Abstract
Myosin storage myopathy (MSM) is a congenital myopathy characterized by the presence of subsarcolemmal inclusions of myosin in the majority of type I muscle fibers, and has been linked to 4 mutations in the slow/cardiac muscle myosin, β-MyHC (MYH7). Although the majority of the >230 disease causing mutations in MYH7 are located in the globular head region of the molecule, those responsible for MSM are part of a subset of MYH7 mutations that are located in the α-helical coiled-coil tail. Mutations in the myosin head are thought to affect the ATPase and actin-binding properties of the molecule. To date, however, there are no reports of the molecular mechanism of pathogenesis for mutations in the rod region of muscle myosins. Here, we present analysis of 4 mutations responsible for MSM: L1793P, R1845W, E1886K, and H1901L. We show that each MSM mutation has a different molecular phenotype, suggesting that there are multiple mechanisms by which MSM can be caused. These mechanisms range from thermodynamic and functional irregularities of individual proteins (L1793P), to varying defects in the assembly and stability of filaments formed from the proteins (R1845W, E1886K, and H1901L). In addition to furthering our understanding of MSM, these observations provide the first insight into how mutations affect the rod region of muscle myosins, and provide a framework for future studies of disease-causing mutations in this region of the molecule.
Myosin is the molecular motor of muscle and is the major component of the thick filament. In vivo, myosin exists as a dimer of myosin heavy chains (MyHC) with 2 globular heads attached to a long, α-helical coiled-coil region known as the myosin rod. Although the head region is responsible for myosin's ATPase and actin binding properties, the rod region functions to incorporate myosin into bipolar thick filaments through charge based interactions between adjacent rods (1–3). β-myosin, which is expressed in the adult heart and slow skeletal muscle fibers, is of particular clinical importance because to date >230 disease causing mutations have been found in the molecule (4). The majority of mutations (65%) are located in the head region and result in cardiomyopathies, many of which have been well characterized (5). However, several mutations have also recently been described in the rod region of myosin that result in a variety of diseases (6, 7). Because of the disparate roles that the head and rod regions of myosin play, these mutations are thought to represent a unique mode of pathogenesis, although no mutations in the rod region of muscle myosins have been characterized.
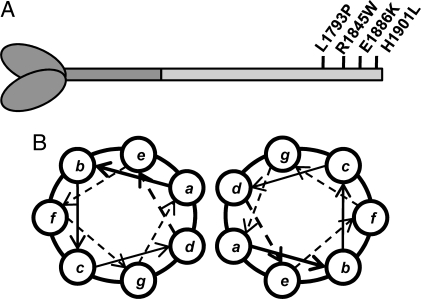
We sought to investigate a subset of mutations located in the myosin rod that are responsible for the disease myosin storage myopathy (MSM). Also known as hyaline body myopathy, MSM is a rare, congenital myopathy with variable inheritance that is characterized by the presence of subsarcolemmal accumulations of myosin in the majority of type-I skeletal muscle fibers (8, 9). Clinically, patients exhibit variable age of onset ranging from birth through childhood, and occasionally middle age. Symptoms also vary, but typically include slowly progressive muscle hypertonia, scapularperoneal weakness, and respiratory insufficiency (6). MSM has been associated with 4 missense mutations in the MYH7 gene, which encodes slow/β-cardiac myosin heavy chain (MyHC). All 4 mutations responsible for MSM, L1793P, R1845W, E1886K, and H1901L, are located in the distal rod region of the protein and have been postulated to result in pathogenesis by perturbing the lateral interactions between coiled-coils (Fig. 1A) (8–10).
Fig. 1.
Location of MSM mutations in the coiled-coil rod region of β-MyHC. (A) Schematic of β-MyHC structure. The globular head region and N-terminal portion of the myosin rod which comprise heavy meromyosin are shown in dark gray. The C-terminal LMM region of the rod is shown in light gray with the relative position of MSM mutations in the distal portion of the rod marked. (B) Diagram of the relative positions of amino acids within the coiled-coil. The position of amino acids within the heptad is denoted by a–g. Three MSM mutations (R1845W, E1886K, and H1901L) are located in the outer f position, whereas 1 (L1793P) is located in the inner d position.
The α-helices that compose the coiled-coil myosin rod are characterized by a repeating heptad of residues (denoted a–g), that in turn is part of a larger 28 aa repeat composed of 14 positively charged residues followed by 14 negatively charged residues (Fig. 1B) (1–3). It is the interaction of these charged repeats, in proper register, which is responsible for formation of the thick filament. Also crucial to the association of the rods is a region of 29 aa known as the assembly competent domain (ACD), which our laboratory has demonstrated to be necessary for the formation of myosin rod assemblies in vitro (11) .
In an α-helical coiled-coil, residues in the a and d position are typically hydrophobic and create a seam along the interface of the coiled-coil, whereas residues in the e and g positions interact electrostatically to stabilize the coiled-coil. Residues in the outer positions such as b, c, and f are typically charged and mediate the interactions between coiled-coils (Fig. 1B) (12–14). Of the mutations responsible for MSM, 3 (R1845W, E1886K, and H1901L) are located in the f position, whereas 1 (L1793P) is located in the d position. Interestingly, L1793P, R1845W, and H1901L are associated with MSM in the absence of any overt cardiomyopathy, even though the protein is abundant in both heart and skeletal muscle. E1886K is the only mutation that is responsible for MSM that also results in hypertrophic cardiomyopathy, and is located in the ACD region of the rod. Given that these mutations have been predicted to alter the ability of myosin to properly assemble into stable thick filaments, we sought to experimentally characterize their effects by performing a wide variety of structural and functional assays. Here, we show that MSM mutations expressed in the C-terminal fragment of the rod, light meromyosin (LMM), cause structural, thermodynamic, and functional differences that correlate with varying phenotypes. This provides the first description of how disease causing mutations affect the rod region of a muscle myosin, and sheds lights on the pathogenesis associated with these mutations.
Results
The entire LMM region of β-MyHC (amino acids 1231–1938) was cloned and tagged with T7 and 6X-His at the N terminus for recombinant expression. WT and mutant proteins were generated and purified as described in Materials and Methods, and had an expected electrophoretic mobility of ≈82 kDa. SDS/PAGE analysis confirmed that all proteins were purified to relative homogeneity.
MSM Mutations Alter Protein Stability Without Affecting Secondary Structure.
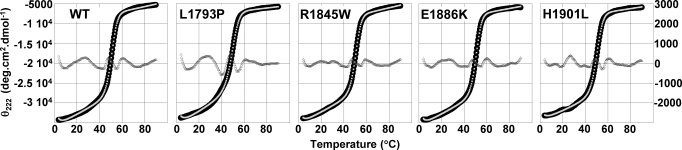
To determine whether the ability of LMM to self-assemble could be affected by structural defects within the molecule, circular dichroism (CD) spectroscopy was used to ascertain whether mutations altered the α-helical coiled-coil secondary structure of LMM. As an α-helical coiled-coil, the LMM region of the myosin rod displays a distinct CD spectra, with characteristic minima at 222 nm and 208 nm (15). The spectrum for wild-type LMM displayed the expected pattern, as did all 4 of the mutant proteins studied (S1). All of the mutant proteins also displayed similar α-helical content to WT, each being ≈88%. The θ222/208 ratios, which report on the ability of the α-helices to form coiled-coils, were all ≈1.11, indicating that coiled-coil formation is not disrupted (Table 1) (13, 14, 16).
Table 1.
Biophysical data for myosin tail constructs
| Construct | [θ]222, deg·cm2·dmol−1 | α-helix, % | [θ]222/208 | Tm, °C | ΔGapp,37, kcal·mol−1 | ΔH, kcal·mol−1 | ΔS, kcal·mol−1·K−1 | ΔCp, kcal·mol−1·K−1 |
|---|---|---|---|---|---|---|---|---|
| Wild type | −32,708 | 87.9 | 1.11 | 50.9 | −2.57 | −80.0 | −0.247 | −2.8 |
| L1793P | −32,352 | 87.0 | 1.11 | 49.4 | −1.95 | −65.2 | −0.202 | −2.3 |
| R1845W | −32,390 | 86.9 | 1.11 | 50.5 | −2.59 | −81.8 | −0.253 | −2.9 |
| E1886K | −33,143 | 88.6 | 1.12 | 50.7 | −2.66 | −83.2 | −0.257 | −2.9 |
| H1901L | −33,153 | 89.1 | 1.11 | 50.2 | −2.31 | −73.7 | −0.228 | −2.5 |
With both WT and mutant LMM coiled-coils shown to be fully folded, we next determined the effect of these mutations on the thermal stability of LMM by monitoring [θ]222 as the temperature was increased from 5 °C to 90 °C. WT and all 4 mutant proteins were modeled as a 2-state transition of monomer from a folded to an unfolded state with a change in heat capacity, ΔCp, between the folded and unfolded forms (17). The calculated thermal midpoints of unfolding for R1845W, E1886K, and H1901L are slightly decreased from WT, whereas L1793P displays a relatively larger decrease in thermal stability (Fig. 2). Additionally, although the biophysical parameters for R1845W and E1886K are nearly indistinguishable from WT, L1793P has a ΔΔG at 37 °C of −0.62 kcal/mol and displays decreases of ≈18% for ΔH, ΔS and ΔCp. H1901L displays an intermediate phenotype, with a ΔΔG at 37 °C of −0.26 kcal/mol and an ≈8% decrease in ΔH, ΔS, and ΔCp (Table 1). Taken together, these data indicate that the secondary structure of LMM is not altered by any of the mutations studied, whereas 2 of the mutations causing MSM (L1793P and H1901L) do alter the thermodynamic stability of the molecule.
Fig. 2.
Thermal denaturation of the myosin rod reveals differences in protein thermodynamics. The transition of LMM from a folded to an unfolded state was followed as a function of temperature by measuring θ222, which monitors α-helical structure, as the temperature was gradually increased. All experiments were performed in the same high salt buffer as for far-UV CD to prevent protein assembly, and were done at 4 °C. Observed θ222 data (black) were fit to a theoretical melting curve (light gray), and are plotted on the left axis. The residuals (dark gray), calculated as the difference between the observed and the fit data at each point, are plotted on the right axis and are ≈0 for each melt.
MSM Mutations Affect Filament Formation.
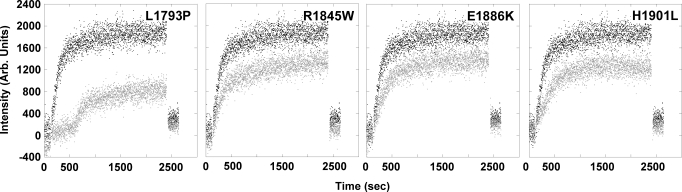
We next sought to examine whether these disease causing mutations do indeed affect the ability of LMM to self-assemble. To monitor this process, LMM was diluted from 300 mM NaCl where it exists stably as a coiled-coil, to 150 mM NaCl where it will self-assemble into ordered structures and the reaction was monitored by 90° light scattering. 90° light scattering is an assay that follows the assembly of LMM in real time based upon the size of the particles scattering light, and represents a tractable model for studying a variety of mutations in the myosin rod (18, 19). By this assay, WT LMM quickly assembled, with the majority of the reaction completed after 8–10 min. R1845W, E1886K, and H1901L appear to assemble at rates comparable to WT, but the extent to which the reactions occur is truncated to ≈60% of WT in each case. L1793P assembles much more slowly than WT, with a lag phase before assembly that effectively doubles the amount of time required for the majority of the protein to self-associate. In addition, the extent to which the reaction occurs reaches <50% of that for WT (Fig. 3).
Fig. 3.
Real-time self-assembly of LMM shows MSM mutants are assembly defective. The amount of 90° light scattering for no salt buffer (10 mM TES, 3.5 mM EDTA, 1 mM TCEP) was observed for 120 s before the addition of protein to obtain a baseline for scattering. An equal volume of either WT (black) or mutant (gray) LMM in high salt buffer was then injected into the sample chamber, diluting the sample to a final concentration of 150 mM NaCl and 100 nM protein to initialize self-assembly. Reactions were followed for 40 min before the addition of 5M salt to return the sample to a final salt concentration of 300 mM to demonstrate the reversibility of the reaction. The intensity of the 90° light scattering is plotted in arbitrary units versus time.
To further investigate the ability of LMM to self-assemble, we tested the ability of WT and each of the mutant proteins to form paracrystals. Paracrystals are well-ordered protein assemblies that provide an established model for thick filament formation (1, 20–26). Similarly to light scattering experiments, proteins were dialyzed from a high salt buffer into a buffer with physiological salt concentration allowing them to assemble into well-ordered arrays. Paracrystals were then analyzed by electron microscopy (EM). WT LMM formed well-ordered paracrystals with a periodicity of 14.0 nm (±0.44 nm for n = 20 paracrystals; S2). This value is consistent with reported values for LMM paracrystal periodicity and corresponds well to the 14.3 nm axial spacing of full-length β-MyHC found in bipolar thick filaments (1, 20, 21, 24, 26). Paracrystals formed from L1793P, R1845W, E1886K, and H1901L LMM all had similar periodicities to WT, indicating that these mutations do not alter the gross morphology of the assemblies (S3).
MSM Mutations Alter Filament Properties.
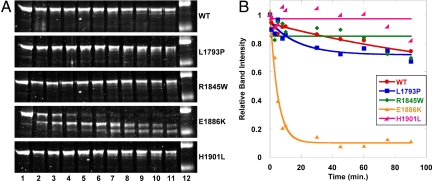
Although the periodicity of paracrystals formed from WT and mutant proteins is similar, we wanted to further examine these assemblies to assess whether their stability is affected by the mutations being studied. Paracrystals were formed as before, and limited proteolysis was carried out by digestion with a low concentration of porcine trypsin. Reactions were quenched at various time points out to 90 min using soybean trypsin inhibitor, and the reactions were analyzed via SDS/PAGE to visualize degradation (Fig. 4A). The band intensity of the full length LMM was then measured and plotted over time, and the curves were fit to an exponential decay model (Fig. 4B). WT protein is shown to be relatively stable over time, with ≈80% of the full length protein still intact after 90 min. L1793P LMM shows a similar pattern to WT, although it is degraded slightly more rapidly. E1886K LMM, however, is much more readily proteolyzed, with the majority of the full length protein digested by the 10-min time point. Interestingly, R1845W and H1901L LMM appear to be more stable than WT. Careful analysis of the gels, however, shows degradation of both R1845W and H1901L at later time points (30% of R1845W and 18% of H1901L are degraded at 90 min compared with 26% of WT). Because degradation begins later for paracrystals formed from these 2 proteins, when these data are fit to an exponential decay curve it results in a straight line. Taken together, these data suggest that the kinetics of R1845W and H1901L LMM proteolysis occur more slowly than WT and that paracrystals formed from these mutant proteins may be slightly more stable, whereas L1793P and E1886K LMM are more readily degraded.
Fig. 4.
MSM mutants affect the proteolytic stability of LMM paracrystals. (A) Time course of limited tryptic proteolysis of LMM. Lanes numbered 1 through 11 represent different time points for digestion (lane 1, 0 min; lane 2, 1 min; lane 3, 3 min; lane 4, 5 min; lane 5, 8 min; lane 6, 10 min; lane 7, 30 min; lane 8, 45 min; lane 9, 60 min; lane 10, 75 min; lane 11, 90 min). Lane 12 is a protein standard where the top band is 125 kDa and the bottom band is 82 kDa. (B) Fitting proteolysis data to an exponential decay curve. Data are plotted as relative intensity of the full length LMM band versus time. The band intensity for each protein at the zero time point is defined as 1, and all other band intensities for the protein are plotted with respect to that value.
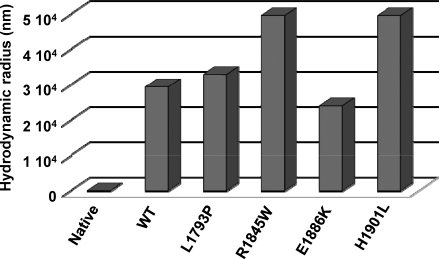
We next carried out molecular sizing experiments, using dynamic light scattering (DLS) to ascertain whether qualitative differences exist between paracrystals formed from WT and mutant LMM. DLS is a technique that measures the size (hydrodynamic radius) of a particle by determining the frequency shift of coherent light directed at that particle. Our DLS experiments show that control protein that was not assembled into paracrystals had a measured hydrodynamic radius of 22 nm, whereas paracrystals formed from WT LMM had hydrodynamic radii of ≈29,500 nm. L1793P and E1886K LMM formed paracrystals that were uniform in size distribution, with a single species present that was similar in size to WT. Paracrystals formed from R1845W and H1901L LMM also had a uniform distribution, but both formed assemblies that were much larger in size than WT, with measured hydrodynamic radii of 49,650 nm (Fig. 5). It should be noted, however, that because this size is at the limit of detection for the device used, the actual size is likely to be larger than the measured size for each mutant.
Fig. 5.
R1845W and H1901L LMM form drastically larger paracrystals. Paracrystals were formed by diluting the protein from high salt buffer to low salt buffer, similarly to static light scattering experiments, but to a final protein concentration of 200 nM. Reactions were then allowed to proceed for 45 min at room temperature to allow for complete assembly before molecular sizing experiments were performed. Native protein denotes WT LMM protein, which was measured before assembly as a control.
Discussion
To date, all reported instances of MSM are associated with 1 of 4 mutations in the LMM region of the β-MyHC rod (8–10, 27–32). By using an array of assays to biochemically and biophysically characterize the effects that these mutations have on the protein, we have found that each mutation possesses a unique molecular phenotype. Because all of these mutations are ultimately responsible for the same disease phenotype, our results suggests that there are several possible ways in which mutations in the LMM region of β-MyHC can adversely affect the ability of the protein to form stable, functional bipolar thick filaments. The assembly of muscle filaments is a multistep process that involves both the proper folding of α-helices into coiled-coils, and the assembly of these coiled-coils, in proper register, into filaments. Our findings indicate that defects in either one of these steps can result in improper filament formation leading to disease.
L1793P.
The L1793P mutation results in the largest thermodynamic differences from WT protein of any of the mutations studied. L1793P LMM has a Tm that is 1.5 °C lower than wild type, and the lower ΔG at 37 °C suggests that under in vivo conditions the mutant protein is appreciably less stable. Surprisingly, our findings show no difference in secondary structure. Proline is known to be detrimental to the secondary structure of α-helices, and is not found at the d position in the rod of any muscle myosin isoform from any species (4, 33, 34). Although some computational algorithms such as COILS and PAIRCOIL predict this mutation to have a negative effect on secondary structure, our CD results reveal no difference in the α-helical content of mutant protein when compared with WT. Although there is no detectable structural difference, the instability of L1793P LMM does appear to have a functional impact. By using 90° light scattering to monitor the reaction, we show that the L1793P mutation alters the ability of LMM to assemble. The lag phase present in the initial stages of L1793P LMM self-assembly suggests that the nucleation of new filaments is less favored, presumably because of the increased instability of the molecule. This interpretation is also consistent with our proteolytic studies, which show that the paracrystals that form, although similar in morphology to WT, are more susceptible to proteolytic cleavage. Taken together, these data suggest that replacing the L1793 amino acid with a proline destabilizes the dimer interface under conditions similar to those found in vivo, and that this instability affects the ability of LMM to properly assemble.
R1845W.
Located in the outer f position of the α-helix heptad, the R1845W mutation is expected to disrupt interactions between coiled-coils without disturbing the structure of the molecule. Predictably, R1845W LMM is nearly indistinguishable from WT in both secondary structural characteristics and biophysical parameters. Functional assays, however, reveal that mutating the normally charged R1845 residue to a bulky, hydrophobic tryptophan results in an interesting phenotype. Our 90° light scattering assay shows that, although it assembles at a similar rate to WT, R1845W LMM does not appear to be able to assemble to the same extent. Because the signal is proportional to the size of the particles responsible for the light scattering, one would expect structures formed from this mutant protein to be smaller in size than those formed from WT. Results, however, from molecular sizing experiments performed by dynamic light scattering show that the structures formed by R1845W LMM are actually much larger that those formed by WT. These data are substantiated by our limited proteolysis experiments, which demonstrate that paracrystals of R1845W LMM are more stable than those of WT, although they appear morphologically similar by EM. This suggests that for R1845W LMM we are observing the formation of fewer assemblies that are much larger in size than those normally formed. These results imply a model whereby the R1845W mutation alters the interactions between filaments such that their assembly is less constrained, causing the formation of abnormally large, degradation resistant structures.
E1886K.
Of the 3 MSM mutations in the f position of the heptad repeat, E1886K is expected to have the most severe phenotype because it is located in a portion of the rod, the ACD, which is known to be critical for the proper assembly of myosin filaments (11). Similarly to R1845W, E1886K LMM displays no discernable differences in secondary structure or biophysical parameters from WT, as would be expected. The mutation does, however, have a strong functional impact on filament formation. Results from 90° light scattering show that, like R1845W, E1886K LMM has a decreased ability to assemble to the same extent as WT LMM, although the initial rates of assembly are similar. Unlike R1845W LMM, although, E1886K LMM forms structures that are measured by dynamic light scattering to have a hydrodynamic radius similar to WT. Interestingly, even though EM of E1886K LMM paracrystals does not reveal any overt morphological phenotype, our limited proteolysis studies show that paracrystals formed from this mutant protein are much more readily degraded than those formed from WT protein. Proteolytic cleavage by trypsin is known to occur at lysine and arginine residues, so it is possible that the increased proteolysis is due to inserting an exogenous cleavage site. However, control experiments performed by inserting a single lysine residue into another site in LMM do not result in a significant difference from WT (data not shown). Additionally, 125 lysine and arginine residues exist in the WT LMM being studied, 31 of which are located in the f position of the coiled-coil. These potential cleavage sites do not appear to be targets of excessive proteolytic cleavage, suggesting that the location of the cleavage site is important. Taken together, our data intimate that mutating the negatively charged E1886 residue to a positively charged lysine disrupts an important charge based interaction in a critical region of the molecule. This appears to cause the protein to form filaments that are packed together less tightly, allowing for proteolytic cleavage by trypsin in our assay. The altered packing of the filaments may also destabilize them, explaining why fewer filament are observed in our 90° light scattering assay.
H1901L.
The third MSM mutation located in the f position, H1901L replaces a positively charged amino acid, presumably involved in electrostatic interactions between adjacent coiled-coils, with a small, neutrally charged amino acid. Although we did not expect any structural or biophysical differences owing to this mutation, the Tm of the protein did decrease by 0.7 °C and this was associated with a ΔΔGat 37 °C of −0.26 kcal/mol, indicating that the mutation does decrease the stability of the molecule. Functionally, the H1901L mutation appears to affect the protein in much the same manner as R1845W. 90° light scattering assays with H1901L LMM show that the initial rate of assembly is unaffected, whereas the extent of assembly is diminished compared with WT. Molecular sizing experiments are also similar, showing that paracrystals formed from H1901L LMM are much larger than those formed from WT, whereas the morphology as visualized by EM is unaffected. Additionally, the H1901L mutation behaves similarly to R1845W in our tryptic digestion assay, with paracrystals formed from H1901L LMM appearing to be more stable than those formed from WT protein. Our interpretation of these data, then, is much the same as for R1845W. Although the H1901L mutation does slightly decrease the stability of the molecule, this does not appear to drastically affect its ability to form filaments. In fact, mutating H1901 to a lysine appears to alter the interactions between filaments such that larger, more stable structures are formed.
The picture that emerges from our work is that there are several possible pathways by which mutations in the rod portion of a muscle myosin can result in the same disease. MSM can be caused by thermodynamic instabilities that drastically decrease the ability of the protein to properly assemble, such as with L1793P, or by charge changes that interrupt the interaction between adjacent proteins and destabilize the filaments, such as with E1886K. In vivo, these proteins may form thick filaments that are less stable or more easily damaged, leading to sarcomeric disarray. MSM can also be caused by the loss of important charged amino acids, as is the case with R1845W and H1901L, which result in the formation of atypically large, unusually stable filaments. In vivo, these proteins may form thick filaments that are not always properly incorporated into the sarcomere or are not properly degraded, resulting in aberrant accumulations of myosin in the muscle fibers. It should be noted that although our data support these hypotheses, the possibility cannot be eliminated that the observed differences are epiphenomena. Our model will only be completely substantiated when these mutations, along with other mutations having similar biophysical profiles, are investigated in vivo. Further work will also be needed to investigate the cause of the tissue specificity of the MSM disease phenotype, but future studies into the pathogenesis of mutations in the myosin rod should benefit from our analysis. Finally, it will be important to determine why the mutations studied here result specifically in MSM whereas other mutations result in a host of other disease phenotypes, and what functional impact these mutations have on the muscle in vivo.
Materials and Methods
LMM Expression Constructs.
Wild type MYH7 LMM (amino acid residues 1231–1938) was cloned into the pUC18 expression vector and N-terminal T7 and 6X-His tags were added. Site-directed mutagenesis was then performed using inverse PCR to generate the L1793P, R1845W, E1886K, and H1901L mutations. Positive clones were verified by sequencing and were then subcloned into the pET3a expression vector (New England Biolabs), using XhoI and SpeI. The cloning process resulted in 3 additional amino acids (TSC) being added to the C terminus of the protein
Purification of Wild-Type and Mutant LMM Proteins.
Wild-type and mutant plasmids were transformed into BL-21 cells and expression was induced with 1 mM IPTG. Cells were harvested and protein was extracted using B-PER (Thermo Scientific). Protein lysate was bound to Ni-NTA agarose at 4 °C for 1 h, and washed with increasing amounts of imidazole up to 50 mM. Proteins were eluted with 0.5 M imidazole and dialyzed overnight at 4 °C into 40 mM sodium pyrophosphate, 100 mM NaCl, 3.5 mM EDTA at pH 8.5. Dialyzed protein was then additionally purified by anion exchange chromatography on an AKTA Purifier with 1 mL of HiTrap Q HP columns (GE Lifesciences). Fractions containing LMM were analyzed via SDS/PAGE gel for purity and pooled. Protein was then concentrated with Amicon Ultracel 50k centrifuge columns (Millipore) and concentrations were determined via uBCA assay (Thermo Scientific).
Circular Dichroism Analysis.
Wild-type and mutant LMM proteins were dialyzed into 10 mM TES, 300 mM NaCl, and 3.5 mM EDTA overnight at 4 °C, diluted to ≈0.3 mg/mL, reduced with 1 mM TCEP and degassed. Circular dichroism (CD) was measured using a Jasco J-810 spectropolarimeter (Jasco) with constant N2 flushing. A Peltier temperature control device was used to perform all measurements at 4 °C and a rectangular 1-mm path length cell was used. Spectra were determined from 250 nm to 190 nm at a sensitivity of 100 mdeg, with a 0.2-nm data pitch, a continuous scanning speed of 50 nm/min, and averaged >6 accumulations for each protein. Buffer spectra were collected in the same manner and subtracted from protein spectra. Exact protein concentrations were determined via uBCA assay, and the mean residue molar ellipticity was calculated using the equation,
where θobs is the observed ellipticity in millidegrees, mrw is the mean residue molecular weight, l is the optical path length of the CD cell (in cm), and c is the peptide concentration (in milligrams per milliliter) (35). The percentage α-helix for each protein was determined using the equation,
where θ208 is the mean residue molar ellipticity at 208 nm.
Thermal Denaturation of LMM Monitored by Circular Dichroism.
Protein stability of wild-type and mutant LMM was monitored at 222 nm to follow α-helical secondary structure during temperature-induced denaturation. Data were collected at 0.5 °C intervals at a scan rate of 60 °C per hour from 4 °C to 90 °C. The change in mean residue ellipticity, θ, as a function of temperature was modeled using a nonlinear least squares algorithm assuming the 2-state transition of a monomer from a folded to an unfolded state with a change in heat capacity, ΔCp, between the folded and unfolded forms (17). A detailed description of the calculations used is given in S3. Residuals were calculated at each data point by taking the difference between the fit mean residue ellipticity and the experimentally determined mean residue ellipticity at that point.
Static Light Scattering.
The ability of wild-type and mutant protein to self-assemble was measured in real-time by monitoring 90° light scattering, using a PTI QM-2000–6SE fluorescence spectrometer (Photon Technology International) at 25 °C. Excitation was at 320 nm with a 2-nm slit width, and emission was measured at 320 nm with an 8-nm slit width. Measurements were taken for buffer (10 mM TES, 3.5 mM EDTA, 1 mM TCEP, pH 7.3) for 2 minutes to obtain a baseline for scattering, at which point an equal volume of 200 nM protein in high salt buffer (10 mM TES, 300 mM NaCl, 3.5 mM EDTA, 1 mM TCEP, pH 7.3) was added, diluting the protein to 150 mM NaCl and allowing for self-assembly at a final protein concentration of 100 nM. Assembly reactions were followed out to 40 min before 5M salt was added to return the concentration to 300 mM. Data were analyzed using Kaleidagraph (Synergy Software) with background buffer scattering subtracted from each reading.
Paracrystal Formation and Visualization.
Paracrystals were formed for wild-type and mutant protein by dialyzing 100 μg of LMM from high salt buffer into 10 mM bis-tris propane, 100 mM NaCl, 3.5 mM EDTA, pH 7.3, overnight at 4 °C. Paracrystals were adsorbed onto glow-discharged, carbon-coated, 400 mesh grids for 60 s, negatively stained with 2% uranyl acetate, and rinsed with dialysis buffer. Electron micrographs were imaged using a Phillips CM10 TEM at 80 kV. Image analysis was performed using GNU Image Manipulation Program software (Free Software Foundation).
Limited Trypsinization.
LMM was assembled as described for paracrystal formation, reduced with 1 mM TCEP, and protein concentrations were determined via μBCA assay. Trypsinization experiments were performed essentially as described in ref. 36. Briefly, paracrystals were treated with porcine trypsin at 0.002 mg/mL and incubated at 30 °C. The reaction was quenched at various times by mixing with 40× excess soybean trypsin inhibitor. Samples were run on 4–20% SDS/PAGE gels, stained with Imperial Protein Stain (Thermo Scientific) and imaged with a Li-COR Odyssey infrared imager (Li-COR).
Dynamic Light Scattering.
Wild-type and mutant protein was self-assembled as described for static light scattering. Proteins were diluted to a final concentration of 200 nM and incubated at room temperature for 45 min to allow for complete assembly. Molecular sizing experiments were performed with a DynaPro (Wyatt Technology) set to 10% laser power at a wavelength of 824.2 nm. All experiments were done at room temperature, using a 5-s acquisition time. Data analysis was performed using the Dynamics software package (Wyatt Technology) and hydrodynamic radii were estimated using the software's Coils model.
Supporting Information.
Detailed methods, along with CD and paracrystal data, can be found in SI.
Supplementary Material
Acknowledgments.
We thank M. Buvoli, B. Thompson, and all members of the L.A.L. laboratory for invaluable support, discussion, and suggestions; S. Kwok and B. Hirsch at the University of Colorado Health Sciences Center Biophysics core for technical advice and support; M. Stowell for editorial assistance and advice; and M. Buvoli for assisting with the preparation of the manuscript. This work was supported by National Institutes of Health Grant 5R01 HL085573-01 (to L.A.L.) and a National Inistitutes of Health Molecular Biophysics Training Grant T32GM065103 (to T.Z.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900107106/DCSupplemental.
References
- 1.Atkinson SJ, Stewart M. Molecular interactions in myosin assembly. Role of the 28-residue charge repeat in the rod. J Mol Biol. 1992;226:7–13. doi: 10.1016/0022-2836(92)90118-4. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson SJ, Stewart M. Molecular basis of myosin assembly: Coiled-coil interactions and the role of charge periodicities. J Cell Sci Suppl. 1991;14:7–10. doi: 10.1242/jcs.1991.supplement_14.2. [DOI] [PubMed] [Google Scholar]
- 3.McLachlan AD, Karn J. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature. 1982;299:226–231. doi: 10.1038/299226a0. [DOI] [PubMed] [Google Scholar]
- 4.Buvoli M, Hamady M, Leinwand LA, Knight R. Bioinformatics assessment of beta-myosin mutations reveals myosin's high sensitivity to mutations. Trends Cardiovasc Med. 2008;18:141–149. doi: 10.1016/j.tcm.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morimoto S. Sarcomeric proteins and inherited cardiomyopathies. Cardiovasc Res. 2008;77:659–666. doi: 10.1093/cvr/cvm084. [DOI] [PubMed] [Google Scholar]
- 6.Oldfors A. Hereditary myosin myopathies. Neuromuscul Disord. 2007;17:355–367. doi: 10.1016/j.nmd.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Laing NG, Sewry CA, Lamont P. Congenital myopathies. Handb Clin Neurol. 2007;86:1–33. doi: 10.1016/S0072-9752(07)86001-5. [DOI] [PubMed] [Google Scholar]
- 8.Tajsharghi H, et al. Myosin storage myopathy associated with a heterozygous missense mutation in MYH7. Ann Neurol. 2003;54:494–500. doi: 10.1002/ana.10693. [DOI] [PubMed] [Google Scholar]
- 9.Bohlega S, et al. Autosomal dominant hyaline body myopathy: Clinical variability and pathologic findings. Neurology. 2003;61:1519–1523. doi: 10.1212/01.wnl.0000096022.09887.9d. [DOI] [PubMed] [Google Scholar]
- 10.Dye DE, Azzarelli B, Goebel HH, Laing NG. Novel slow-skeletal myosin (MYH7) mutation in the original myosin storage myopathy kindred. Neuromuscul Disord. 2006;16:357–360. doi: 10.1016/j.nmd.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Sohn RL, et al. A 29 residue region of the sarcomeric myosin rod is necessary for filament formation. J Mol Biol. 1997;266:317–330. doi: 10.1006/jmbi.1996.0790. [DOI] [PubMed] [Google Scholar]
- 12.Kwok SC, Hodges RS. Stabilizing and destabilizing clusters in the hydrophobic core of long two-stranded alpha-helical coiled-coils. J Biol Chem. 2004;279:21576–21588. doi: 10.1074/jbc.M401074200. [DOI] [PubMed] [Google Scholar]
- 13.Zhou NE, Kay CM, Hodges RS. The role of interhelical ionic interactions in controlling protein folding and stability. De novo designed synthetic two-stranded alpha-helical coiled-coils. J Mol Biol. 1994;237:500–512. doi: 10.1006/jmbi.1994.1250. [DOI] [PubMed] [Google Scholar]
- 14.Zhou NE, Kay CM, Hodges RS. Synthetic model proteins. Positional effects of interchain hydrophobic interactions on stability of two-stranded alpha-helical coiled-coils. J Biol Chem. 1992;267:2664–2670. [PubMed] [Google Scholar]
- 15.Holzwarth G, Doty P. The Ultraviolet Circular Dichroism of Polypeptides. J Am Chem Soc. 1965;87:218–228. doi: 10.1021/ja01080a015. [DOI] [PubMed] [Google Scholar]
- 16.Hodges RS, Zhou NE, Kay CM, Semchuk PD. Synthetic model proteins: Contribution of hydrophobic residues and disulfide bonds to protein stability. Pept Res. 1990;3:123–137. [PubMed] [Google Scholar]
- 17.Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinard JH, Stafford WF, Pollard TD. The mechanism of assembly of Acanthamoeba myosin-II minifilaments: Minifilaments assemble by three successive dimerization steps. J Cell Biol. 1989;109(4 Pt 1):1537–1547. doi: 10.1083/jcb.109.4.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinard JH, Pollard TD. The effect of heavy chain phosphorylation and solution conditions on the assembly of Acanthamoeba myosin-II. J Cell Biol. 1989;109(Pt 1):1529–1535. doi: 10.1083/jcb.109.4.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King MV, Young M. Selective non-enzymic cleavage of the myosin rod. Electron-microscopic studies on crystals and paracrystals of light meromyosin-C. J Mol Biol. 1970;50:491–507. doi: 10.1016/0022-2836(70)90207-x. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura A, Sreter F, Gergely J. Comparative studies of light meromyosin paracrystals derived from red, white, and cardiac muscle myosins. J Cell Biol. 1971;49:883–898. doi: 10.1083/jcb.49.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Safer D, Pepe FA. Axial packing in light meromyosin paracrystals. J Mol Biol. 1980;136:343–358. doi: 10.1016/0022-2836(80)90394-0. [DOI] [PubMed] [Google Scholar]
- 23.Bennett PM. The structure of spindle-shaped paracrystals of light meromyosin. J Mol Biol. 1981;146:201–221. doi: 10.1016/0022-2836(81)90432-0. [DOI] [PubMed] [Google Scholar]
- 24.Ward R, Bennett PM. Paracrystals of myosin rod. J Muscle Res Cell Motil. 1989;10:34–52. doi: 10.1007/BF01739855. [DOI] [PubMed] [Google Scholar]
- 25.Ward R, Murray JM. Three-dimensional structure of frozen-hydrated paracrystals of myosin rod. J Muscle Res Cell Motil. 1990;11:403–418. doi: 10.1007/BF01739761. [DOI] [PubMed] [Google Scholar]
- 26.Atkinson SJ, Stewart M. Expression in Escherichia coli of fragments of the coiled-coil rod domain of rabbit myosin: Influence of different regions of the molecule on aggregation and paracrystal formation. J Cell Sci. 1991;99(Pt 4):823–836. doi: 10.1242/jcs.99.4.823. [DOI] [PubMed] [Google Scholar]
- 27.Bohlega S, et al. Mutation of the slow myosin heavy chain rod domain underlies hyaline body myopathy. Neurology. 2004;62:1518–1521. doi: 10.1212/01.wnl.0000123255.92062.37. [DOI] [PubMed] [Google Scholar]
- 28.Laing NG, et al. Myosin storage myopathy: Slow skeletal myosin (MYH7) mutation in two isolated cases. Neurology. 2005;64:527–529. doi: 10.1212/01.WNL.0000150581.37514.30. [DOI] [PubMed] [Google Scholar]
- 29.Oldfors A, Tajsharghi H, Thornell LE. Mutation of the slow myosin heavy chain rod domain underlies hyaline body myopathy. Neurology. 2005;64:580–581. doi: 10.1212/wnl.64.3.580-a. [DOI] [PubMed] [Google Scholar]
- 30.Rafay MF, Halliday W, Bril V. Hyaline body myopathy: Adulthood manifestations. Can J Neurol Sci. 2005;32:253–256. doi: 10.1017/s0317167100004078. [DOI] [PubMed] [Google Scholar]
- 31.Shingde MV, et al. Myosin storage (hyaline body) myopathy: A case report. Neuromuscul Disord. 2006;16:882–886. doi: 10.1016/j.nmd.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Tajsharghi H, Oldfors A, Swash M. Myosin storage myopathy with cardiomyopathy. Neuromuscul Disord. 2007;17(9–10):725. doi: 10.1016/j.nmd.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 34.McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: Improved prediction of coiled coils from sequence. Bioinformatics. 2006;22:356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- 35.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sumida JP, Wu E, Lehrer SS. Conserved Asp-137 imparts flexibility to tropomyosin and affects function. J Biol Chem. 2008;283:6728–6734. doi: 10.1074/jbc.M707485200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.