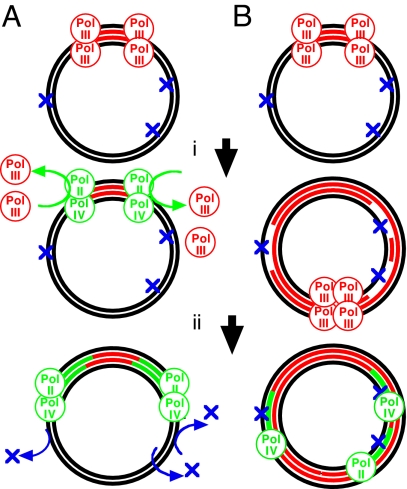
We used to think cells could get by with just a few DNA polymerases. One processive polymerase in Escherichia coli [polymerase III (Pol III)] was needed to make the long trip around the genome, and another one (Pol I) was needed to replace Okazaki fragment primers or damaged nucleotides. This view changed radically after 2 seminal studies by Nelson, Lawrence, and Hinkle (1, 2) in which 2 yeast enzymes, Rev1 and Rev3-Rev7, were found to incorporate nucleosides or polymerize past template sites with missing or damaged bases. These new polymerases helped to explain a rich history of mutational phenomena and led to the realization that organisms have several of these specialized DNA polymerases; E. coli has 3 (Pol II, Pol IV, and Pol V). Yeast have 5. Humans have >10. Each appears specialized for polymerization through different structural classes of DNA damage (3). Although there is no shortage of polymerases from which to choose, the question of how and when they act in the cell has proved difficult to answer. Perhaps this is not so surprising when one considers how long they went unnoticed. A novel and provocative function is proposed in this issue of PNAS in a study by Indiani et al. (4). They demonstrate that either Pol II or Pol IV can replace Pol III at an active replication fork. When this occurs, the new polymerases shift the replisome into a “lower gear,” reducing the speed of replication. That observation complements a recent in vivo study by Uchida et al. (5) in which the rate of DNA synthesis could be slowed or inhibited by overexpression of Pol IV. Indiani et al. (4) found that this was also true when Pol II was overexpressed. Both studies speculate that the slower Pol II or Pol IV replisomes are biologically relevant, serving a checkpoint-like function that allows more time for damaged DNA to be repaired before it is replicated (Fig. 1A).
Fig. 1.
Models for translesion DNA polymerase function during replication in the presence of DNA damage. (A) Translesional polymerases (green circles) replace Pol III (red circles) at replication forks (i), slowing the speed of replication and allowing more time for the damage (blue Xs) to be repaired (ii). (B) After replication encounters unrepaired damage (i), the translesion DNA polymerases function to fill in the gaps left in the DNA at these sites (ii).
The concept that translesion polymerases function to reduce replication speed derives from their observation that as polymerases are swapped in and out of the replisome, DNA synthesis continues at the rate of the active polymerase, which is much slower for translesion Pol II or Pol IV than it is for the replicative Pol III (4). In vitro, polymerase switching occurs spontaneously when Pol II or Pol IV are added to the reaction at high concentrations, and it reverses when their concentrations drop (4). Both the Uchida et al. (5) and Indiani et al. (4) studies also show that overexpression of translesion Pol II or IV can inhibit replication in vivo. They argue that the high polymerase concentrations may mimic cellular conditions after DNA damage, when all 3 E. coli translesion polymerases are up-regulated as part of the SOS regulon (6).
The replicational-slowing model is attractive, but it also raises some questions and suggests experiments that deserve further examination. E. coli mutants that constitutively overexpress Pol II and Pol IV as part of the SOS regulon appear healthy and do not grow poorly (7, 8). The observation suggests that if Pol II and Pol IV overexpression slows DNA synthesis then the effect may be transient or even prevented by other SOS gene products.
A second observation worth considering with respect to a polymerase-checkpoint model is that the presence or absence of the translesion polymerases does not have a dramatic effect on survival. Mutants lacking Pol II or Pol IV are not hypersensitive to many forms of DNA damage and recover DNA synthesis after damage with kinetics very similar to that for wild-type cells (9). A checkpoint function that slows replication and allows more time for repair might be predicted to provide a more general protective effect against a broad spectrum of DNA damage.
Curiously, although the effect of translesion polymerases on viability is relatively minor, their effect on mutagenesis can be comparatively dramatic. Pol V mutants were originally isolated based on their mutational effects after DNA damage (10). Similarly, the presence or absence of Pol II or Pol IV can alter the mutation frequency after DNA damage, even when survival is unaffected (3). Analogously in humans, patients with the variant form of xeroderma pigmentosum (XP) lack a polymerase, Pol η, that efficiently bypasses UV-induced lesions. These patients are as prone to developing cancer and appear clinically similar to those XP patients who are defective in repairing UV-induced damage (11). Remarkably, however, unlike cells from any of the repair-deficient forms of XP, viability is not significantly compromised in XP variant cells exposed to UV (11).
The distinction between the mutational phenotype and the lethal phenotype has led other investigators to propose alternative models, in which the translesion polymerases function to fill in gaps left at damaged sites after replication, somewhat like touching up the missed spots after painting a room (Fig. 1B). This type of model is based on observations that the presence or absence of these polymerases does not alter the rate at which replication recovers after DNA damage in E. coli or the ability of cells to complete replication in yeast (9, 12, 13). Also consistent with this type of model is the observation that Rev-1, a central regulator of translesion synthesis in yeast, is up-regulated after S phase and just before the G2–M transition in the cell cycle (13).
In addition to the novelty of the model suggested, the Indiani et al. study (4) contributes to an emerging view that replication is a far more dynamic process than previously appreciated. The dynamics appear to center around the ability of multiple polymerases to interact with the replication machinery's processivity clamp, a protein complex that encircles the DNA to keep the polymerase tethered to its template (14–16). The plasticity of these interactions is highlighted in the Indiani et al. study by their observation that Pol II or Pol IV can gain access to the processivity clamp within an active replisome, engage the primed template, and then continue to extend the nascent strand without interruption. Impressively, all of this occurs without disrupting either the processivity clamp or the helicase operating at the replication fork.
Other recent studies from this group have revealed additional plasticity within the replisome. In addition to exchanging polymerases, the polymerase in the replisome is able to release and reengage with a different primer during elongation (17). As with the polymerase exchange, the primer exchange also occurs without disrupting the clamp or helicase of the replisome. Potentially extending the dynamics of replication even further is their observation that the replication holoenzyme can accommodate 3 core polymerases, rather than 2 (18). Although the biological significance of these observations remains to be established, they are exciting and challenge us to rethink some fundamental aspects of how the genome is copied.
Acknowledgments.
Work in my laboratory is supported by National Science Foundation Career Award MCB0551798.
Footnotes
The author declares no conflict of interest.
See companion article on page 6031.
References
- 1.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 2.Nelson JR, Lawrence CW, Hinkle DC. Thymine–thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 3.Wagner J, Etienne H, Janel-Bintz R, Fuchs RP. Genetics of mutagenesis in E. coli: Various combinations of translesion polymerases (Pol II, IV, and V) deal with lesion/sequence context diversity. DNA Repair. 2002;1:159–167. doi: 10.1016/s1568-7864(01)00012-x. [DOI] [PubMed] [Google Scholar]
- 4.Indiani C, Langston LD, Yurieva O, Goodman MF, O'Donnell M. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc Natl Acad Sci USA. 2009;106:6031–6038. doi: 10.1073/pnas.0901403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uchida K, et al. Overproduction of Escherichia coli DNA polymerase DinB (Pol IV) inhibits replication fork progression and is lethal. Mol Microbiol. 2008;70:608–622. doi: 10.1111/j.1365-2958.2008.06423.x. [DOI] [PubMed] [Google Scholar]
- 6.Walker GC. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mount DW. A mutant of Escherichia coli showing constitutive expression of the lysogenic induction and error-prone DNA repair pathways. Proc Natl Acad Sci USA. 1977;74:300–304. doi: 10.1073/pnas.74.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krueger JH, Elledge SJ, Walker GC. Isolation and characterization of Tn5 insertion mutations in the lexA gene of Escherichia coli. J Bacteriol. 1983;153:1368–1378. doi: 10.1128/jb.153.3.1368-1378.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courcelle CT, Belle JJ, Courcelle J. Nucleotide excision repair or polymerase V-mediated lesion bypass can act to restore UV-arrested replication forks in Escherichia coli. J Bacteriol. 2005;187:6953–6961. doi: 10.1128/JB.187.20.6953-6961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977;156:121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- 11.Cleaver JE. Xeroderma pigmentosum: Variants with normal DNA repair and normal sensitivity to ultraviolet light. J Invest Dermatol. 1972;58:124–128. doi: 10.1111/1523-1747.ep12538913. [DOI] [PubMed] [Google Scholar]
- 12.Courcelle CT, Chow KH, Casey A, Courcelle J. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc Natl Acad Sci USA. 2006;103:9154–9159. doi: 10.1073/pnas.0600785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G2/M phase rather than S phase. Proc Natl Acad Sci USA. 2006;103:8971–8976. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becherel OJ, Fuchs RP, Wagner J. Pivotal role of the β-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair. 2002;1:703–708. doi: 10.1016/s1568-7864(02)00106-4. [DOI] [PubMed] [Google Scholar]
- 15.Maul RW, Ponticelli SK, Duzen JM, Sutton MD. Differential binding of Escherichia coli DNA polymerases to the beta-sliding clamp. Mol Microbiol. 2007;65:811–827. doi: 10.1111/j.1365-2958.2007.05828.x. [DOI] [PubMed] [Google Scholar]
- 16.Furukohri A, Goodman MF, Maki H. A dynamic polymerase exchange with Escherichia coli DNA polymerase IV replacing DNA polymerase III on the sliding clamp. J Biol Chem. 2008;283:11260–11269. doi: 10.1074/jbc.M709689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pomerantz RT, O'Donnell M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature. 2008;456:762–766. doi: 10.1038/nature07527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McInerney P, Johnson A, Katz F, O'Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]