Abstract
To look for a direct role of ultraviolet radiation (UV) exposure in cutaneous melanoma induction, we studied xeroderma pigmentosum (XP) patients who have defective DNA repair resulting in a 1000-fold increase in melanoma risk. These XP melanomas have the same anatomic distribution as melanomas in the general population. We analyzed laser capture microdissection samples of skin melanomas from XP patients studied at the National Institutes of Health. The tumor suppressor gene PTEN was sequenced and analyzed for UV-induced mutations. Samples from 59 melanomas (47 melanomas in situ and 12 invasive melanomas) from 8 XP patients showed mutations in the PTEN tumor suppressor gene in 56% of the melanomas. Further, 91% of the melanomas with mutations had 1 to 4 UV type base substitution mutations (occurring at adjacent pyrimidines) (P < 0.0001 compared to random mutations). We found a high frequency of amino-acid-altering mutations in the melanomas and demonstrated that these mutations impaired PTEN function; UV damage plays a direct role in induction of mutations and in inactivation of the PTEN gene in XP melanomas including in situ, the earliest stage of melanoma. This gene is known to be a key regulator of carcinogenesis and therefore these data provide solid mechanistic support for UV protection for prevention of melanoma.
Keywords: DNA repair, ultraviolet radiation, PTEN, skin cancer, laser capture microdissection
Melanoma mortality for Caucasians in the U.S. has increased at an annual rate of 1.5% from 1950 to 2005. For 2008, more than 110,000 new cases of melanoma [about 50,000 melanoma in situ (MIS) and 60,000 invasive melanomas (IM)] are estimated with more than 8,000 deaths (1, 2). The causative relationship between UV exposure and non-melanoma skin cancer is well documented (3–6), and important molecular targets have been identified in basal cell carcinoma (patched protein in the hedgehog pathway) and squamous cell carcinoma (SCC) (p53) in normal and xeroderma pigmentosum (XP) patients (7–9). In contrast, the relationship between sunlight exposure and melanoma is less well understood but still apparent (10, 11). The role of UV exposure in melanoma pathogenesis is complex and has some paradoxical features (5, 10, 12). In the U.S. Caucasian population and in XP patients, the site distribution of melanomas is similar. However, this anatomic distribution is different from that of basal cell carcinoma and squamous cell carcinoma and does not correspond to the most sun exposed areas of the body (face, head, and neck) (5, 13). Similarly, melanoma is more closely associated with intermittent intense sun exposure rather than long term, constant exposure as in SCC (5, 6). On the other hand, melanomas can be induced in some animals by exposure to UV (14–16).
Solar UV (including UVB and UVA) induces photoproducts at adjacent pyrimidines, predominately cyclobutane dimers and 6–4 pyrimidine-pyrimidone photoproducts (17), which, if not repaired, can lead to base substitution mutations at the site of damage (17, 18). XP patients, who have defective DNA repair, have a 1000-fold increase in melanoma and individual patients often have multiple melanomas (4, 19). XP thus provides a powerful model for the study of the molecular pathogenesis of melanoma in humans and permits evaluation of a large number of melanomas with the same exposure history and genetic background in a small number of patients. While the mechanism of induction of UV damage in cells from normal individuals and in XP patients is the same, the effects of UV damage are amplified in XP since the damage is not repaired.
The functional loss of tumor suppressor genes is a fundamental cause of cancer progression. The p53 tumor suppressor gene has been shown to have a high frequency of UV type base substitution mutations in human cutaneous SCC and precancerous lesions, a feature of UV-induced carcinogenesis (7–9). However, p53 is mutated in only a small proportion of melanomas (20–22). The tumor suppressor gene PTEN (phosphatase and tensin homologue) is one of the most frequently mutated genes in human cancer (23). Other studies have described PTEN loss or mutation in cancer specimens, cancer cell lines, and inherited cancer predisposition syndromes (20, 22–28). We hypothesized that analysis of base substitution mutations in this gene in melanomas from XP patients could provide evidence for UV induction of the mutations and thereby demonstrate a role of UV in causation of melanomas.
Results
Melanomas and Mutations.
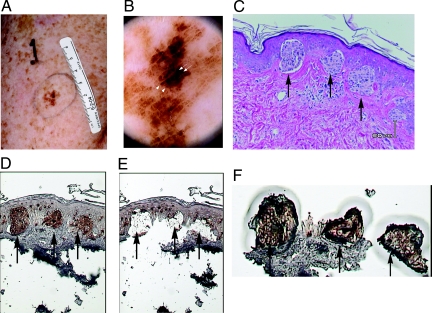
We studied melanomas from XP patients who were examined at the National Institutes of Health Clinical Center from 1971 to 2008. Pigmented lesions in the patients were evaluated clinically (Fig. 1A). Lesions in patients seen recently were evaluated by dermatoscopy, a technique that increases melanoma detection by use of detailed examination of magnified pigmented lesions (29) (Fig. 1B). Melanoma samples were processed, diagnosed, and archived by the National Cancer Institute Laboratory of Pathology. After pathological diagnosis, additional histological sections were prepared for use in laser capture microdissection (Fig. 1 C-F). Melan-A immunohistochemistry (30) was used to localize the tumor cells (Fig. 1D).
Fig. 1.
Melanoma identification and laser capture microdissection of melanoma cells. (A) A pigmented lesion on the left upper arm of a 52-year-old XP patient, XP295BE, showing the melanoma features of asymmetry, border irregularity, color variation, and large diameter. (B) Dermatoscopic image of A (20× magnified) showing a central, asymmetric, homogeneously hyperpigmented area with irregularly distributed black dots (arrows) and irregular streaks (arrowheads). (C) Atypical melanocytes are present singly and in nests at the dermal-epidermal junction (arrows) and show focal extension into the superficial dermis (hematoxyalin and eosin stain). (D) Melan-A staining showing melanoma cells expressing Melan-A (arrows). (E) Tissue remaining after capture of the melanoma cells showing high efficiency of capture (arrows). (F) Nests of melanoma cells captured (arrows) along with small amount of adjacent tissue.
We studied 59 melanomas from 8 XP patients (5 females and 3 males) ranging in age from 28 to 63 years (Table 1). There were 47 MIS, the earliest lesion that can be histologically identified as melanoma, and 12 invasive melanomas (IM). Table 2 summarizes the pathological and mutational features of all melanoma samples. None were metastatic. The anatomic distribution of XP melanomas in our series was similar to the distribution in the U.S. general population, in agreement with a previous report (13). Of the 59 melanomas 54% were on the lower extremity, 19% on the face or scalp, 14% on the upper extremities and 14% on the shoulder or back. There were no melanomas on the palms or soles (Table S1). Fifty-six percent (33) of the melanomas had PTEN mutations and 91% (30) of the melanomas with mutations had UV type mutations occurring at dipyrimidine sites (Tables 1 and 2). PTEN mutations were found in 57% (27) of the MIS.
Table 1.
XP patients, melanomas, and PTEN mutations
| Patient | Age*/sex | Number of melanomas with PTEN sequencing | Number of melanomas with PTEN mutations | Number of melanomas with UV type PTEN mutations | Number of PTEN mutations | Number of UV type PTEN mutations |
|---|---|---|---|---|---|---|
| XP295BE†,‡ | 49/F | 5 (0)§ | 3 (0)¶ | 3 (0) | 5 (0) | 5 (0) |
| XP86BE†,‡ | 52/F | 2 (0) | 1 (0) | 1 (0) | 2 (0) | 1 (0) |
| XP376BE†,‡ | 44/F | 3 (0) | 2 (0) | 2 (0) | 2 (0) | 2 (0) |
| XP21BE† | 28/F | 9 (4) | 5 (1) | 3 (1) | 7 (1) | 5 (1) |
| XP24BE† | d35/F | 6 (0) | 2 (0) | 2 (0) | 3 (0) | 2 (0) |
| XP29BE‖ | d37/M | 18 (5) | 10 (3) | 9 (3) | 15 (6) | 13 (6) |
| XP31BE** | 63/M | 2 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| XP1BE† | d49/F | 14 (2) | 10 (2) | 10 (2) | 20 (4) | 20 (4) |
| Total | 59 (12) | 33 (6) | 30 (6) | 54 (11) | 48 (11) | |
| Frequency (% of total number of melanomas (n = 59(12)§)) | 100% (100%) | 56%†† (50%)‡‡ | 91%§§ (100%)¶¶ | |||
| Frequency (% of total number of PTEN mutations (n = 54(11)§)) | 100% (100%) | 89% (100%) |
*Age at last melanoma or age at death (d)
†Complementation group C
‡Members of same kindred
§Number of invasive melanomas
¶Number of invasive melanomas with PTEN mutations
‖Complementation group D
**XP variant
††% of melanomas
‡‡% of invasive melanomas
§§% of melanomas with mutations
¶¶% of invasive melanomas with mutations
Table 2.
Characteristics of 33 XP melanomas with PTEN mutations
| Patient* | Tumor no. | Tumor |
Mutated codon |
PTEN gene |
||||
|---|---|---|---|---|---|---|---|---|
| Histologic type† | Location | Mutation‡ | UV type mut?§ | LOH?¶ | Amino acid sub | |||
| XP295BE | 1 | MIS | ARM | 95 | cCc283cTc | + | − | Pro/Ser‖ |
| XP295BE | 2 | MIS | ABD | 72 | cTg216cCg | + | − | - |
| XP295BE | 2 | 74 | gAc222 gGc | + | − | - | ||
| XP295BE | 2 | 245 | aGc735aTc | + | − | Gln/His | ||
| XP295BE | 3 | MIS | ANKLE | 267 | aGg801aTg | + | − | Leu/Asn |
| XP86BE | 4 | MIS | BACK | 301 | aTa903aCa | − | + | - |
| XP86BE | 4 | 332 | aAg996aGg | + | + | - | ||
| XP376BE | 5 | MIS | SHIN | 154 | tCt462tGt | + | + | Phe/Leu‖ |
| XP376BE | 6 | MIS | ARM | intron 4 | aGt→aAt | + | + | Splice |
| XP21BE | 7 | MIS | LEG | 230 | aGg688aTg | + | − | Gly/Stop |
| XP21BE | 7 | 316 | tCt946tTt | + | + | - | ||
| XP21BE | 8 | MIS | LEG | 312 | aCa936aTa | − | − | - |
| XP21BE | 9 | MIS | THIGH | 250 | gTg750 gCg | − | − | - |
| XP21BE | 10 | IM | LEG | 362 | tCa1085tTa | + | − | Ser/Leu |
| XP21BE | 11 | MIS | LEG | 98 | gCt292 gTt | + | + | - |
| XP21BE | 11 | 104 | tTt311tCt | + | − | Phe/Ser | ||
| XP24BE | 12 | MIS | LEG | 98 | gCt292 gTt | + | + | - |
| XP24BE | 13 | MIS | ANKLE | 95 | cAc285cTc | − | + | - |
| XP24BE | 13 | 110 | cAa329cGa | + | + | Gln/Arg | ||
| XP29BE | 14 | IM | SCALP | 95 | cCc283cTc | + | − | Pro/Ser‖ |
| XP29BE | 14 | 103 | cCt309cTt | + | − | - | ||
| XP29BE | 14 | 114 | aAg342aTg | + | − | Glu/Asp | ||
| XP29BE | 15 | MIS | BACK | 362 | tTc1084tCc | + | + | Ser/Leu |
| XP29BE | 16 | MIS | CHEEK | 236 | gAc707 gGc | + | − | Asp/Gly |
| XP29BE | 17 | MIS | SHOULDER | 243 | tTc728tCc | + | + | Phe/Ser |
| XP29BE | 17 | 223 | aAg668aGg | + | + | Lys/Arg | ||
| XP29BE | 18 | MIS | SHOULDER | 98 | gCt292 gTt | + | − | - |
| XP29BE | 19 | MIS | Cheek | 134 | aTg401aCg | − | − | Met/Thr |
| XP29BE | 20 | IM | Chin | 98 | gCt292 gTt | + | − | - |
| XP29BE | 21 | MIS | ARM | 272 | cAc815cGc | − | − | His/Gln |
| XP29BE | 21 | 291 | gAg872 gGa | + | − | Glu/Gly | ||
| XP29BE | 22 | MIS | SCALP | 325 | tCt973tTt | + | + | Leu/Phe |
| XP29BE | 23 | IM | Chin | 98 | gCt292 gTt | + | + | - |
| XP29BE | 23 | 149 | aAg447aCg | + | − | Gln/His | ||
| XP1BE | 24 | MIS | LEG | 55 | aGg164aAg | + | − | Arg/Lys |
| XP1BE | 24 | 235 | gGa703 gAa | + | − | Glu/Lys | ||
| XP1BE | 25 | MIS | LEG | 25 | cTt73cCt | + | − | - |
| XP1BE | 26 | MIS | LEG | 248 | cTg744cCg | + | − | - |
| XP1BE | 27 | MIS | SHIN | 9 | gTt26 gCt | + | − | Val/Ala |
| XP1BE | 28 | MIS | FOOT | 301 | cGa901cAa | + | + | Asp/Asn |
| XP1BE | 29 | MIS | FOOT | 23 | cTt67cCt | + | − | - |
| XP1BE | 29 | MIS | FOOT | 16 | aTc48aAc | + | − | Tyr/Stop |
| XP1BE | 29 | MIS | FOOT | 44 | gGc131 gAc | + | + | Gly/Asp |
| XP1BE | 30 | IM | LEG | 70 | cTt209cCt | + | − | Leu/Pro |
| XP1BE | 30 | 301 | cGa901cAa | + | + | Asp/Asn | ||
| XP1BE | 30 | intron 8 | aAg→aGg | + | + | Splice | ||
| XP1BE | 31 | IM | LEG | 83 | tTg247tCg | + | − | Cys/Arg |
| XP1BE | 32 | MIS | LEG | 24 | gAc71 gGc | + | − | Asp/Gly |
| XP1BE | 32 | 44 | gGc131 gAc | + | + | Gly/Asp | ||
| XP1BE | 32 | 84 | cAg250cGg | + | − | Arg/Gly | ||
| XP1BE | 33 | MIS | BACK | 95 | cCc283cTc | + | − | Pro/Ser‖ |
| XP1BE | 33 | 301 | cGa901cAa | + | − | Asp/Asn | ||
| XP1BE | 33 | 388 | aGa1162aAa | + | + | Glu/Lys | ||
| XP1BE | 33 | 403 | aGt1207aAt | + | + | Val/Iie | ||
*Patients are indcated by XP numbers
†MIS Melanoma in situ, IM invasive melanoma
‡Base substitution mutations are indicated as capital letters with adjacent nucleotides and cDNA location on the coding strand
§+dipyrimidine, − not dypyrimidine
¶+ loss of heterozygosity, − no loss of heterozygosity
‖Cancer associated mutation listed in Sanger database Catalogue of Somatic Mutations in Cancer
High Frequency of UV-Type Mutations.
We found 54 PTEN base substitution mutations in the 33 melanomas (Tables 1 and 2). Forty-two percent (14) of the melanomas with PTEN mutations showed more than one mutation, a feature of UV mutagenesis in tumors (7, 9) and in UV-treated plasmids (18). Eight melanomas had 2 mutations, 5 melanomas showed 3 mutations, and 1 carried 4 mutations (Table S2). Among these 54 mutations 89% were UV type mutations occurring at adjacent pyrimidines. This frequency of mutations at dipyrimidines is significantly greater (P < 0.0001) than that predicted by random mutagenesis based on the frequency of dipyrimidines in the PTEN region sequenced (Table 3). Forty-eight of the mutations were UV type occurring at adjacent pyrimidines, and 6 were non-UV type (Tables 2 and 3). Loss of heterozygosity (LOH) (32) was found in 48% of the 33 melanomas with PTEN mutations. All of these melanomas had UV type mutations. There was a predominance of transition type base substitution mutations (85%) (Table 4), a feature of UV mutagenesis (17, 33). The equal frequency of G:C to A:T and A:T to G:C mutations is different from what was previously reported in studies of non-melanoma skin cancers in XP and non-XP patients (7–9) and in UV-treated plasmids (33) where G:C to A:T mutations predominated. The observed equal frequency of C to T and T to C transitions is suggestive of the mutation spectrum described for the HPRT gene in UV-treated human or mouse cells lacking error prone polymerase eta (31, 34). Of the 6 non-UV mutations (at alternating pyrimidines and purines) 5 involved A:T base pairs and 4 resulted in A:T to G:C mutations (Table 2). Oxidative damage has been reported to produce lesions such as cycloadenines which are poorly repaired by XP cells (35) and might result in mutagenic lesions at A:T base pairs.
Table 3.
Frequency of UV type base substitution mutations in PTEN gene in melanomas compared to internal tumors
| Number of PTEN base substitution mutations (frequency) |
|||
|---|---|---|---|
| UV type | Non-UV type | Total | |
| XP Melanoma | 48 (89%) | 6 (11%) | 54 (100%) |
| Non XP Melanoma* | 20 (87%) | 3 (13%) | 23 (100%) |
| Cancer of endometrium* | 86 (48%)† | 93 (52%) | 179 (100%) |
| Cancer of central nervous system* | 171 (63%)† | 101 (37%) | 272 (100%) |
| PTEN sequence (expected freq) | 746 bp (54%)†,‡ | 641 bp (46%)§ | 1387 bp (100%) |
*from non-XP patients in Sanger database Catalogue of Somatic Mutations in Cancer http://www.sanger.ac.uk/perl/genetics/CGP/cosmic including 2 primary melanomas, 11 metastatic melanomas and 10 melanoma cultures.
†P < 0.0001 vs XP Melanoma
‡Frequency of adjacent pyrimidines or purines
§Frequency of alternating pyrimidines/purines
Table 4.
Types of PTEN mutations and alterations of amino acids in XP melanomas
| XP Melanoma (number of PTEN mutations) | % | AA changed* (number) | |
|---|---|---|---|
| Transitions | |||
| G:C to A:T | 23 | 42.5 | 15 |
| A:T to G:C | 23 | 42.5 | 15 |
| Transversions | |||
| G:C to T:A | 3 | 5.5 | 3 |
| G:C to C:G | 1 | 2 | 1 |
| A:T to T:A | 3 | 5.5 | 2 |
| A:T to C:G | 1 | 2 | 1 |
| Total | 54 | 100 | 37 (69%) |
*including 33 missense, 2 nonsense and 2 splicing mutations (from Table 2)
Location and Effect of PTEN Mutations.
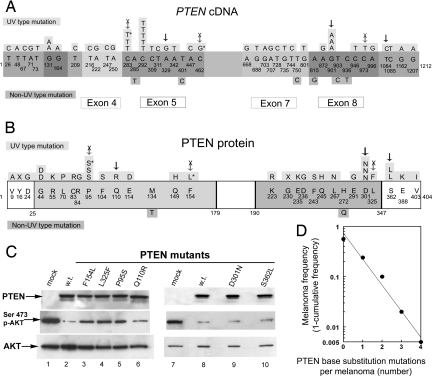
Figure 2 illustrates the distribution of mutations in the cDNA and protein domains of PTEN. In our study, 52 base substitution PTEN mutations were located in 8 of the 9 exons. Overall, 37 (69%) of the base substitution mutations resulted in changes of amino acids; these were found for all classes of mutations (Table 4). Inactivation of the PTEN tumor suppressor gene would be expected to result from the 2 nonsense mutations (p.Y16X and p.G230X) and the 2 splice mutations (Table 2). Thirty-one UV type and 2 non-UV type base substitutions resulted in missense mutations (Tables 2 and 4). Four missense mutations occurred more than once (c.131G>A; c.283C>T; c.292C>T; and c.901G>A). Twenty-seven of these missense mutations were located inside the phosphatase and C2 calcium/lipid-binding domains of PTEN protein. Of the 33 melanomas with PTEN mutations, 25 (76%) had one or more changed amino acid (Table 2). There were 17 synonymous (silent) base substitution mutations (13 UV type and 4 non-UV type) that would not be expected to alter PTEN function, 8 of these were present in melanomas that also had one or more missense mutations that could affect PTEN function (Table 2).
Fig. 2.
Location of PTEN mutations in melanomas and effect of mutations on PTEN function. (A) Sites of 52 PTEN cDNA base substitution mutations found in 33 melanomas from 8 XP patients. The 9 exons of the 1212 bp PTEN cDNA are indicated. Mutated bases are numbered. UV type mutations are indicated above the exons and non-UV type mutations are indicated below the exons. An * indicates cancer-associated mutation listed in the Sanger COSMIC database (22). Vertical arrows indicate mutations tested for alteration of PTEN function. Arrows with X indicate mutations that result in decreased PTEN function. (B) Sites of 33 PTEN nonsynonymous amino acid substitution mutations and 2 nonsense mutations found in 33 melanomas from 8 XP patients. The 404 aa protein has a dual specificity protein phosphatase domain from amino acid 25 to 179, a tyrosine specific protein phosphatase region from amino acid 123 to 134 and a C2 calcium/lipid-binding region, (CaLB) from amino acid 190 to 347. The altered amino acids are numbered. UV type mutations are indicated above the sequence and non-UV type mutations are indicated below. An * indicates cancer associated mutation listed in Sanger COSMIC database (22). Vertical arrows indicate mutations tested for alteration of PTEN function. Arrows with X indicate mutations that result in decreased PTEN function. (C) Functional assay of phosphorylation of Akt by selected PTEN mutants. NCI-H1155 PTEN-null cells were transfected with pCMV5 HA-PKB/Akt (containing hemagglutinin (HA) tagged PKB/Akt) plus pCMV5 with wild type (w.t.) (lanes 2 and 8) or mutated (lanes 3–6, 9–10) PTEN or empty vector (mock) (lanes 1 and 7). After 24 h, the cells were lysed and immunoprecipitated with anti-HA antibodies. This HA-PKB/Akt was analyzed by Western blotting with an anti-Akt phosphoserine 473 antibody (Middle). The membrane was then stripped and analyzed using an antibody against total Akt (Lower). Expression of transfected PTEN proteins was confirmed by anti-PTEN Western blot analysis of cell lysates (Upper). The phosphorylation of Akt by mutants p.F154L (lane 3), p.L325F (lane 4), and p.P95S (lane 5) indicates loss of PTEN suppressor function while the low level of phosphorylation of Akt by mutants p.Q110R (lane 6), p.D301N (lane 9), and p.S362L (lane 10) indicates preservation of PTEN suppressor function. (D) Relationship of number of PTEN base substitution mutations per melanoma to melanoma frequency (1- cumulative frequency). The data are consistent with a random (Poisson) distribution of mutations.
PTEN functions as a tumor suppressor through its lipid phosphatase function (36), and it is responsible for phosphatidylinositol triphosphate (PIP3) dephosphorylation and clearance. This process is required to antagonize the PI3K-dependent activation of Akt (37) and is a prognostic marker for melanoma progression (38). To determine the effect of missense mutations on PTEN function, we investigated the phosphorylation of Akt in cells lacking PTEN. We selected isolated PTEN mutations that occurred in association with LOH (Table 2 and Fig. 2 A and B). We transfected the cells with expression vectors for 6 UV type missense mutants and a wild type control. Western blot analysis showed that the expression of wild-type PTEN dramatically reduced the level of Serine 473 Akt phosphorylation (Fig. 2C, lanes 2 and 8). In contrast, expression of the PTEN p.F154L, p.L325F, and p.P95S (Fig. 2C, lanes 3, 4 and 5) mutants did not reduce AKT phosphorylation to the same extent indicating that these mutations impaired PTEN function. Two of these mutations (p.P95S and p.F154L) were previously reported to be associated with cancers (24, 39) (Fig. 2B). In contrast, PTEN mutants p.Q110R, p.D301N, and p.S362L reduced AKT phosphorylation to a similar extent as wild-type PTEN (Fig. 2C, lanes 6, 9 and 10) indicating that these mutations did not alter this PTEN function.
Discussion
Because XP patients have defective DNA repair, UV damage more frequently leads to mutations, and therefore it is easier to detect their consequences in a shorter time and with fewer patients than in the general population (4, 17, 19). In this study, we found UV type mutations in the tumor suppressor gene PTEN, a gene that is a key regulator of carcinogenesis (20, 22–28, 36), providing direct molecular evidence of UV involvement in melanoma induction in humans.
Tumor suppressor genes lead to cancer by being inactivated in contrast to oncogenes, where mutations must cause activation to cause cancer. Since a wider range of mutations can inactivate genes, an effect of UV on tumor suppressors can be more easily detected compared to mutations that activate oncogenes. The PTEN tumor suppressor is one of the most frequently mutated genes in human cancer (23) and PTEN mutations have been detected in up to 40% of cutaneous melanoma cell lines (20, 22, 40). While many prior studies looked for deletions, promoter methylations and immunohistochemical evidence of PTEN inactivation (41–44), we looked for and found base substitution mutations as an indicator of UV damage.
Evidence for UV as Cause of Mutations.
If the dipyrimidine (UV type) base substitution mutations that we observed had occurred randomly, they would be expected to occur at the same frequency (54%) as the dipyrimidine sites present in the PTEN gene. However, we found mutations in XP melanomas to occur at dipyrimidines at a significantly greater frequency (89%, P < 0.0001) (Table 3), a feature characteristic of UV mutagenesis. The frequency of UV type mutations (occurring at dipyrimidines) in XP melanomas (89%) was similar to that in non-XP melanomas (87%) (22) (Table 3) indicating that the role of UV in inducing melanomas is similar in both. In contrast, the overall frequency of PTEN base substitution mutations in exons in the non-XP melanomas was 6% (23/381—including primary, metastatic, and cultured melanomas) (40, 45–51). This is about 10-fold lower than the 56% we found in the primary XP melanomas (Table 1), probably reflecting the UV hypermutability of XP cells (17) and the 1000-fold increase in melanoma frequency in XP patients (13, 52). We did not perform DNA sequencing of the PTEN introns in the XP melanomas, and thus it is possible that there were additional mutations in the introns as reported in non-XP melanomas (49).
The frequency of dipyrimidine mutations in internal tumors in non-UV-exposed sites such as endometrium cancers (48%) and central nervous system cancers (63%) was significantly lower than that in XP or non-XP melanomas and was similar to that predicted by random mutations (54%) (Table 3), thus showing a large contrast between internal tumors and melanomas.
Loss of heterozygosity in a cell represents the loss of one allele of a gene. It is a common occurrence in cancer where it indicates selection related to the absence of a functional tumor suppressor gene in the lost region (32). We found 48% of the melanomas with PTEN mutations had LOH. All of the XP melanomas with LOH had UV type mutations in the remaining allele (88% with missense mutations) indicating a major role of UV in induction of these melanomas.
The occurrence of multiple mutations in a single gene is another feature of UV mutagenesis. Multiple mutations of the p53 tumor suppressor gene were common in human sunlight-related skin tumors in normal (8, 9) and XP patients (7). We found 42% (14) of the 33 XP melanomas with PTEN mutations had as many as 4 different mutations (Table S2). In 6 of the XP melanomas, all of the 200–300 cells removed by laser capture microdissection came from a single microscopic nest of tumor cells and had 2, 3, or 4 mutations, thus suggesting that they were derived from a single clone with multiple mutations. However, it is possible that molecular heterogeneity was present within a single microscopic nest of melanoma cells. The number of PTEN base substitution mutations per melanoma is consistent with a Poisson distribution (Fig. 2D), suggesting that these mutations accumulated randomly at sites of unrepaired UV damage. Seven of the XP melanomas had one or more mutations that altered amino acids in association with one or more silent mutations (Table 2). It is likely that the amino acid altering mutations accumulated and provided positive selection which would account for the persistence of the silent mutations. In contrast to these skin melanomas, multiple mutations were very rarely reported in PTEN in internal (UV-protected) tumors (4% of 503 central nervous system and 13% of 512 endometrial cancers) (22).
The tumor suppressor functions of PTEN are thought to be mediated by maintaining downstream Akt in a dephosphorylated state (36); however, other functions have been proposed (23, 37, 53). The 2 nonsense and 2 splice mutations in the PTEN gene in the XP melanomas (Table 2) are very disruptive and would be expected to impair protein function. We found 33 missense mutations and many of these were UV type mutations within exons 5 and 8 that control phosphatase activity and protein turnover, respectively. To determine if the missense mutations altered PTEN function, 6 PTEN missense mutations were analyzed in an in vitro assay for the ability to inhibit the activation of the proto-oncogene Akt, one of the downstream activities of PTEN (36). In 3 of these, the ability of PTEN mutant protein to inhibit the activation of Akt was impaired, demonstrating functional impairment of PTEN. Two of these mutations have previously been reported in other cancers (24, 39). The missense mutations may also alter other PTEN functions (23, 37, 53).
UV Type PTEN Mutations in Melanomas in XP and in the General Population.
We found that different melanomas in the same patient may have different PTEN mutations (Table 2), indicating that each melanoma arose independently. The importance of these mutations in the pathogenesis of the melanomas is indicated by the finding that PTEN mutations were present in 57% of 47 MIS, signifying that PTEN mutation occurs at an early stage in melanoma induction. Furthermore, 52% of the MIS with PTEN mutations also had LOH indicating absence of the normal allele and suggesting a functional role for those mutations. Finding these UV type mutations in early melanomas in association with functional impairment of PTEN substantiates the direct role of UV in the development of melanomas in XP patients. The similarity in anatomic site distribution (13) and predominance of UV type base substitution PTEN mutations (Table 3) in XP melanomas and melanomas in the general population indicates a similar role for UV induction in both, although the overall mutation rate is much greater in the XP patients. Direct molecular evidence of UV type mutations in the PTEN tumor suppressor gene provides a clear mechanistic framework for the role of UV in the induction of melanomas and a sound rationale for UV protective measures for melanoma prevention.
Methods
We studied XP patients from 1971 to 2008 at the NIH Clinical Center under protocols approved by the National Cancer Institute Institutional Review Board. The patients provided informed consent. Laser-capture microdissection was performed by use of an Arcturus PixCell II microscope (Arcturus Engineering) to separate melanoma cells from normal cells. About 300 cells were collected on CapSure LCM Caps (Arcturus Engineering) in each case from serial tissue sections. DNA was extracted using PicoPure DNA Extraction Kit as per the manufacturer's protocol (Arcturus Engineering). Genomic DNA from each patient's cultured fibroblasts was analyzed as a control.
All 9 exons of the PTEN gene were amplified as described (54). The PCR products were treated with 0.5 U shrimp alkaline phosphatase (Promega) and 5 U exonuclease I (New England Biolabs) and both strands were sequenced directly using a Prism Model 3700 Capillary Array sequencer and Big Dye Terminator Chemistry (Applied Biosystems).
We compared the mutations of PTEN in the XP melanomas to mutations in non-XP melanomas, cancers of endometrium, and cancers of the central nervous system listed in the Sanger COSMIC database (22). The Fisher's exact test was applied to compare frequency of the UV type mutations (located at adjacent pyrimidines on either strand) in the cancers to the frequency of adjacent pyrimidines on either strand in the regions of the PTEN gene sequenced.
The effect of mutations identified in the PTEN gene was examined as described (55), except we used the PTEN-null cell line NCI-H1155 (56) and expression vectors for Glu-Glu-tagged PTEN wild-type cDNA and HA-tagged AKT wild-type cDNA in pCMV5. Mutations in PTEN cDNA were introduced with the QuikChange site-directed mutagenesis kit (Stratagene). We used EzviewTM Red Anti-HA Affinity Gel (Sigma), antiphospho-active-PKB/Akt (Ser-473) antibody (Cell Signaling), anti-PKB/Akt (Cell Signaling) and anti-PTEN antibodies (Cell Signaling) in immunoblotting experiments.
Supplementary Material
Acknowledgments.
We thank Drs. Glenn Merlino and Yanlin Yu for helpful comments on the conduct of this study. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812401106/DCSupplemental.
References
- 1.Ries LAG, et al. SEER Cancer Statistics Review. [Accessed October 2008];2008 Available at 1975–2005 http://seer.cancer.gov/csr/1975_2005/
- 2.American Academy of Dermatology. Melanoma Fact Sheet. [Accessed October 2008];2008 Available at http://www.aad.org/media/background/factsheets/fact_melanoma.html.
- 3.De Gruijl FR, Van Kranen HJ, Mullenders LH. UV-induced DNA damage, repair, mutations and oncogenic pathways in skin cancer. J Photochem Photobiol B. 2001;63:19–27. doi: 10.1016/s1011-1344(01)00199-3. [DOI] [PubMed] [Google Scholar]
- 4.Kraemer KH. Sunlight and skin cancer: Another link revealed. Proc Natl Acad Sci USA. 1997;94:11–14. doi: 10.1073/pnas.94.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armstrong BK, Kricker A. The epidemiology of UV induced skin cancer. J Photochem Photobiol B. 2001;63:8–18. doi: 10.1016/s1011-1344(01)00198-1. [DOI] [PubMed] [Google Scholar]
- 6.Scotto J, Fears TR, Kraemer KH, Fraumeni JF., Jr . In: Cancer Epidemiology and Prevention. Schottenfeld D, Fraumeni JF Jr., editors. New York: Oxford Univ Press; 1996. pp. 1313–1330. [Google Scholar]
- 7.D'Errico M, et al. UV mutation signature in tumor suppressor genes involved in skin carcinogenesis in xeroderma pigmentosum patients. Oncogene. 2000;19:463–467. doi: 10.1038/sj.onc.1203313. [DOI] [PubMed] [Google Scholar]
- 8.Ziegler A, et al. Mutation hotspots due to sunlight in the p53 gene of nonmelanoma skin cancers. Proc Natl Acad Sci USA. 1993;90:4216–4220. doi: 10.1073/pnas.90.9.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziegler A, et al. Sunburn and p53 in the onset of skin cancer. Nature. 1994;372:773–776. doi: 10.1038/372773a0. [DOI] [PubMed] [Google Scholar]
- 10.Gilchrest BA, Eller MS, Geller AC, Yaar M. The pathogenesis of melanoma induced by ultraviolet radiation. N Engl J Med. 1999;340:1341–1348. doi: 10.1056/NEJM199904293401707. [DOI] [PubMed] [Google Scholar]
- 11.Houghton AN, Polsky D. Focus on melanoma. Cancer Cell. 2002;2:275–278. doi: 10.1016/s1535-6108(02)00161-7. [DOI] [PubMed] [Google Scholar]
- 12.Whiteman DC, et al. Anatomic site, sun exposure, and risk of cutaneous melanoma. J Clin Oncol. 2006;24:3172–3177. doi: 10.1200/JCO.2006.06.1325. [DOI] [PubMed] [Google Scholar]
- 13.Kraemer KH, Lee MM, Andrews AD, Lambert WC. The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer. The xeroderma pigmentosum paradigm. Arch Dermatol. 1994;130:1018–1021. [PubMed] [Google Scholar]
- 14.Kusewitt DF, Applegate LA, Ley RD. Ultraviolet radiation-induced skin tumors in a South American opossum (Monodelphis domestica) Vet Pathol. 1991;28:55–65. doi: 10.1177/030098589102800108. [DOI] [PubMed] [Google Scholar]
- 15.Setlow RB, Woodhead AD, Grist E. Animal model for ultraviolet radiation-induced melanoma: Platyfish-swordtail hybrid. Proc Natl Acad Sci USA. 1989;86:8922–8926. doi: 10.1073/pnas.86.22.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noonan FP, et al. Neonatal sunburn and melanoma in mice. Nature. 2001;413:271–272. doi: 10.1038/35095108. [DOI] [PubMed] [Google Scholar]
- 17.Friedberg EC, et al. DNA Repair and Mutagenesis. Washington, D.C.: ASM Press; 2006. [Google Scholar]
- 18.Seidman MM, Bredberg A, Seetharam S, Kraemer KH. Multiple point mutations in a shuttle vector propagated in human cells: Evidence for an error-prone DNA polymerase activity. Proc Natl Acad Sci USA. 1987;84:4944–4948. doi: 10.1073/pnas.84.14.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruenger TM, DiGiovanna JJ, Kraemer KH. In: Fitzpatrick's Dermatology in General Medicine. Wolff K, Goldsmith LA, Katz SI, Gilchrest BA, Paller AS, Leffell DJ, editors. New York: McGraw Hill; 2008. pp. 1311–1325. [Google Scholar]
- 20.Hocker T, Tsao H. Ultraviolet radiation and melanoma: A systematic review and analysis of reported sequence variants. Hum Mutat. 2007;28:578–588. doi: 10.1002/humu.20481. [DOI] [PubMed] [Google Scholar]
- 21.Lübbe J, Reichel M, Burg G, Kleihues P. Absence of p53 gene mutations in cutaneous melanoma. J Invest Dermatol. 1994;102:819–821. doi: 10.1111/1523-1747.ep12381544. [DOI] [PubMed] [Google Scholar]
- 22.Sanger Institute. Catalogue of Somatic Mutations in Cancer. [Accessed July 2008];2008 Available at http://www.sanger.ac.uk/perl/genetics/CGP/cosmic.
- 23.Baker SJ. PTEN enters the nuclear age. Cell. 2007;128:25–28. doi: 10.1016/j.cell.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 24.Abdel-Rahman MH, et al. High frequency of submicroscopic hemizygous deletion is a major mechanism of loss of expression of PTEN in uveal melanoma. J Clin Oncol. 2006;24:288–295. doi: 10.1200/JCO.2005.02.2418. [DOI] [PubMed] [Google Scholar]
- 25.Bonneau D, Longy M. Mutations of the human PTEN gene. Hum Mutat. 2000;16:109–122. doi: 10.1002/1098-1004(200008)16:2<109::AID-HUMU3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 26.Mikhail M, et al. PTEN expression in melanoma: Relationship with patient survival, Bcl-2 expression, and proliferation. Clin Cancer Res. 2005;11:5153–5157. doi: 10.1158/1078-0432.CCR-05-0397. [DOI] [PubMed] [Google Scholar]
- 27.Slipicevic A, et al. Expression of activated Akt and PTEN in malignant melanomas: Relationship with clinical outcome. Am J Clin Pathol. 2005;124:1–9. doi: 10.1309/YT58WWMTA6YR1PRV. [DOI] [PubMed] [Google Scholar]
- 28.Curtin JA, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 29.Carli P, et al. Improvement of malignant/benign ratio in excised melanocytic lesions in the “dermoscopy era”: A retrospective study 1997–2001. Br J Dermatol. 2004;150:687–692. doi: 10.1111/j.0007-0963.2004.05860.x. [DOI] [PubMed] [Google Scholar]
- 30.Sztramska A, Dymerska D, Chwirot BW. Skin layer-specific Melan-A expression during progression of human cutaneous melanoma: Implications for diagnostic applications of the marker. Melanoma Res. 2008;18:259–267. doi: 10.1097/CMR.0b013e328303beac. [DOI] [PubMed] [Google Scholar]
- 31.Dumstorf CA, et al. Participation of mouse DNA polymerase iota in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc Natl Acad Sci USA. 2006;103:18083–18088. doi: 10.1073/pnas.0605247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bishop JM. The molecular genetics of cancer. Science. 1987;235:305–311. doi: 10.1126/science.3541204. [DOI] [PubMed] [Google Scholar]
- 33.Levy DD, Saijo M, Tanaka K, Kraemer KH. Expression of a transfected DNA repair gene (XPA) in xeroderma pigmentosum group A cells restores normal DNA repair and mutagenesis of UV-treated plasmids. Carcinogenesis. 1995;16:1557–1564. doi: 10.1093/carcin/16.7.1557. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, et al. Evidence that in xeroderma pigmentosum variant cells, which lack DNA polymerase eta, DNA polymerase iota causes the very high frequency and unique spectrum of UV-induced mutations. Cancer Res. 2007;67:3018–3026. doi: 10.1158/0008-5472.CAN-06-3073. [DOI] [PubMed] [Google Scholar]
- 35.Brooks PJ. The case for 8,5′-cyclopurine-2′-deoxynucleosides as endogenous DNA lesions that cause neurodegeneration in xeroderma pigmentosum. Neuroscience. 2007;145:1407–1417. doi: 10.1016/j.neuroscience.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myers MP, et al. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sulis ML, Parsons R. PTEN: From pathology to biology. Trends Cell Biol. 2003;13:478–483. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 38.Dai DL, Martinka M, Li G. Prognostic significance of activated Akt expression in melanoma: A clinicopathologic study of 292 cases. J Clin Oncol. 2005;23:1473–1482. doi: 10.1200/JCO.2005.07.168. [DOI] [PubMed] [Google Scholar]
- 39.Kanaya T, et al. Association of mismatch repair deficiency with PTEN frameshift mutations in endometrial cancers and the precursors in a Japanese population. Am J Clin Pathol. 2005;124:89–96. doi: 10.1309/PAACLG8DXDK0X2B1. [DOI] [PubMed] [Google Scholar]
- 40.Guldberg P, et al. Disruption of the MMAC1/PTEN gene by deletion or mutation is a frequent event in malignant melanoma. Cancer Res. 1997;57:3660–3663. [PubMed] [Google Scholar]
- 41.Tsao H, Zhang X, Benoit E, Haluska FG. Identification of PTEN/MMAC1 alterations in uncultured melanomas and melanoma cell lines. Oncogene. 1998;16:3397–3402. doi: 10.1038/sj.onc.1201881. [DOI] [PubMed] [Google Scholar]
- 42.Tsao H, Mihm MC, Jr, Sheehan C. PTEN expression in normal skin, acquired melanocytic nevi, and cutaneous melanoma. J Am Acad Dermatol. 2003;49:865–872. doi: 10.1016/s0190-9622(03)02473-3. [DOI] [PubMed] [Google Scholar]
- 43.Goel VK, Lazar AJ, Warneke CL, Redston MS, Haluska FG. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J Invest Dermatol. 2006;126:154–160. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- 44.Mirmohammadsadegh A, et al. Epigenetic silencing of the PTEN gene in melanoma. Cancer Res. 2006;66:6546–6552. doi: 10.1158/0008-5472.CAN-06-0384. [DOI] [PubMed] [Google Scholar]
- 45.Birck A, Ahrenkiel V, Zeuthen J, Hou-Jensen K, Guldberg P. Mutation and allelic loss of the PTEN/MMAC1 gene in primary and metastatic melanoma biopsies. J Invest Dermatol. 2000;114:277–280. doi: 10.1046/j.1523-1747.2000.00877.x. [DOI] [PubMed] [Google Scholar]
- 46.Celebi JT, Shendrik I, Silvers DN, Peacocke M. Identification of PTEN mutations in metastatic melanoma specimens. J Med Genet. 2000;37:653–657. doi: 10.1136/jmg.37.9.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daniotti M, et al. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene. 2004;23:5968–5977. doi: 10.1038/sj.onc.1207780. [DOI] [PubMed] [Google Scholar]
- 48.Jonsson G, et al. Genomic profiling of malignant melanoma using tiling-resolution arrayCGH. Oncogene. 2007;26:4738–4748. doi: 10.1038/sj.onc.1210252. [DOI] [PubMed] [Google Scholar]
- 49.Poetsch M, Dittberner T, Woenckhaus C. PTEN/MMAC1 in malignant melanoma and its importance for tumor progression. Cancer Genet Cytogenet. 2001;125:21–26. doi: 10.1016/s0165-4608(00)00353-8. [DOI] [PubMed] [Google Scholar]
- 50.Reifenberger J, et al. Allelic losses on chromosome arm 10q and mutation of the PTEN (MMAC1) tumour suppressor gene in primary and metastatic malignant melanomas. Virchows Arch. 2000;436:487–493. doi: 10.1007/s004280050477. [DOI] [PubMed] [Google Scholar]
- 51.Tsao H, Zhang X, Fowlkes K, Haluska FG. Relative reciprocity of NRAS and PTEN/MMAC1 alterations in cutaneous melanoma cell lines. Cancer Res. 2000;60:1800–1804. [PubMed] [Google Scholar]
- 52.Kraemer KH, Lee MM, Scotto J. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch Dermatol. 1987;123:241–250. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- 53.Shen WH, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 54.Muhr D, Wagner T, Oefner PJ. Polymerase chain reaction fidelity and denaturing high-performance liquid chromatography. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;782:105–110. doi: 10.1016/s1570-0232(02)00695-5. [DOI] [PubMed] [Google Scholar]
- 55.Leslie NR, Gray A, Pass I, Orchiston EA, Downes CP. Analysis of the cellular functions of PTEN using catalytic domain and C-terminal mutations: Differential effects of C-terminal deletion on signaling pathways downstream of phosphoinositide 3-kinase. Biochem J. 2000;346(Pt 3):827–833. [PMC free article] [PubMed] [Google Scholar]
- 56.Brognard J, Clark AS, Ni Y, Dennis PA. Akt/protein kinase B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer Res. 2001;61:3986–3997. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.