genotypes during the course of the study.
As a cooperative behavior, QS is amenable to exploitation by noncooperators (cheats), which do not contribute to the production of public goods (17). Indeed, noncooperating lasR mutants were shown to emerge in vitro under conditions requiring QS for growth (18). To determine whether the fitness advantage of lasR mutants in the colonizing populations is due to social exploitation of QS wild-type, rather than other direct benefits, we compared bacterial population densities between patients in which only wild-type lasR alleles (n = 10) or only lasR mutants (n = 11) were present. There was no difference in bacterial load between these 2 groups (Fig. 2C) [repeated measures analysis using residual maximum likelihood (REML): P > 0.2], demonstrating that the fitness advantage of QS noncooperators is only apparent in the presence of QS wild-type isolates. Consistent with theory (23–26) and in vitro work (17), these data suggest that social exploitation of exoproducts might play a crucial role in determining fitness of QS nonresponders. More surprising is the lack of growth advantage of QS responders over QS nonresponders when in isolation from each other, given the clear and consistent benefit of QS for colonization and infection in animal models (27). It is possible that QS has both costs and benefits during initial colonization of intubated patients and that some of the beneficial exoproducts can be exploited by QS nonresponders. The benefits of QS may however be more apparent once infections are established.
Within-Host Population Dynamics.
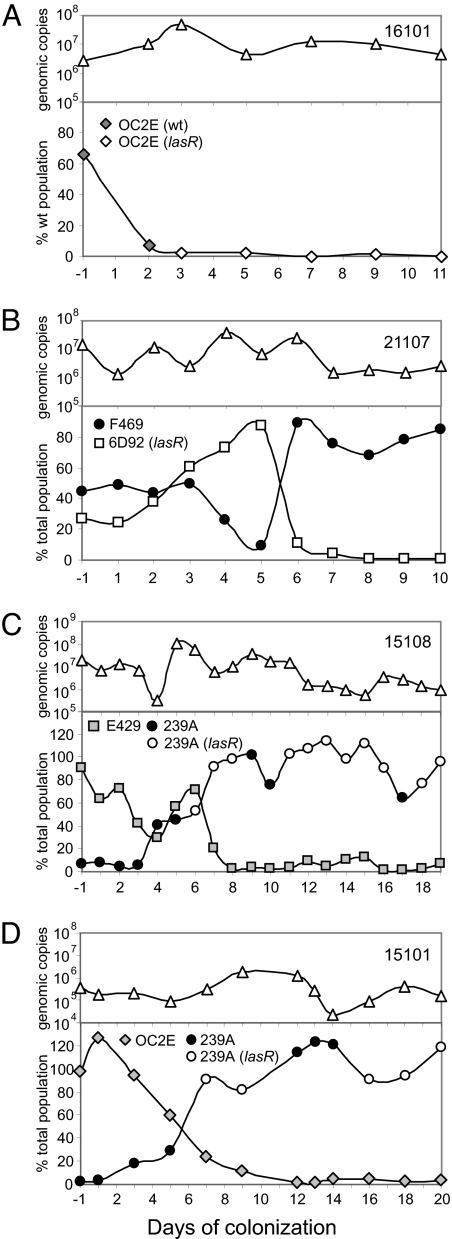
To further explore the selective advantage of lasR mutants, we followed the within-host dynamics of P. aeruginosa populations. We extracted total genomic DNA from the daily tracheal aspirates of 4 patients, 3 of whom were colonized by 2 genotypes (patients 21107, 15108, and 15101). The proportion of each genotype was determined by qRT-PCR by using genotype specific primer pairs as detailed in the legend of Fig. 3. In 3 cases (patients 15108, 15101, and 16101), lasR mutants evolved from wild type, which rapidly decreased in frequency (Fig. 3 A, C, D). In the 2 mixed genotype colonizations, the competing clones, both of which had wild-type lasR alleles, decreased in frequency with the increase in the lasR mutant (Fig. 3 C and D).
Fig. 3.
P. aeruginosa population dynamics in intubated patients. Total P. aeruginosa genomic copy numbers were determined by qRT-PCR on total genomic DNA preparations from tracheal aspirates by using the rpsL primer pair and are expressed as genomic copies/g aspirate (A–D, Upper). (A) QS population dynamics in patient 16101 colonized by a single genotype. To discriminate in the genomic DNA preparations between lasR wild-type and mutant populations, a lasR primer pair was designed which amplifies a 200 bp DNA fragment that is absent in the lasR deletion mutants (isolates from days 3 to 11). The amount of total P. aeruginosa copies was determined in the same DNA samples by using the rpsL primer pair. (Lower) The graph shows the percentage of lasR wild-type copies among the total P. aeruginosa population. (B, C, D) QS population dynamics in patients colonized by 2 genotypes. In patient 21107, populations of clone F469 (lasR wild-type, exoS+, exoU−) and clone 6D92 (lasR mutant, exoS−, exoU+) were quantified by using primer pairs specific for exoS and exoU, respectively. The relative proportions of the 2 populations are expressed as a percentage of the total number of P. aeruginosa genomic copies determined in the same DNA preparations by using the rpsL primer pair (B Lower). The proportions of clones E429 (PA0636+, PA0722−) and 239A (PA0636−, PA0722+) in patient 15108 were quantified by using primer pairs specific for the variable genes PA0636 and PA0722, respectively, and expressed as percentage of total P. aeruginosa genomic copies (C Lower). The proportions of clone OC2E (PA0636+, PA0728−) and clone 239A (PA0636−, PA0728+) in patient 15101 were quantified similarly by using primer pairs specific for variable genes PA0636 and PA0728, respectively (D Lower) (22). Because only 1 isolate was available per day, which usually corresponded to the most abundant clone, the lasR allele of the isolate from the minor population could not be assessed.
In the fourth patient (21107) (Fig. 3B) one initially colonizing clone was already a lasR mutant (Fig. S3). Despite a rapid increase during the first 5 days of colonization, this clone rapidly decreased in frequency and the cocolonizing wild-type lasR genotype increased in frequency (Fig. S3). It is not clear what caused this sudden reversion in frequencies, but it suggests that the outcome of short-term competition between wild-type and lasR mutants becomes much less predictable when the bacteria have different genetic backgrounds.
Another striking feature of the within-host dynamics is the apparent fluctuations in genotype frequencies of the wild-type and mutant lasR genotypes (Fig. 3 C and D, Fig. S1 and Fig. S2). The lasR mutants rapidly increased in frequency but generally failed to completely eliminate the wild-type. In the one patient where such fluctuations did not occur (16101) (Fig. 3A), the sampling period was terminated before potential fluctuations were observed. This result is consistent with our observation in the whole patient cohort, which showed a steady increase in the proportion of lasR mutants over the first 11 days, followed by a plateau or even a decrease (Fig. 2A). Such fluctuations are consistent with interactions between public good producers and exploiters in spatially structured environments (28) but not with a fitness advantage of lasR mutants that is independent of social exploitation. The occurrence of spatially structured microcolonies in the current study is further strongly supported by the observation that patients colonized by a single genotype sometimes contained multiple lasR alleles (patients 13117, 13121, 15102) (Table 1).
Virulence of QS Populations.
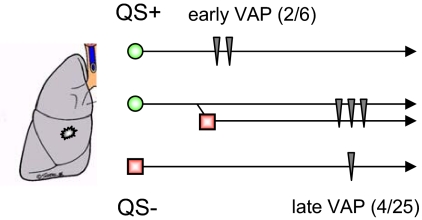
Finally, we considered how QS affects virulence. Of the 31 patients, 6 developed P. aeruginosa (VAP). Two of them (13111, 13128) were colonized exclusively by lasR/rhlR wild-type isolates; 3 patients (16101, 21107, 24101) harbored both lasR wild-type and mutant isolates, and one patient (13116) carried only lasR/rhlR double mutants (Fig. 1A). The proportion of patients who developed VAP did not differ between patients harboring exclusively QS wild-type or mutant isolates (Fishers Exact Test, P > 0.2). However, VAP occurred significantly earlier (day 4–5) in the 2 patients exclusively colonized by QS wild-type isolates, but later (day 9–11) in patients also harboring lasR mutant isolates (t test; t = 11.4, P = 0.001) (Fig. 4). The bacterial factors that result in the progression from colonization to acute infection are unclear, but a positive relationship has been observed between bacterial density and the probability of an infection developing (29). This relationship suggests that the cooperative production of public goods through QS might in fact result in more efficient longer term exploitation of hosts, and hence increased virulence. A decrease in virulence with increasing within-host diversity (and hence competition) where extracellular public goods are important for within-host growth is consistent with recent theory (30, 31). However, in circumstances where extracellular products are less important for growth, within-host competition may favor genotypes that exploit the host most rapidly, resulting in the evolution of increased virulence (32). The relationship between within-host competition and virulence is further complicated where anticompetitor toxins are produced (33).
Fig. 4.
QS and virulence of clinical P. aeruginosa populations. Intubated patients were colonized by either only QS wild-type isolates (QS+, 6 patients), only QS mutant isolates (QS, 5 patients) or mixed QS wild-type and mutant populations (20 patients). VAP (vertical arrows) occurred earlier (days 4–5) in patients harboring only QS wild-type populations and later (days 9–11) in patients cocolonized by QS mutant isolates.
Conclusions
In summary, we have shown that P. aeruginosa isolates colonizing intubated patients present a remarkable diversity of QS phenotypes, most of which result from the rapid emergence of lasR mutants during the first 10 days of colonization. The most convincing explanation for this rapid adaptation is the massive fitness advantage of the lasR mutants gained by exploiting the cooperating wild-type population without contributing to the public good. This result suggests that QS plays an important role in bacterial social interactions in clinical contexts, as has been frequently suggested, and argues against the recent hypothesis that the production of QS molecules primarily provide a direct benefit to the producing bacteria through detecting when exoproducts are likely to diffuse away (diffusion sensing) (34, 35). It is therefore possible that genetic or ecological manipulation of the social environment (by inhibiting, for instance, cooperative systems such as QS) may impact on the severity of P. aeruginosa infections.
Materials and Methods
Patients and Clinical Collection.
The patients included in this study were part of the placebo control group of a multicenter European trial (ClinicalTrials.gov ID# NCT00610623) that compared azithromycin to placebo for the prevention of pneumonia in intubated patients colonized by P. aeruginosa. We obtained approval for this study from the local ethical committees of the 13 participating hospitals and written consent from all patients or their legal representatives. We screened mechanically ventilated patients for respiratory tract colonization by P. aeruginosa every other day. Patients with ongoing P. aeruginosa infection, or having received therapies against the bacteria during the last 14 days, were not included. Starting the first day of proven colonization (D-1), we collected tracheal aspirates (usually 0.3 to 5 ml) and one P. aeruginosa isolate (collection period: 3–20 days) every day for a maximum of 20 days. Samples were frozen on site at −80 °C within 15 min. Reasons for early discontinuation included extubation, death, or proven P. aeruginosa VAP (as defined by strict clinical and laboratory data and confirmed by an external blind panel of experts).
Genotype and Mutation Analyses.
For intrapatient comparison of isolates, we performed random amplification of polymorphic DNA (RAPD) (Fig. S1 and Fig. S2). Primer 207 was used for routine analysis. When distinct band patterns (>2 bands difference) were observed, RAPD was repeated with a second primer (primer 272) (36). The SNP type of one representative isolate from each RAPD type was determined by using the Clondiag array (22). Mutations in the lasR and rhlR genes identified initially by DNA sequencing, were confirmed in the other isolates from the same patients by high resolution melting (HRM) analysis by using the SensiMix Kit and Evagreen as the intercalating agent according to the manufacturer's instructions (Quantace Ltd. ). Portions of the lasR and rhlR genes were amplified by using specifically designed primer pairs (all primer sequences are available on request). PCR amplifications and melt curves (0.1 °C increment) were performed in a RotorGene 6000 RealTime PCR machine (Corbett Research) and analyzed by using the integrated HRM software.
Exoproduct Assays.
Elastase activity (Fig. S1 and Fig. S2) was determined in supernatants of cultures grown for 7 h in PB medium (37) at 37 °C, by using the Elastin Congo Red (ECR) assay (38). Rhamnolipid production was assessed on modified SW-Blue plates as described (39). PAO1 was used as the reference strain in each assay. Results of duplicate elastase determinations are expressed as the mean absorption ratios OD495/OD600.
Extraction of DNA from Tracheal Aspirates.
Tracheal aspirates and one P. aeruginosa isolate from 31 intubated colonized patients were collected prospectively. Genomic DNA was extracted by using the DNAzol solution (Invitrogen). The final DNA pellet was dissolved in 0.2 ml 8 mM NaOH by heating at 56 °C for 10 min. Genomic DNA could be detected (>5 × 104 genomic copies/g aspirate) in 98% of the aspirates. All extractions were done in duplicate.
Determination of Bacterial Load in the Lung.
The number of P. aeruginosa in tracheal aspirates was determined by qRT-PCR of genomic DNA preparations. The rpsL F/R primer pair (40) was tested for specificity against genomic DNA preparations from clinical isolates of Streptococcus pneumoniae, Klebsiella pneumoniae, Escherichia coli and Acinetobacter baumannii. An amplification signal was obtained only with genomic DNA isolated from P. aeruginosa. Aliquots of genomic DNA preparations were diluted 10 fold into H2O and 3 μl of this dilution were added to the PCR mix containing 1× Quantitect Sybr Green Master Mix (Qiagen) and 600 nM primers in a total volume of 15 μl. PCR conditions were as recommended by the manufacturer. A standard curve was obtained by addition of 10-fold dilutions of a P. aeruginosa culture to an aspirate collected from a patient not colonized by this organism. Genomic DNA was then isolated as described above and quantified by qRT-PCR. Under these conditions, we detected 104 genomic copies/g aspirate. Standard curves yielded reproducible values during the 3 month analysis period. P. aeruginosa was found in 301 of the 308 aspirates analyzed at levels varying from 5 × 104 to 1.8 × 108 genomic copies/g aspirate. The linearity and specificity of our assay was tested by adding serial dilutions of a P. aeruginosa laboratory strain culture to a tracheal aspirate that contained Gram-positive and Gram-negative bacteria but no P. aeruginosa. A linear correlation over at least 3 orders of magnitude was observed between the amount of bacteria determined on the genomic DNA extracts and the number of cfu added.
Statistical Analyses.
Logistic regression was used to analyze the change in the proportion of lasR and rhlR mutants across patients through time; the data were slightly overdispersed and were therefore corrected by scaling factor to equalize the error deviance and degrees of freedom. To analyze the genomic copy data, we used REML to fit a linear mixed model with patient (random factor), time and lasR mutant/wild-type (both fixed factors). All analyses were carried out by using GenStat 10.
Supplementary Material
Acknowledgments.
We thank the responsible physicians in the participating centers, the nurses and medical staff involved in this study, and J.-L. Dumas and R. Comte for excellent technical assistance. We thank Dr. G. Erba (Biolabo SA, Châtel-St. Denis, Switzerland) for help with the HRM analysis. The clinical study was financed by Anbics Corporation. Work of our teams was supported by the Swiss National Science Foundation Grants 4049-063239 (to T.K. and C.v.D.) and 320000-108106 (to C.v.D.) and the Royal Society and Leverhulme Trust (A.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811741106/DCSupplemental.
References
- 1.Williams P, Winzer K, Chan WC, Camara M. Look who's talking: Communication and quorum sensing in the bacterial world. Philos Trans R Soc Lond Ser B. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: Effects of growth phase and environment. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Delden C, Iglewski BH. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearson JP, Feldman M, Iglewski BH, Prince A. Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect Immun. 2000;68:4331–4334. doi: 10.1128/iai.68.7.4331-4334.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rumbaugh KP, Griswold JA, Iglewski BH, Hamood AN. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect Immun. 1999;67:5854–5862. doi: 10.1128/iai.67.11.5854-5862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lesprit P, et al. Role of the quorum-sensing system in experimental pneumonia due to Pseudomonas aeruginosa in rats. Am J Respir Crit Care Med. 2003;167:1478–1482. doi: 10.1164/rccm.200207-736BC. [DOI] [PubMed] [Google Scholar]
- 8.Singh PK, et al. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 9.Favre-Bonte S, et al. Detection of Pseudomonas aeruginosa cell-to-cell signals in lung tissue of cystic fibrosis patients. Microb Pathog. 2002;32:143–147. doi: 10.1006/mpat.2001.0487. [DOI] [PubMed] [Google Scholar]
- 10.Denervaud V, et al. Characterization of cell-to-cell signaling-deficient Pseudomonas aeruginosa strains colonizing intubated patients. J Clin Microbiol. 2004;42:554–562. doi: 10.1128/JCM.42.2.554-562.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaber JA, et al. Analysis of quorum sensing-deficient clinical isolates of Pseudomonas aeruginosa. J Med Microbiol. 2004;53:841–853. doi: 10.1099/jmm.0.45617-0. [DOI] [PubMed] [Google Scholar]
- 12.Cabrol S, Olliver A, Pier GB, Andremont A, Ruimy R. Transcription of quorum-sensing system genes in clinical and environmental isolates of Pseudomonas aeruginosa. J Bacteriol. 2003;185:7222–7230. doi: 10.1128/JB.185.24.7222-7230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamood AN, Griswold J, Colmer J. Characterization of elastase-deficient clinical isolates of Pseudomonas aeruginosa. Infect Immun. 1996;64:3154–3160. doi: 10.1128/iai.64.8.3154-3160.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith EE, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Argenio DA, et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol. 2007;64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heurlier K, et al. Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J Bacteriol. 2005;187:4875–4883. doi: 10.1128/JB.187.14.4875-4883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 18.Sandoz KM, Mitzimberg SM, Schuster M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci USA. 2007;104:15876–15881. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 20.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 22.Wiehlmann L, et al. Population structure of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2007;104:8101–8106. doi: 10.1073/pnas.0609213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamilton WD. The genetic evolution of social behaviour, I & II. J Theor Biol. 1964;7:1–52. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 24.Brown SP, Johnstone RA. Cooperation in the dark: Signalling and collective action in quorum-sensing bacteria. Proc Biol Sci. 2001;268:961–965. doi: 10.1098/rspb.2001.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maynard Smith J, Harper D. Animal Signals. Oxford: Oxford Univ Press; 2003. [Google Scholar]
- 26.Keller L, Surette MG. Communication in bacteria: An ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 27.Bjarnsholt T, Givskov M. Quorum-sensing blockade as a strategy for enhancing host defences against bacterial pathogens. Philos Trans R Soc Lond Ser B. 2007;362:1213–1222. doi: 10.1098/rstb.2007.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross-Gillespie A, Gardner A, West SA, Griffin AS. Frequency dependence and cooperation: Theory and a test with bacteria. Am Nat. 2007;170:331–342. doi: 10.1086/519860. [DOI] [PubMed] [Google Scholar]
- 29.Pugin J, et al. Diagnosis of ventilator-associated pneumonia by bacteriologic analysis of bronchoscopic and nonbronchoscopic “blind” bronchoalveolar lavage fluid. Am Rev Respir Dis. 1991;143:1121–1129. doi: 10.1164/ajrccm/143.5_Pt_1.1121. [DOI] [PubMed] [Google Scholar]
- 30.Brown SP, Hochberg ME, Grenfell BT. Does multiple infection select for raised virulence? Trends Microbiol. 2002;10:401–405. doi: 10.1016/s0966-842x(02)02413-7. [DOI] [PubMed] [Google Scholar]
- 31.West SA, Buckling A. Cooperation, virulence and siderophore production in bacterial parasites. Proc Biol Sci. 2003;270:37–44. doi: 10.1098/rspb.2002.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank SA. Models of parasite virulence. Q Rev Biol. 1996;71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- 33.Buckling A, Brockhurst MA. Kin selection and the evolution of virulence. Heredity. 2008;100:484–488. doi: 10.1038/sj.hdy.6801093. [DOI] [PubMed] [Google Scholar]
- 34.Redfield RJ. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 2002;10:365–370. doi: 10.1016/s0966-842x(02)02400-9. [DOI] [PubMed] [Google Scholar]
- 35.Hense BA, et al. Does efficiency sensing unify diffusion and quorum sensing? Nat Rev Microbiol. 2007;5:230–239. doi: 10.1038/nrmicro1600. [DOI] [PubMed] [Google Scholar]
- 36.Mahenthiralingam E, Campbell ME, Foster J, Lam JS, Speert DP. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:1129–1135. doi: 10.1128/jcm.34.5.1129-1135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Essar DW, Eberly L, Crawfords IP. Identification and characterization of genes for a second anthranilate synthetase in Pseudomonas aeruginosa: Interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tateda K, et al. Azithromycin Inhibits Quorum Sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2001;45:1930–1933. doi: 10.1128/AAC.45.6.1930-1933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Köhler T, Kocjancic-Curty L, Barja F, Van Delden C, Pechère JC. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol. 2000;182:5990–5996. doi: 10.1128/jb.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dumas JL, Van Delden C, Perron K, Köhler T. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol Lett. 2006;254:217–225. doi: 10.1111/j.1574-6968.2005.00008.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.