Abstract
Notch has been linked to β-catenin-dependent tumorigenesis; however, the mechanisms leading to Notch activation and the contribution of the Notch pathway to colorectal cancer is not yet understood. By microarray analysis, we have identified a group of genes downstream of Wnt/β-catenin (down-regulated when blocking Wnt/β-catenin) that are directly regulated by Notch (repressed by γ-secretase inhibitors and up-regulated by active Notch1 in the absence of β-catenin signaling). We demonstrate that Notch is downstream of Wnt in colorectal cancer cells through β-catenin-mediated transcriptional activation of the Notch-ligand Jagged1. Consistently, expression of activated Notch1 partially reverts the effects of blocking Wnt/β-catenin pathway in tumors implanted s.c. in nude mice. Crossing APCMin/+ with Jagged1+/Δ mice is sufficient to significantly reduce the size of the polyps arising in the APC mutant background indicating that Notch is an essential modulator of tumorigenesis induced by nuclear β-catenin. We show that this mechanism is operating in human tumors from Familial Adenomatous Polyposis patients. We conclude that Notch activation, accomplished by β-catenin-mediated up-regulation of Jagged1, is required for tumorigenesis in the intestine. The Notch-specific genetic signature is sufficient to block differentiation and promote vasculogenesis in tumors whereas proliferation depends on both pathways.
Keywords: beta-catenin, APC, intestine, crosstalk
The Wnt signaling pathway plays a crucial role during development of different tissues and organisms. A multiprotein complex including adenomatosis polyposis coli (APC), axin and Glycogen Synthase Kinase-3β (GSK3β) is responsible for regulating β-catenin (ctnnb1) protein levels. Activation of canonical Wnt signaling results in GSK3β inhibition, stabilization and nuclear accumulation of β-catenin and subsequent activation of lymphoid enhancer factor/T cell factor (LEF1/TCF) target genes (1).
Constitutive activation of Wnt is one of the best-characterized events in several types of cancer (2). Familial Adenomatous Polyposis (FAP), a disease characterized by the presence of hundreds of colorectal polyps, is usually associated with APC germ-line mutations. Colorectal cancer (CRC) develops upon mutational damage or loss of the wild-type allele resulting in further increased β-catenin/TCF activity (3, 4). Similarly, mice carrying germ-line mutations in APC, APCMin (Multiple intestinal neoplasia), are predisposed to the formation of intestinal adenomas (5, 6). Canonical Wnt target genes such as c-myc, cyclinD1, MMP7, TCF1, and EphB2 but also the Notch-target gene hes1 have an increased expression in the tumors of these mice (7–9). Overexpression of hes1 and other Notch targets has been associated with sporadic colorectal tumors (10), human medulloblastomas (11), melanomas (12), meningiomas (13), and T cell leukemias (14, 15).
Notch is a family of transmembrane receptors that plays important roles in regulating cell fate decisions. After ligand binding, Notch protein undergoes a proteolytic cleavage dependent on γ-secretase activity that releases the active intracellular domain, Notch-IC (16, 17). Once activated, Notch translocates into the nucleus to bind RBPjκ and to activate specific gene transcription (for review, see ref. 18). Different Notch receptors with specific functions have been identified but, interestingly, they all trigger the activation of the same downstream cascade. In the intestine, Notch signaling is required for the maintenance of the proliferative compartment (8, 19) and a recent report shows that both Notch1 and 2 participate in the regulation of intestinal cell differentiation (20). Different Notch ligands (Jagged1, 2 and Delta 1–4) can also confer specificity to the Notch receptors (21, 22); however, distinct functions for each ligand remain unknown on many tissues.
The cross-talk between Wnt and Notch pathways including genetic interactions in Drosophila (23–25), the physical binding of Notch to β-catenin (26) or their association to common cofactors (27) have been described. In mammalian cells, GSK3β directly phosphorylates the Notch protein thus modulating its transcriptional activity (28). Moreover, β-catenin activates Jagged1 transcription thus leading to Notch activation during murine hair follicle differentiation (29). Conversely, in different types of tumor cells, Notch activates the Wnt pathway stabilizing β-catenin by unknown mechanisms (12) or by transcriptional activation of slug (30).
In APC mutant mice that generate multiple intestinal tumors because of β-catenin activation, treatment with a γ-secretase/Notch inhibitor promotes differentiation of the adenoma cells into goblet cells, similar to the effect of Notch signaling deficiency in the intestine (8). How Notch is activated in β-catenin-dependent tumors and what is the contribution of the Notch pathway to Wnt-dependent intestinal tumorigenesis is largely unknown.
We have now demonstrated that β-catenin/TCF is responsible for activating Notch in CRC cells through direct regulation of Jagged1 expression. By microarray screening of colorectal cancer cells, we have identified several genes downstream of TCF/β-catenin pathway that are directly regulated by the Jagged1/Notch pathway including hes1, CD44, KLF5, NOX1, EpHB3, and SOX9. Consequent with a crucial role for Notch in the tumorigenesis induced by β-catenin, N1IC reverts the growth arrest imposed by dnTCF4 expression in Ls174T cells whereas deletion of a single Jagged1 allele reduces the size of tumors arising in the intestine of APCMin/+ mice. Finally, we show correlative evidence that this mechanism operates in human adenomas of APC-associated FAP patients.
Results
Inhibiting Notch or Wnt Pathways Results in the Down-Regulation of a Common Genetic Program in Ls174T Cells.
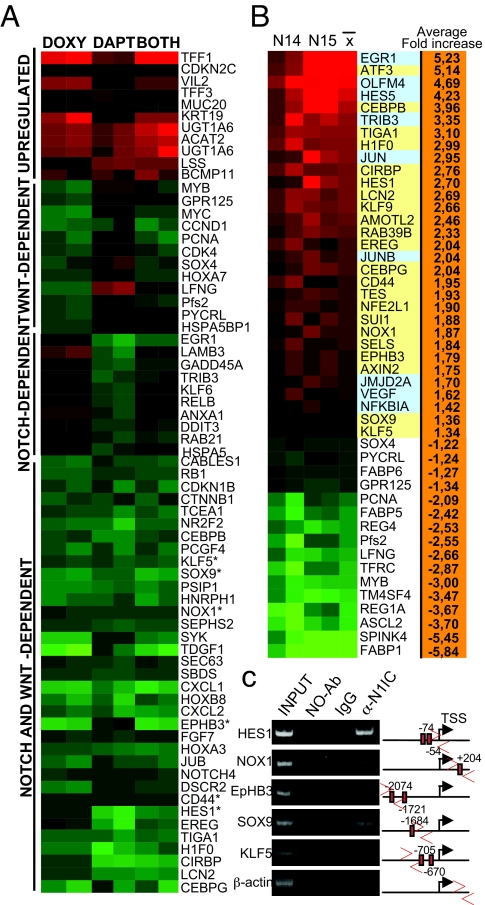
To determine the contribution of the Notch pathway to β-catenin/TCF-dependent tumorigenesis, we first compared the transcriptional effects of blocking Wnt, Notch or both pathways in the Ls174T CRC cells that contain active Notch1 (see Fig. S1a) and nuclear β-catenin (9). We took advantage of a published cell line carrying a doxycycline-inducible plasmid encoding a dominant negative TCF4 that does not bind β-catenin (Ls174T/dnTCF4) (9). We performed microarray analysis comparing Ls174T/dnTCF4 cells untreated, treated for 48 h with doxycycline and/or with the Notch/γ-secretase inhibitor DAPT (31), using a whole human genome oligonucleotide microarray from Agilent (G4112A). We identified 366 genes that were simultaneously down-regulated (>1.3-fold) in response to doxycycline and/or DAPT treatments (Table S1 and Fig. 1A) corresponding to 34% of the genes down-regulated by doxycycline. Some of the genes in this group have been associated with cancer such as CD44, hes1, EpHB3, SOX9, NOX1, or KLF5 in different systems (32–40) and a number of them had been identified as β-catenin/TCF targets (9). We designed specific primers (Table S2) for some of these genes to confirm the microarray data by qRT-PCR, including the use of two different γ-secretase inhibitors, DAPT and L685,458 (Fig. S1 b and c). In addition, blocking Notch and/or Wnt pathways, resulted in an increase in the expression levels of several differentiation markers such as villin2, muc20 or TFF1 (Fig. 1A).
Fig. 1.
Inhibition of Wnt and Notch leads to down-regulation of a common gene program. (A) Gene expression profile from Ls174T/dnTCF4 cells treated for 48 h with doxycyline (to inhibit Wnt signaling) or DAPT (Notch inhibitor) or both compared with untreated cells. Some top significant genes ranked by Student's t test are shown for each group. *, genes further studied in this work. (B) Microarray analysis of 2 doxycycline-treated Ls174T/dnTCF4/N1IC clones (N14, N15) compared with doxycycline-treated Ls174T/dnTCF4. Color codes represent only-Notch targets (blue) and the Notch-targets downstream of Wnt (yellow). (C) ChIP experiment in Ls174T cells and PCR of the indicated promoters. Scheme of the 2-kb proximal promoter showing the position of the primers used, the RBP-binding sites and TSS.
Many of the TCF4-dependent genes identified in our screening were also inhibited by the γ-secretase treatment, suggesting that at least two different mechanistical explanations: (i) Notch is downstream of β-catenin/TCF signaling or (ii) both β-catenin and Notch are required to properly activate a specific gene signature in CRC cells. To test this, we first studied the transcriptional signature of cells expressing dnTCF4 with or without the constitutively active N1IC [control (C) versus N1IC (N) clones, see Fig. S1d]. We found that N1IC expression is sufficient to reactivate 31% of the genes identified as common Wnt-Notch-dependent genes including hes1, SOX9, CD44, KLF5, or NOX1 in a β-catenin/TCF-deficient background (Fig. 1b, Fig. S1d, and Table S3). Using the Genomatix software, we identified Notch/RBPjκ-binding consensus in the regulatory regions of all of the analyzed genes (hes1, Nox1, EpHB3, SOX9, KLF5, CD44) (Fig. 1c and Table S4). By ChIP experiments, we found that Notch1 associated to the promoters of these genes in Ls174T cells (Fig. 1c and Fig. S1e), but also in other CRC cell lines concomitant with increased transcriptional activity (Fig. S1f). Moreover, we found that DAPT treatment reduces the recruitment of Notch to these promoters (Fig. S1e). As expected, several genes involved in intestinal differentiation, such as FABP1 and FABP5, were down-regulated in N1IC-expressing clones (Fig. 1B) indicating the effects of Notch in inhibiting differentiation in these cells.
We next studied whether the Wnt-Notch-dependent genes were overrepresented in any specific functional category. Our analysis revealed that these genes were highly enriched into functional categories related to proliferation, differentiation and DNA and RNA metabolism, including transcription and replication. Interestingly, the putative direct targets of Notch downstream of β-catenin (those that are reexpressed in the N1IC clones) are specifically enriched for cell cycle arrest and differentiation categories(Fig. S1g).
Together, these results strongly suggest that Notch is a direct regulator of specific gene transcription downstream of β-catenin/TCF nevertheless most of the identified Wnt-Notch-dependent genes likely require the cooperative effects of Notch and β-catenin pathways.
Activated Notch1 Blocks Differentiation and Promotes Vasculogenesis in Vivo in the Absence of β-Catenin/TCF Signaling.
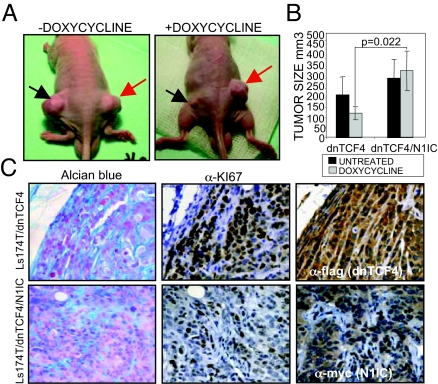
We next studied whether N1IC conferred any malignant capacity to Ls174T cells in the absence of β-catenin/TCF signaling. We found that doxycycline-treated Ls174T/dnTCF4/N1IC clones displayed an increased clonogenic capacity in soft agar cultures compared with Ls174T/dnTCF4 clones (Fig. S2a), together with a blockage in goblet-cell differentiation (Fig. S2b). In contrast, N1IC expression did not revert the antiproliferative effects of dnTCF4 as indicate the cell cycle profiles (Fig. S2c). To test the tumorigenic capacity of these cells in vivo, we injected 1.5 × 106 cells from different control Ls174T/dnTCF4 (C1, C5) or Ls174T/dnTCF4/N1IC (N14, N15, N17, N18) s.c. in nude mice and treated or not the animals with doxycycline for 4 weeks. We found that N1IC expressing clones (Ls174T/dnTCF4/N1IC) generated tumors that were significantly larger compared with the doxycycline-treated control group (P = 0.022) (Fig. 2 A and B) although we did not detect significant differences in the proliferation ratio in the growing areas as measured by Ki67 staining (Fig. 2C). However, we observed that tumors expressing N1IC showed a strong reduction of Alcian blue staining indicating a blockage in mucosecreting cell differentiation (Fig. 2C) together with an increase in the proportion of growing areas within the tumor concomitant with the presence of vascularization as detected by α-SMA staining (11/11 in N1IC tumors versus 3/10 in control tumors). This is in contrast with the extensive necrotic areas observed in most of the control tumors (7/10) (Fig. S2d). Importantly, we detected high levels of endogenous Notch activity in the untreated Ls174T/dnTCF4 tumors as detected by a specific antibody recognizing cleaved Notch1 (N1ICv antibody) that was greatly inhibited after doxycycline treatment (Fig. S3g).
Fig. 2.
Notch1 restores tumor growth in the absence of β-catenin/TCF signaling. (A and B) A total of 1.5 × 106 cells were implanted s.c. in nude mice [Ls174T/dnTCF4 (black arrow) and Ls174T/dnTCF4/N1IC (red arrow) clones] untreated or treated with doxycycline for 4 weeks (2 independent experiments, n = 20 mice each). Representative animals for each group (A) and average and SEM of the tumor volume (in cubic millimeters) (B) are shown. P values are based on a nonparametric ANOVA applying a rank transformation on the dependent variable. (C) Alcian blue staining and IHC with α-Ki67 in serial sections of representative tumors. α-flag or α-myc staining shows the expression of dnTCF4 and N1IC (400×).
Together, these results indicate that activated Notch1 exert a direct effect in regulating goblet-cell differentiation and tumor vasculogenesis whereas regulation of cell proliferation requires the contribution of β-catenin/TCF signaling pathway.
Notch Is Downstream of Wnt Through Transcriptional Activation of Jagged1 by β-Catenin/TCF.
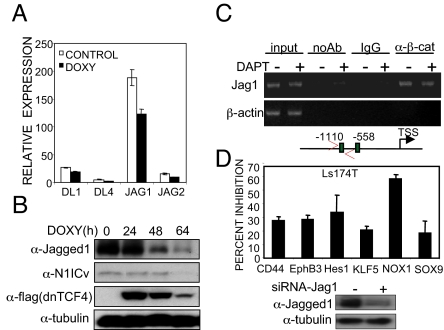
To determine whether β-catenin/TCF is responsible for the high Notch activity that is found in CRC cells (Figs. S1a and S3d), we searched in the microarray data for the presence of Notch ligands and found that Jagged1 and Delta-like1 were down-regulated 1.3-fold after doxycycline treatment in Ls174T cells (Table S1). Using qRT-PCR, we confirmed that jagged1 is highly expressed in Ls174T/dnTCF4 and its mRNA levels decreased after doxycycline treatment (Fig. 3A). Most important, expression of dnTCF4 or inhibition of β-catenin with PKF115–584 (41) results in a huge reduction in the protein levels of Jagged1 concomitant with a decrease in the amount of activated Notch1 (Fig. 3B and Fig. S3b), similarly to that found in s.c. tumors (Fig. S3g). Of note, the disappearance of Jagged1 was delayed with respect to dnTCF4 expression likely because of the extended half-life of Jagged1 protein (24 h) (Fig. S3a).
Fig. 3.
Notch is downstream of Wnt/β-catenin through activation of Jagged1. (A) qRT-PCR of different Notch ligands in Ls174T/dnTCF4 cells untreated or treated for 48 h with doxycycline. (B) Western blot with the indicated antibodies of Ls174T/dnTCF4 cells treated with doxycycline for the indicated times. Tubulin is shown as loading control. (C) Recruitment of β-catenin to Jagged1 promoter in Ls174T cell line untreated or treated with DAPT. β-actin gene is shown as a control. (D) Percent transcriptional inhibition in Ls174T cells transfected with siRNA-Jag1 compared with scramble siRNA as determined by qRT-PCR. Down-regulation of Jagged1 by siRNA was determined by Western blot analysis (Lower).
By sequence analysis, we identified 2 TCF-binding consensus in the human Jagged1 promoter and showed the recruitment of β-catenin by ChIP experiments in Ls174T (Fig. 3C and Fig. S3c). We next asked whether β-catenin regulates Jagged1 and Notch activation in other CRC cell lines. We found that Jagged1 transcription was highly increased in cells carrying nuclear β-catenin, such as HCT116, SW480, and Ls174T, compared with the nontransformed HS27 control cells (Fig. S3d) and β-catenin was recruited to the Jagged1 promoter to a different extent, in all these cancer cell lines (Fig. S3e). Moreover, stabilization of β-catenin in a nontumorigenic cell line (NIH 3T3) by a 16-h treatment with the GSK3β inhibitor LiCl (30 mM) was sufficient to increase both mRNA and protein levels of Jagged1 (Fig. S3f). By cell fractionation followed by Western blot analysis, we detected activated Notch1 and Notch2 (cleaved Notch2 fragment of 110 kDa) in the nuclear fraction of CRC cell lines with high Jagged1 levels (Fig. S3d).
To demonstrate that Jagged1 was responsible for regulating Notch-targets downstream of β-catenin, we treated Ls174T cells with specific siRNA against Jagged1 and found that all of the selected genes were down-regulated (from 20 to 60% inhibition) compared with cells treated with scrambled siRNA (Fig. 3D). In addition, ectopic expression of Jagged1 is sufficient to block differentiation in ≈85% of the transfected Ls174T/dnTCF4 cells when Wnt/β-catenin signaling is switched off, similar to N1IC expression in these conditions (Fig. S3h).
Deletion of a Single Jagged1 Allele Reduces Tumor Growth in the APCMin/+ Intestine.
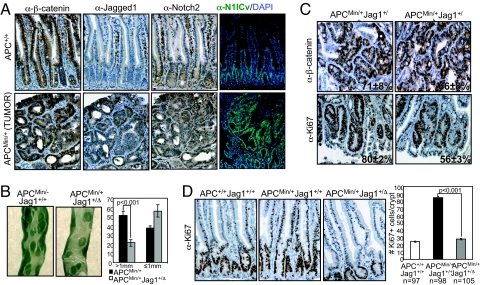
To investigate whether the expression of Jagged1 is altered by the activation of the Wnt pathway in vivo, we determined the levels of this protein in intestinal tumors arising in the APCMin/+ mice compared with normal mucosa. By IHC, we found a strong overexpression of Jagged1 in the tumor tissue of these animals compared with the normal crypts that was concomitant with Notch1 and Notch2 activation (Fig. 4A).
Fig. 4.
Jagged1 is essential in APCMin/+ intestinal tumorigenesis. (A) IHC of serial section of wild type intestine and APCMin/+ tumor stained with the indicated antibodies (100×). (B) Stereoscopical image of the methylene blue staining. The average number of visible polyps in the small intestines (>1 mm or ≤1 mm) from the different genotypes at 16 weeks of age is represented. Error bars are SEM. P values are based on a Mann–Whitney U test. (C) (Upper) Immunostaining with α-β-catenin of representative tumors from different genotypes. The average percentage of cells showing nuclear β-catenin is indicated. (Lower) Representative images of α-Ki67 staining and average percentage and SEM of Ki67+ cells per tumor from 5 APCMin/+Jag1+/+ (n = 51) and 4 APCMin/+Jag1+/Δ (n = 27). (D) Sections of normal crypts from different genotypes stained with α-Ki67 antibody (Left). The average and SEM of Ki67+ cells per crypt from 10 APC+/+Jag1+/+, 5 APCMin/+Jag1+/+ and 4 APCMin/+Jag1+/Δ is represented (Right). n, number of crypts counted. P values are based on a Mann–Whitney U test.
To test the importance of Jagged1-dependent activation of Notch downstream of β-catenin, we crossed the APCMin/+ mice with the Jagged1 hemizygous (Jag1+/Δ) mice, which are phenotypically normal (whereas Jag1Δ/Δ are lethal) (42). A total of 23 animals of the different genotypes were killed at 4 months of age and analyzed for the presence of macroscopic intestinal tumors. We found that deletion of one Jagged1 allele is sufficient to significantly reduce the size of tumors in the APC mutant background (P = 0.0001) (Fig. 4B) concomitant with a reduction in the amount of active Notch1 (Fig. S4), whereas the nuclear levels of β-catenin in the tumors from the different genotypes were equivalent (Fig. 4C Upper). This result suggests that Jagged1 deficiency confers a growing disadvantage to β-catenin-dependent tumors. In agreement with this, we found a reduction in the number of Ki67 positive cells in the tumors of the APCMin/+Jag1+/Δ double mutants compared with the ones from the APCMin/+ littermates (from 80.4 ± 2% to 55.6 ± 3%, P < 0.001) (Fig. 4C Lower), whereas no differences were found in the number of apoptotic cells as measured by TUNEL assay or in the number of infiltrating lymphocytes in the tumors (Fig. S4). Moreover, the expansion of the proliferative compartment in the morphologically normal crypts of APC mutant mice described in ref. 43, is reverted in the APCMin/+Jag1+/Δ mice (P < 0.001) (Fig. 4D). These results demonstrate that activation of Notch by Jagged1 confers a proliferative advantage to the tumors with APC mutations.
High Levels of Jagged1 Correlate with Activated Notch1 and Notch2 in Human Colorectal Tumors Containing Nuclear β-Catenin.
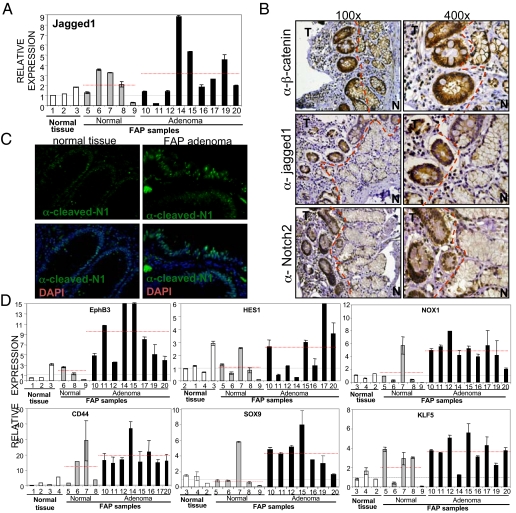
We next investigated whether this mechanism for Notch activation was relevant in human colorectal adenomas arising in Familial Adenomatous Polyposis (FAP) patients harboring APC germ line mutations. By qRT-PCR we found that Jagged1 mRNA levels were significantly increased in most FAP adenomas compared with normal intestinal tissue (P = 0.05). Interestingly, some increase in Jagged1 expression was also detected in the normal colonic mucosa of FAP patients, compared with normal controls (Fig. 5A). By IHC, we found high levels of the Jagged1 protein confined in the tumor areas containing nuclear β-catenin staining (n = 6) that were not detected in the normal adjacent tissue (Fig. 5B and Fig. S5a). This was concomitant with the presence of nuclear Notch2 (Fig. 5B and Fig. S5a) and active Notch1 (Fig. 5C and Fig. S5a). Further demonstrating the importance of Notch transcriptional activity in human tumors carrying active β-catenin, we found increased expression of CD44 (P = 0.0002), SOX9 (P = 0.005), NOX1 (P = 0.002), KLF5 (P = 0.01) and hes1 (not significant) (Fig. 5D), identified in our microarray screening as Wnt-dependent Notch targets (see Fig. 1), in the adenoma samples from FAP patients. Some of these genes were also up-regulated in the normal colonic mucosa from these patients compared with the normal controls (Fig. 5D) likely because of the presence of 1 mutated APC allele (Fig. S5b). These results indicate that Notch, downstream of Jagged1, acts as an essential mediator of β-catenin-dependent intestinal tumorigenesis (i.e., FAP) and is responsible for regulating a specific transcriptional cancer signature.
Fig. 5.
Notch1 and Notch2 are activated in colorectal tumors. (A) Levels of Jagged1 mRNA as measured by qRT-PCR. The red line indicates the average value for each group. (B) Serial sections of colorectal adenoma from a FAP patient stained with the indicated antibodies. Dashed lines indicate the boundary between the normal adjacent (N) and the tumor (T) tissue. Hematoxylin (blue) was used for nuclear staining. (C) Immunoflouresence with α-N1ICv (green) of normal or adenoma tissue from FAP sample. Nuclei were stained with DAPI (blue) (600×). (D) qRT-PCR to determine the levels of the indicated genes in normal tissue compared with FAP samples. The red line indicates the average value for each group. Statistical significance of the differences between the adenoma and normal samples are calculated based on 2-sided Student's t test. P values are: EphB3, P = 0.014; CD44, P = 0.0002; SOX9, P = 0.005; NOX1, P = 0.002; KLF5, P = 0.01; and hes1, not significant.
Discussion
Previous work has shown that inactivating mutations of the Wnt/β-catenin pathway eliminate the stem/proliferative compartment of intestinal cells (44). Similarly inactivation of the Notch pathway leads to an exhaustion of the stem cell compartment concomitant with increased goblet cell differentiation (8, 19). Here, we define the mechanism that mediates the cross-talk between Notch and Wnt pathways in CRC and determine its functional relevance. In these tumors, the Notch ligand Jagged1 is directly regulated by β-catenin, thus leading to aberrant activation of Notch1 and 2. We identified a total of 366 genes that are simultaneously regulated by both Notch and Wnt in Ls174T CRC cells and 31% are reactivated by active N1IC in the presence of dnTCF4, indicating that are directly dependent on Notch. This group of genes was sufficient to revert some of the anti-tumorigenic effects of dnTCF4 both in vitro and in vivo, because N1IC expression inhibits differentiation and promotes vasculogenesis in the absence of Wnt/β-catenin. However, N1IC is not able to regulate proliferation in these conditions, similar to that observed by Fre et al. (52). As expected, one of the genes identified as direct Notch target in our arrays is hes1. Hes1 regulates intestinal differentiation (45) by inhibiting math1 thus favouring enterocytic differentiation (46). It is overexpressed in CRC (10) and in tumors arising in APC mutant mice (8). However, and in contrast to the effects of N1IC, we found that hes1 was not sufficient to induce tumor growth in vivo in the absence of β-catenin/TCF activity indicating that other Notch-targets downstream of β-catenin participate in regulating colorectal tumorigenesis. Consistently, several of the identified Notch-target genes in our screening have been involved in different aspects of intestinal differentiation or cancer including SOX9 (38, 39) KLF5 (47), CD44 (32), or NOX1 (48).
Mutations in the APC gene have been found in most sporadic colorectal tumors and in those arising in FAP patients. We have found that Jagged1 is up-regulated in the tumors but also, in a lesser extent, in the normal adjacent tissue of FAP patients suggesting that inactivation of 1 APC allele could be sufficient to increase Notch activity. In this sense, others and we have observed that Ki67-positive cells are aberrantly distributed throughout the intestinal glands in APC mutant mice, a phenotype that is abolished in the Jag1Δ/+APCMin/+ mice. This indicates that Notch is involved in the crypt/villus compartmentalization. Most important, tumor growth is reduced in Jag1Δ/+APCMin/+ mice indicating that partial inhibition of Notch in an active β-catenin background may be therapeutically relevant for FAP patients.
Because we identified 3 different genetic signatures associated with CRC cells that depend on Notch, β-catenin or both pathways that are likely complementary for tumor development, we propose that using of a combination of Notch and β-catenin inhibitors will enhance the antitumoral effects of the individual drugs. However, this strategy needs to be tested.
In summary, our results provide an explanation for the contribution of Notch to β-catenin-dependent tumorigenesis and the mechanism for Notch activation through Jagged1. Specific inhibitors for Jagged1-mediated Notch activation [i.e., inhibitors of glycosil-transferases that modulate Notch/Notch-ligand interaction (21)] would be a new promising strategy for CRC therapy.
Materials and Methods
Animals.
APCMin/+ mice (Jackson Laboratories) were from homogenous outbred C57BL/6J background. Jagged1 mutant mice (Jag1Δ/+) are described in ref. 42. Jag1Δ/+ were backcrossed into C57BL6/J background (n > 4) and crossed with APCMin/+. In our experiments, cohorts of age-matched APCMin/+Jag1+/Δ were compared with APCMin/+Jag+/+ and APC+/+Jag+/+. All mice were genotyped by PCR. Animals were kept under pathogen-free conditions and all procedures were approved by the Animal Care Committee.
Cell Lines and Reagents.
NIH 3T3, HS27, HCT116, SW480, DLD1, and Ls174T/dnTCF4 [kindly provided by H. Clevers (Hubrecht Laboratories, Utrecht, Netherlands)]. SB216763 (Sigma) was used at 10 μM/mL. Doxycycline (Sigma) was used at 1 μg/mL. DAPT and L685,458, γ-secretase inhibitors, were obtained from Calbiochem, PKF115–584 (41) was a gift from Novartis. Ls174T/dnTCF4 cells were transfected with Notch1IC-pCDNA3/TO using PPEI (Polysciences). Cells were selected with 1 mg/mL G418, 5 μg/mL Blasticidin, and 100 μg/mL Zeocin.
Antibodies.
α-cleaved Notch1 (N1ICv) (Cell Signaling), α-Notch2 (49), α-Jagged1 (sc-6011), α-α-tubulin and α-β-catenin (C2206) from Sigma. Secondary antibodies conjugated to Horse Radish Peroxidase (HRP) were from DAKO (with AlexaFluor488 (A-11055) and 546 (A-11056) from Molecular Probes (Invitrogen) and Cy3-coupled tyramide from PerkinElmer.
Immunofluorescence.
Cells were fixed in 3% paraformaldehyde, permeabilized in 0.3% Triton X-100 (Pierce). Primary antibodies were incubated overnight and secondary antibodies for 90 min. Slides were mounted in Vectashield plus DAPI (Vector) and visualized in an Olympus BX-60 or BX-61 microscope. Protocol for cleaved-Notch1 detection is described in ref. 50. For immunohistochemistry (IHC), secondary antibodies were developed with the DAB peroxidase substrate kit (Dako Cytomation).
RT-PCR.
Total RNA was extracted with Qiagen kit, and RT-First Strand cDNA Synthesis kit (Amersham Pharmacia Biotech) was used. The primers used for RT-PCR analysis are listed in Table S2. qRT-PCR was performed in LightCycler480 system using SYBR Green I Master kit (Roche).
Chromatin Immunoprecipitation.
ChIP assay are described in ref. 51. Briefly, Chromatin from cross-linked cells was sonicated, incubated overnight with the indicated antibodies in RIPA buffer and precipitated with protein G/A–Sepharose. Cross-linkage of the coprecipitated DNA-protein complexes was reversed, and DNA was used as a template for the PCR. α-cleaved Notch1 (ab8925; Abcam), and α-β-catenin (BD Bioscience; catalog no. 61054) antibodies were used. The primers used are listed in Table S2.
Human Colorectal Samples.
Samples from FAP patients were obtained from the Tumor Bank of ICO-IDIBELL. All patients gave written consent to donate the tumor specimen. The study was approved by the Ethics Committee of our institution.
In Vivo Tumor-Growth Assay.
Twenty male nu/nu Swiss mice per experiment (Harlam) were housed in a sterile environment. Animals were injected s.c. with 1.5 × 106 Ls174T/dnTCF4 cells in the left leg and 1.5 × 106 Ls174T/dnTCF4/N1IC cells in the right leg. The control group drank 1% sucrose and the treated group 1% sucrose, 2 mg/mL doxycycline in water ad libitum. Tumor volumes were calculated as (length × width2)π/6.
Statistical Analysis.
SAS version 9.1.3 (SAS Institute Inc.) and SPSS version 12 softwares were used.
Supplementary Material
Acknowledgments.
We thanks to H. Clevers, A. García de Herreros (Institut Municipal d'Investigaciones Mèdiques, Barcelona, Spain), E. Batlle (Institute for Research in Biomedicine, Barcelona, Spain), and E. Sancho (Institute for Research in Biomedicine, Barcelona, Spain) for reagents; Novartis for PKF115-584; M.E. López for technical assistance; and J. Inglés-Esteve for helpful discussions. This work was supported by an Institut d'Investigació Biomèdica de Bellvitge (V.R.) and Department d'Universitats i Recerca 2005-FI00458 (to V.F.M.) fellowships; Ministerio Educación y Ciencia Grants SAF2004-03198 and SAF2007-60080, Instituto de Salud Carlos III Grant PI041890, Fundació la Marató de TV3 Grant 051730, Fundación Mutua Madrileña; and RTICC Red Temática de Investigación Cooperativa en Cáncer Grant RD06/0020/0098. L.E. is an Investigator of the Carlos III program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813221106/DCSupplemental.
References
- 1.Willert K, Nusse R. Beta-catenin: A key mediator of wnt signaling. Curr Opin Genet Dev. 1998:95–102. doi: 10.1016/s0959-437x(98)80068-3. [DOI] [PubMed] [Google Scholar]
- 2.Korinek V, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 3.Groden J, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 4.Nishisho I, et al. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science. 1991;253:665–669. doi: 10.1126/science.1651563. [DOI] [PubMed] [Google Scholar]
- 5.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 6.Su LK, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 7.Batlle E, et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 8.van Es JH, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 9.van de Wetering M, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Majada V, et al. Nuclear IKK activity leads to dysregulated notch-dependent gene expression in colorectal cancer. Proc Natl Acad Sci USA. 2007;104:276–281. doi: 10.1073/pnas.0606476104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan X, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64:7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 12.Balint K, et al. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. J Clin Invest. 2005;115:3166–3176. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuevas IC, et al. Meningioma transcript profiles reveal deregulated Notch signaling pathway. Cancer Res. 2005;65:5070–5075. doi: 10.1158/0008-5472.CAN-05-0240. [DOI] [PubMed] [Google Scholar]
- 14.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 17.Mumm JS, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 18.Lai EC. Notch signaling: Control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 19.Fre S, et al. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 20.Riccio O, et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27(Kip1) and p57(Kip2) EMBO Rep. 2008;9:377–383. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicks C, et al. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat Cell Biol. 2000;2:515–520. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- 22.Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. MolCell Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- 23.Couso JP, Martinez Arias A. Notch is required for wingless signaling in the epidermis of Drosophila. Cell. 1994;79:259–272. doi: 10.1016/0092-8674(94)90195-3. [DOI] [PubMed] [Google Scholar]
- 24.Giraldez AJ, Cohen SM. Wingless and Notch signaling provide cell survival cues and control cell proliferation during wing development. Development. 2003;130:6533–6543. doi: 10.1242/dev.00904. [DOI] [PubMed] [Google Scholar]
- 25.Axelrod JD, Matsuno K, Artavanis-Tsakonas S, Perrimon N. Interaction between Wingless and Notch signaling pathways mediated by dishevelled. Science. 1996;271:1826–1832. doi: 10.1126/science.271.5257.1826. [DOI] [PubMed] [Google Scholar]
- 26.Hayward P, Balayo T, Martinez Arias A. Notch synergizes with axin to regulate the activity of armadillo in Drosophila. Dev Dyn. 2006;235:2656–2666. doi: 10.1002/dvdy.20902. [DOI] [PubMed] [Google Scholar]
- 27.Alves-Guerra MC, Ronchini C, Capobianco AJ. Mastermind-like 1 Is a specific coactivator of beta-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res. 2007;67:8690–8698. doi: 10.1158/0008-5472.CAN-07-1720. [DOI] [PubMed] [Google Scholar]
- 28.Espinosa L, Ingles-Esteve J, Aguilera C, Bigas A. Phosphorylation by glycogen synthase kinase-3 beta down-regulates Notch activity, a link for Notch and Wnt pathways. J Biol Chem. 2003;278:32227–32235. doi: 10.1074/jbc.M304001200. [DOI] [PubMed] [Google Scholar]
- 29.Estrach S, Ambler CA, Lo Celso C, Hozumi K, Watt FM. Jagged 1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development. 2006;133:4427–4438. doi: 10.1242/dev.02644. [DOI] [PubMed] [Google Scholar]
- 30.Leong KG, et al. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J Exp Med. 2007;204:2935–2948. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinmaster G, Kopan R. A garden of Notch-ly delights. Development. 2006;133:3277–3282. doi: 10.1242/dev.02515. [DOI] [PubMed] [Google Scholar]
- 32.Dalerba P, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baba I, et al. Involvement of deregulated epiregulin expression in tumorigenesis in vivo through activated Ki-Ras signaling pathway in human colon cancer cells. Cancer Res. 2000;60:6886–6889. [PubMed] [Google Scholar]
- 34.Kim JH, et al. The Bmi-1 oncoprotein is overexpressed in human colorectal cancer and correlates with the reduced p16INK4a/p14ARF proteins. Cancer Lett. 2004;203:217–224. doi: 10.1016/j.canlet.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 35.Pardal R, Molofsky AV, He S, Morrison SJ. Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harb Symp Quant Biol. 2005;70:177–185. doi: 10.1101/sqb.2005.70.057. [DOI] [PubMed] [Google Scholar]
- 36.Blache P, et al. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J Cell Biol. 2004;166:37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batlle E, et al. EphB receptor activity suppresses colorectal cancer progression. Nature. 2005;435:1126–1130. doi: 10.1038/nature03626. [DOI] [PubMed] [Google Scholar]
- 38.Bastide P, et al. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol. 2007;178:635–648. doi: 10.1083/jcb.200704152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori-Akiyama Y, et al. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology. 2007;133:539–546. doi: 10.1053/j.gastro.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 40.Laurent E, et al. Nox1 is over-expressed in human colon cancers and correlates with activating mutations in K-Ras. Int J Cancer. 2008;123:100–107. doi: 10.1002/ijc.23423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lepourcelet M, et al. Small-molecule antagonists of the oncogenic Tcf/beta-catenin protein complex. Cancer Cell. 2004;5:91–102. doi: 10.1016/s1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 42.Xue Y, et al. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet. 1999;8:723–730. doi: 10.1093/hmg/8.5.723. [DOI] [PubMed] [Google Scholar]
- 43.Sansom OJ, et al. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 44.Korinek V, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki K, et al. Hes1-deficient mice show precocious differentiation of Paneth cells in the small intestine. Biochem Biophys Res Commun. 2005;328:348–352. doi: 10.1016/j.bbrc.2004.12.174. [DOI] [PubMed] [Google Scholar]
- 46.Jensen J, et al. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 47.Nandan MO, et al. Kruppel-like factor 5 mediates cellular transformation during oncogenic KRAS-induced intestinal tumorigenesis. Gastroenterology. 2008;134:120–130. doi: 10.1053/j.gastro.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szanto I, et al. Expression of NOX1, a superoxide-generating NADPH oxidase, in colon cancer and inflammatory bowel disease. J Pathol. 2005;207:164–176. doi: 10.1002/path.1824. [DOI] [PubMed] [Google Scholar]
- 49.Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci USA. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robert-Moreno A, et al. Impaired embryonic haematopoiesis yet normal arterial development in the absence of the Notch ligand Jagged1. EMBO J. 2008;27:1886–1895. doi: 10.1038/emboj.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aguilera C, Hoya-Arias R, Haegeman G, Espinosa L, Bigas A. Recruitment of IkappaBalpha to the hes1 promoter is associated with transcriptional repression. Proc Natl Acad Sci USA. 2004;101:16537–16542. doi: 10.1073/pnas.0404429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fre S, et al. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci USA. 2009 Feb 27; doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.