Abstract
RBX1 (RING box protein-1) or ROC1 (regulator of cullins-1) is the RING component of SCF (Skp1, Cullins, F-box proteins) E3 ubiquitin ligases, which regulate diverse cellular processes by targeting various substrates for degradation. However, the in vivo physiological function of RBX1 remains uncharacterized. Here, we show that a gene trap disruption of mouse Rbx1 causes embryonic lethality at embryonic day (E)7.5, mainly due to a failure in proliferation; p27, a cyclin dependent kinase inhibitor, normally undetectable in the early embryos, accumulates at high levels in the absence of Rbx1. Although mice heterozygous for the Rbx1 gene trap appear viable and fertile without obvious abnormalities, the Rbx1+/Gt MEFs do show retarded growth with G1 arrest and p27 accumulation. Simultaneous loss of p27 extended the life span of Rbx1Gt/Gt embryos from E6.5 to E9.5, indicating that p27-mediated cell cycle inhibition contributes to the early embryonic lethality in the Rbx1-deficient embryos. Our study demonstrates that the in vivo physiological function of RBX1 is to ensure cell proliferation by preventing p27 accumulation during the early stage of embryonic development.
Keywords: SCF E3 ubiquitin ligase, protein degradation, growth suppression
The SCF (Skp1, Cullins, F-box proteins) E3 ubiquitin ligases, consisting of Skp1, Cullins/Cdc53, F-box proteins, and the RING domain containing protein RBX1 (RING box protein-1)/ROC1 (regulator of cullins-1) (1–6), are the largest family of E3 ligases (7). Structurally, cullin-1 acts as a scaffold protein, where the N terminus binds to Skp1-F-box complex, and the C terminus binds to RBX1 (8). The core SCF E3 ubiquitin ligase is a complex of RBX1-cullins (9), whereas the substrate specificity of the SCF complex is determined by F-box proteins that bind to Skp1 and cullins through its F-box domain, and to substrates through its WD40, or LRR domains (8). By timely targeting various substrates for degradation, SCF E3 ligases regulate diverse cellular processes, including cell cycle progression, signal transduction, gene transcription, DNA replication, viral modulation, development, as well as circadian clock and protein quality control (10–12).
RBX1 is a 14-kDa protein with a RING finger domain (C3H2C3) required for ubiquitin ligation. RBX1 mediates the neddylation of CUL1, which activates SCF E3 ligase activity by facilitating the ubiquitin transfer from E2 to substrates (4, 13, 14). Two RBX family members, RBX1/ROC1 and RBX2/ROC2/SAG, were identified in human and mouse, and both are evolutionarily conserved in mammals (15). Until now, the known differences between RBX1/ROC1 and RBX2/ROC2/SAG are that Rbx1 is constitutively expressed, whereas Rbx2/Sag is stress inducible (16), and that RBX1 prefers to bind with Cul2/VHL, whereas RBX2/SAG with Cul-5/SOCS (17).
Although RBX1 is the key component of SCF E3 ligase and likely involves the regulation of many cellular processes, the in vivo physiological function of RBX1 in mouse remains uncharacterized. Here, we use a gene trap allele of Rbx1 to generate Rbx1Gt/Gt embryos, and found that these embryos fail to adequately proliferate and die at embryonic day (E)7.5 with a remarkable accumulation of p27. Simultaneous deletion of p27 partially rescues the early lethality of Rbx1 disrupted embryos and extends their life span from E6.5 to E9.5. Thus, RBX1 is required for mouse embryogenesis. By blocking p27 accumulation, RBX1 ensures proper cell proliferation at the early gastrulation stage of embryonic development.
Results
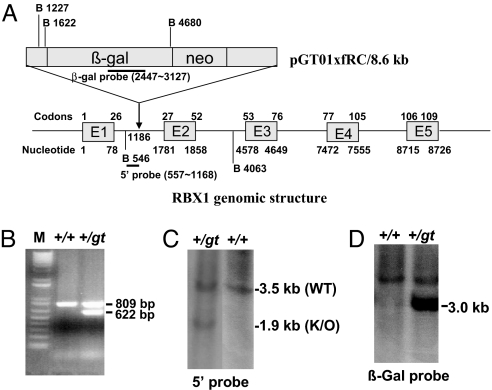
Generation of Rbx1 Gene Trap Mice from a Baygenomic ES Clone, XB674.
We searched the BayGenomics database (now maintained by the International Gene Trap Consortium, www.genetrap.org) for ES cells containing a gene trap of the Rbx1 gene, and identified 3 such clones. Clone XB674 has an insertion in the first intron (Fig. 1A), whereas clones RRS032 and RRS048 have insertions in the third intron. Mouse Rbx1 gene contains 5 exons and 4 introns, and the RING domain, required for E3 ligase activity, consists of codons 42 to 97. Insertion at intron 1 will abolish the RING domain, whereas in intron 3 it will destroy the RING (Fig. 1A). All 3 clones were characterized first by RT-PCR analysis, then by PCR walking to identify the vector insertion site. Only one clone, XB674, was confirmed with insertion site defined at the nucleotide 1186 in intron 1, relative to the translational initiation site of mouse Rbx1 gene. Using primers around the insertion site, we PCR-genotyped this ES clone with the wild-type allele of a 809-bp band and the gene trap allele of a 622-bp band (Fig. 1B). The identity of this clone was further confirmed by 5′end RACE. Clone XB674 was used to generate Rbx1+/Gt heterozygous mice. Resulting agouti chimeric male mice were crossed with C57BL/6 female mice. One male mouse had germ-line transmission with 1 copy of the potentially trapped Rbx1 gene. This Rbx1+/Gt mouse was backcrossed with C57BL/6 female mice to generate more Rbx1+/Gt mice. The identity of Rbx1+/Gt mouse was further confirmed by genomic Southern analysis using 5′-end probe to detect a 3.5-kb wild-type band and 1.9-kb mutant band, respectively (Fig. 1C), and β-Gal probe to detect a 3.0-kb mutant band (Fig. 1D). These Rbx1+/Gt mice develop normally without any obvious signs of abnormality up to 1.5 years of age.
Fig. 1.
Generation of Rbx1 gene trap mice. (A) Genomic structure of mouse Rbx1 gene; 5 exons (E1-E5) and 4 introns (between exons) are shown, along with the insertion site for targeting vector, pGT01xfRC, in ES cell line XB674. The site for target vector insertion is indicated, relatively to translational initiation site of mouse Rbx1 gene, setting arbitrarily as 1. The size and location of probes for Southern blotting are indicated. B, BamHI. Drawing is not to scale. (B) PCR-based genotyping. Genomic DNA was isolated from ES cells and subjected to PCR amplification, as described in Materials and Methods. Shown are 2 fragments with size of 809 and 622 bp, respectively, representing WT and Rbx1Gt. (C and D) Genomic Southern blotting to confirm the genotype. Genomic DNA was isolated from mouse tails, digested with BamHI, and subjected to Southern blot analysis, using 5′-end probe (C) and β-gal probe (D).
Rbx1Gt/Gt Mice Are Embryonic Lethal.
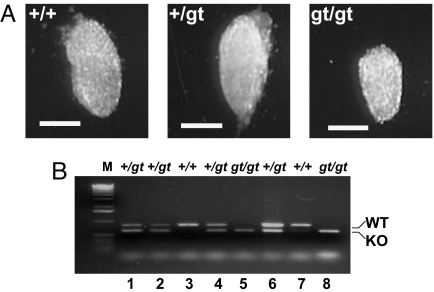
The confirmed heterozygous mice were intercrossed to generate homozygous mice. Among a total of 303 offspring genotyped, none were homozygous for the gene trapped allele (Table 1). The ratio of heterozygous to wild-type mice was ≈2:1, as expected when homozygous disruptions were lethal. To define at which stage the Rbx1 disrupted embryos die during embryonic development, we isolated embryos beginning at E13.5 and worked backwards until we were able to identify embryos homozygous for the gene trapped allele. As shown in Table 1, no viable embryos can be detected at E7.5 or later. Also, no homozygous gene trap embryos were identified by PCR screening of mouse embryonic fibroblast (MEF) lines generated from >40 embryos aged E10.5 to E13.5. However, when E6.5 embryos were isolated, we were able to identify smaller, but viable embryos (Fig. 2A), which were subsequently confirmed to be Rbx1Gt/Gt by PCR-genotyping (Fig. 2B, lanes 5 and 8). A Mendelian ratio of 1:2:1 (+/+:+/Gt:Gt/Gt) was achieved among a total of 16 embryos dissected (Table 1). Thus, we concluded that Rbx1Gt/Gt causes embryonic lethality at E7.5. Also, we collected a total of 43 blastocysts (E3.5) for PCR genotyping, and identified 12 of them to be Rbx1Gt/Gt, a ≈25% ratio (Table 1). However, we were unable to establish Rbx1Gt/Gt ES cell lines from the Rbx1Gt/Gt blastocysts. Among >100 blastocysts collected for this purpose, we were able to establish a total of 32 ES cell lines with 9 as WT (28%) and 23 as heterozygotes (72%). Thus, Rbx1 appears essential for both ES cell propagation and embryonic viability.
Table 1.
Genotypes of RBX1 mutant mice or embryos
| Age | RBX1+/+ | RBX1+/gt | RBX1gt/gt | Total |
|---|---|---|---|---|
| Mice | ||||
| >3 wk | 118 (39%) | 185 (61%) | 0 | 303 |
| Embryos | ||||
| E13.5 | 18 | 25 | 0 | 43 |
| E10.5 | 3 | 12 | 0 | 15 |
| E9.5 | 14 | 26 | 0 | 40 |
| E8.5 | 5 | 11 | 0 | 16 |
| E7.5 | 4 | 12 | 0 | 16 |
| E6.5 | 4 | 8 | 4 | 16 |
| E3.5 | 10 | 21 | 12 | 43 |
Fig. 2.
A Rbx1Gt/Gt embryo at E6.5 is viable but smaller. (A) Appearance of E6.5 embryo with 3 genotypes. The embryos at E6.5 days were dissected from pregnant mice and photographed. (Scale bar, 100 μm) (B) PCR-based genotyping. The embryos were subsequently subjected to genotyping by using PCR-based method, as described in Materials and Methods. Shown are 2 fragments, representing WT and Rbx1Gt, respectively.
Remarkable Reduction of Cell Proliferation in E6.5 Rbx1Gt/Gt Embryos.
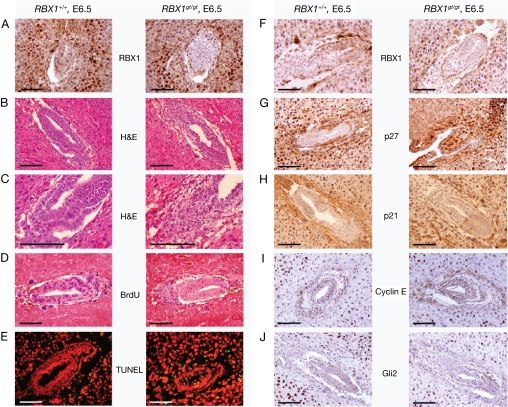
We next sought to determine the cause of the embryonic lethality for Rbx1Gt/Gt embryos at E6.5 day, the time at which gastrulation initiates in the mouse embryo (18). Immuno-histochemical staining using RBX1 specific antibody confirmed that RBX1 is expressed in wild-type, but not Rbx1Gt/Gt embryos (Fig. 3 A and F), suggesting that Rbx1gt is likely a null allele. Unfortunately, we were unable to further validate this finding by Western blot analysis due to lack of sufficient materials. H&E staining showed a general hypocellularity in the Rbx1Gt/Gt embryos (Fig. 3 B and C), suggesting a potential defect in cell proliferation. To pursue this lead, we measured the incorporation of BrdU into DNA during the S phase of the cell cycle. As shown in Fig. 3D, wild-type embryo contains many BrdU positive (blue) staining cells. In contrast, the Rbx1Gt/Gt embryos show a dramatic reduction in number of cells with BrdU staining. Also, the level of BrdU incorporation in the Rbx1Gt/Gt embryo was significantly lower than in wild-type embryo. Thus, cell proliferation, as measured by BrdU incorporation, is remarkably reduced in the Rbx1Gt/Gt embryo. We also measured potential involvement of apoptosis and observed a slight increase of apoptosis in the Rbx1Gt/Gt embryo by the TUNEL assay (Fig. 3E). Thus, the failure in cellular proliferation, essential for initiating gastrulation beginning ≈E6.5, is likely the cause of the embryonic lethality at E7.5 due to Rbx1 disruption.
Fig. 3.
Reduced proliferation and p27 accumulation in Rbx1Gt/Gt embryos. Mouse embryos at E6.5 were dissected from pregnant mice, fixed in 4% PFA-PBS, and embedded in paraffin; 5-μm thick sections were cut for immuno-staining with antibody against RBX1 (A and F), p27 (G), p21 (H), cyclin E (I), and Gli2 (J), or for H&E staining (B and C). Pregnant mice were injected i.p with BrdU labeling reagent 2 h before killing. Embryos were dissected and fixed in 4% PFA-PBS. The BrdU incorporation assay (D) or TUNEL assay (E) was performed, as described in Materials and Methods. (Scale bar, 200 μm.)
Accumulation of p27 in E6.5 Rbx1Gt/Gt Embryos.
We next sought to determine the potential molecular mechanism by which Rbx1 disruption causes the suppression of proliferation, with a focus on proteins that are known to be in vitro substrates of RBX1-SCF E3 ubiquitin ligases or that regulate cell cycle progression. Among the few proteins tested, RbxGt/Gt embryos have a dramatic accumulation of p27, a well-known in vitro substrate of RBX1-SCF (19–21), whose expression was not detectable in wild-type embryos (Fig. 3G). No significant changes were found in other known substrates of SCF E3 ligases that regulate cell proliferation, including p21 (Fig. 3H), cyclin E (Fig. 3I), and Gli2 (Fig. 3J), a mouse ortholog of Drosophila Cubitus interruptus (Ci) protein, which accumulates on deletion of Rbx1a in Drosophila (22). Thus, it appears that at the early stage of embryogenesis, Rbx1 disrupted embryos fail to degrade p27, a cyclin dependent kinase inhibitor that blocks G1 to S phase progression (23). Accumulation of p27 blocks cell proliferation and leads to embryonic lethality.
MEF Cells from Rbx1+/Gt Mice Are Growth Retarded with G1 Arrest and p27 Accumulation.
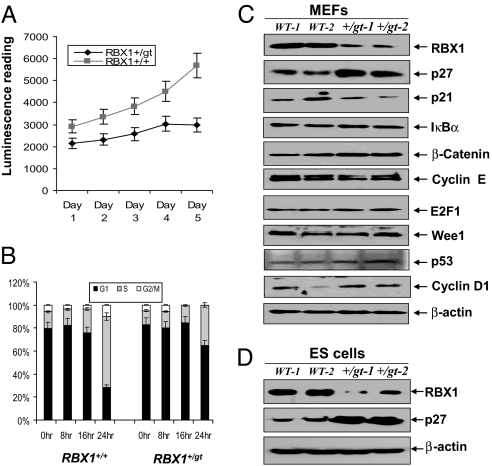
We further determined the growth status of primary MEFs cells from wild-type and Rbx1 heterozygotes. As shown in Fig. 4A, Rbx1+/Gt MEFs grow in a monolayer culture, but with a much slower growth rate, as compared with wild-type MEFs. FACS analysis revealed that although wild-type MEF cells have 65% of their population entering the S phase 24 h after the release from serum starvation, a majority of Rbx1+/Gt MEF cells are still arrested at the G1 phase of cell cycle with only 35% of the population progressing into the S phase (Fig. 4B). Accordingly, accumulation of p27, but not other substrates of RBX1-SCF E3 ubiquitin ligases, including p21, IκBα, β-catenin, cyclin E, E2F1, and Wee1, was readily seen in 2 independent primary Rbx1+/Gt MEF cultures (Fig. 4C). The levels of other growth regulatory proteins, such as p53 and cyclin D1, were not changed. Also, accumulation of p27 was observed in 2 independent Rbx1+/Gt ES clones with reduced RBX1 expression, as compared with the wild-type controls (Fig. 4D). Thus, reduced RBX1 expression in Rbx1 heterozygous cells caused the accumulation of p27, which contributed to growth suppression of Rbx1+/Gt MEF cells.
Fig. 4.
Rbx1+/Gt MEFs are growth retarded and arrested in G1 with p27 accumulation. Primary MEFs cells were prepared from embryo at E13.5, and grown in DMEM containing 15% sera. Every day up to 5 days, 1 dish was harvested and subjected to growth assay by using ATP-lite kit (A). Primary MEF cells were subjected to serum starvation (0.5%) for 24 h. Samples were collected after indicated hours post addition of 15% serum, and subjected to FACS analysis. The percentage cells in each phase of cell cycle were plotted (B). Primary MEF cells (C) or ES cells (D) were grown at 15% serum, and subjected to Western blot analysis using indicated antibodies.
Partial Rescue of Rbx1 Embryonic Lethality by Simultaneous Deletion of p27.
It has been previously shown that p27−/− (also known as Cdkn1b −/−) mice are viable with increased body size (24–26). We reasoned that if p27 accumulation causes early embryonic lethality, then simultaneous deletion of p27 would be able to rescue the lethality. Therefore, we performed an intercross between Rbx1+/Gt and p27−/− mice (from Jackson Laboratory) to generate mice heterozygous to both Rbx1 and p27 (Rbx1+/Gt; p27+/−). Intercrossing of these mice was performed. According to Mendel's distribution, 6.25% of offspring mice should have a genotype of Rbx1Gt/Gt;p27−/−, if simultaneous p27 deletion fully rescues the embryonic lethality. As summarized in Table 2, out of a total of 168 mice genotyped at weaning, none of them were double homozygous null for Rbx1 and p27, indicating that p27 deletion cannot fully rescue the Rbx1Gt/Gt phenotype. We next determined whether p27 deletion could partially rescue the Rbx1Gt/Gt embryos. Out of 98 embryos genotyped at E9.5, we identified 5 Rbx1/p27 double homozygous null embryos, a ratio of 5.1%, which is close to expected Mendelian ratio of 6.25%. Examples of PCR genotyping of Rbx1/p27 double null embryos are shown in Fig. 5A, lanes 2 and 3. No embryos that were Rbx1Gt/Gt;p27+/− were identified, indicating that homozygous, but not heterozygous, deletion of p27 can partially rescue the early embryonic lethality at E7.5 by Rbx1 disruption. Also, no double homozygous null embryos out of 126 genotyped were identified at E10.5 after a crossing of Rbx1+/Gt; p27+/− and Rbx1+/Gt;p27−/− (Table 3), suggesting that RBX1 is required for later stage of embryogenesis as well, independent of p27.
Table 2.
Partial rescue of early embryonic lethality of Rbx1-deficient mice by p27 loss
| Genotype | No. adults | No. E9.5 embryos |
|---|---|---|
| p27+/+ | ||
| Rbx1+/+ | 11 | 8 |
| Rbx1+/gt | 36 | 10 |
| Rbx1gt/gt | 0 | 0 |
| p27+/− | ||
| Rbx1+/+ | 31 | 18 |
| Rbx1+/gt | 62 | 36 |
| Rbx1gt/gt | 0 | 0 |
| p27−/− | ||
| Rbx1+/+ | 9 | 10 |
| Rbx1+/gt | 19 | 11 |
| Rbx1gt/gt | 0 | 5 |
| Total | 168 | 98 |
Fig. 5.
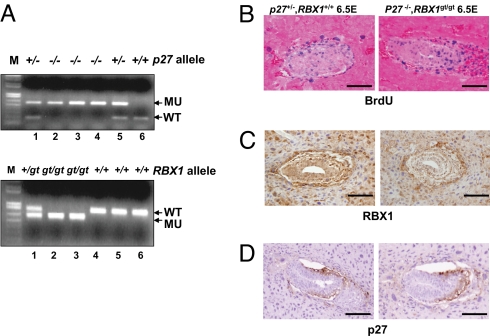
Developmental stage dependent rescue of proliferation defect in Rbx1Gt/Gt embryos by simultaneous deletion of p27. Rbx1+/Gt mice were mated with p27−/− mice to generate mice heterozygous to both Rbx1 and p27 (Rbx1+/Gt;p27+/−). Intercrossing of these mice was performed. The embryos at E9.5 were dissected, and yolk sacs genotyped for Rbx1 and p27 alleles (A). Pregnant mice were injected i.p with BrdU labeling reagent 2 h before killing. Embryos at E6.5 were dissected and fixed in 4% PFA-PBS, and embedded in paraffin; 5-μm thick sections were cut for BrdU staining (B) and immuno-staining with antibodies against RBX1 (C) or p27 (D). (Scale bar, 200 μm.)
Table 3.
No viable Rbx1/p27 double null embryos at E10.5
| Genotype | No. E10.5 embryos |
|---|---|
| p27+/− | |
| Rbx1+/+ | 21 |
| Rbx1+/gt | 47 |
| Rbx1gt/gt | 0 |
| p27−/− | |
| Rbx1+/+ | 21 |
| Rbx1+/gt | 37 |
| Rbx1gt/gt | 0 |
| Total | 126 |
To confirm that simultaneous deletion of p27 indeed rescues the proliferation defect in Rbx1Gt/Gt embryos at E6.5, we performed the BrdU incorporation assay in the embryos. Unlike the Rbx1Gt/Gt embryo, which showed a remarkable reduction of proliferation, as reflected by low BrdU incorporation (Fig. 3D), Rbx1Gt/Gt;p27−/− embryos show a high proliferation rate with a high level of BrdU incorporation, indistinguishable from the wild-type control (Fig. 5B). Expression of RBX1 was detectable in Rbx1+/+;p27+/−, but not in double null embryos (Fig. 5C); p27 is not expressed at E6.5 in the wild-type embryo (Fig. 5D), but is expressed when Rbx1 is ablated (Fig. 3G). The double null condition prevents accumulation of p27 at E6.5 (Fig. 5D), allowing for development to proceed. Thus, accumulation of p27 on Rbx1 disruption is the cause of early embryonic death (E7.5), as a result of dramatically reduced cellular proliferation in the Rbx1Gt/Gt embryos.
Discussion
In this study, we used a mouse gene trap model to elucidate the in vivo physiological function of RBX1, a RING component of SCF E3 ubiquitin ligases, which is constitutively expressed during mouse embryogenesis with the peak of expression at E7.0 (27). Our data clearly demonstrated that Rbx1 is a growth essential gene during mouse embryogenesis, and its disruption causes early embryonic lethality at E7.5 due to a proliferation failure as a result of p27 accumulation. The mouse embryo begins gastrulation ≈E6.5, at which time a dramatic increase in cell proliferation (Fig. 3D) is required for initiating gastrulation that forms the 3 germ layers of an embryo to establish the body plan of the mature organs (18); p27, a known inhibitor of cell cycle progression at the G1 phase (28), is normally not detected at this stage of embryonic development (Fig. 3G), until E13.5-E14.5 (29, 30). Abnormal accumulation of p27, as a result of Rbx1 disruption, could induce proliferation failure, leading to embryonic death. It is certainly the case here, as confirmed by simultaneous deletion of p27, which rescues the proliferation failure and early embryonic lethality at E6.5, and extends the embryo life span from E6.5 to E9.5. Thus, elimination of p27 by RBX1 is critical for rapid proliferation of embryos at the gastrulation.
Earlier studies have shown that Hrt1, the yeast homologue of RBX1, is a growth essential gene whose targeted disruption causes yeast death, which can be rescued by either human RBX1 (3, 6) or its family member, RBX2/SAG (31). In Caenorhabditis elegans, siRNA knockdown of RBX1 causes pronounced defects in the first meiotic division, in mitotic chromosomal condensation, and in germ cell proliferation, indicating its role in chromosome metabolism and cell cycle control (32). In Drosophila, disruption of ROC1a, the homologue of RBX1, causes the lethality due to a proliferation failure as a result of accumulation of Ci (a Drosophila ortholog of mouse Gli2), a transcription factor that regulates Hedgehog signaling (22). This observation is strikingly different from the disruption of Rbx1 in mouse; no Gli accumulation was found (Fig. 3J). Also, targeted deletion of either Cul1−/− (33) or Cul3−/− (34) caused embryonic lethality at E6.5–7.5 with cyclin E accumulation. Unfortunately, none of these studies performed the rescue experiment to confirm that accumulation of Ci or cyclin E is causally related to embryonic lethality. Also, the Cul4A−/− embryos died between E4.5-E7.5 (35), whereas Cul7−/− mice died immediately after birth because of respiratory distress resulting from abnormal vascular morphogenesis (36). Thus, targeted disruption of Rbx1 in different species or of different cullins in mice all induces embryonic lethality (except Cul-7), but with apparently different mechanism. Accumulated p27, which blocks cell proliferation, is the cause of embryo lethality at the early gastrulation stage of Rbx1Gt/Gt development. It is worthy noting that mice lacking SKP2, an F-box protein known to target p27 for degradation by SCF E3 ligase (19–21), were viable, although p27 was accumulated in the MEF cells and fetal liver tissues derived from these mice (37). However, the lack of embryonic death in Skp2−/− mice, despite p27 accumulation, can be logically explained by a simultaneous accumulation of cyclin E (37), another substrate of SCFskp2 E3 ligase, whose accumulation was not seen in Rbx1Gt/Gt embryos (Fig. 3). Thus, under Skp2 disrupted, but not Rbx1 disrupted situation, the negative effect of p27 on cell cycle progression is likely neutralized by positive effect of cyclin E (23), leading to a proper cell proliferation at the stage of gastrulation and beyond, for the entire period of Skp2−/− mouse embryogenesis.
RBX1 is an essential component of SCF E3 ubiquitin ligases for timely ubiquitination and degradation of many cellular proteins involved in growth regulation (10–12). Inability of simultaneous deletion of p27 and Rbx1 to fully rescue embryonic development indicates that the lethality induced by Rbx1 disruption is likely due to a multifactorial process, in analogous to a partial rescue of Brca15-6 early embryonic lethality by p53 or p21 null mutation (38). However, it is known that mouse genome contains 2 RBX family members (RBX1 and RBX2/SAG), and RBX2/SAG is also expressed during embryogenesis (15). The fact that Rbx1 disruption produces an embryonic lethal phenotype in the presence of RBX2/SAG indicates that, like what was observed in Drosophila (39), these 2 RBX1 family members are not functionally redundant in mouse either, and are likely to target different sets of substrates during embryonic development.
In summary, our in vivo study presented here clearly demonstrates that RBX1 is essential for mouse embryonic development, and that early lethality of Rbx1Gt/Gt embryos is caused by proliferation failure at gastrulation as a result of p27 accumulation.
Materials and Methods
ES Cell Culture.
Baygenomics ES cells were cultured in 1× GMEM medium (Sigma) supplemented with 2 mM glutamine, 1 mM sodium pyruvate, 1× nonessential amino acids, 15% (vol/vol) FBS, 1 mM β-mercaptoethanol, and 500–1,000 units per milliliter of leukocyte inhibitory factor (LIF) (Chemicon).
Generation of RBX1 Gene Trapped Mice.
All procedures were approved by the University of Michigan Committee on Use and Care of Animals. Animal care was provided in accordance with the principles and procedures outlined in the National Research Council Guide for the Care and Use of Laboratory Animals. XB674 ES cells were trypsinized, and 16 ES cells were microinjected into blastocysts. Blastocysts were collected from superovulated C57BL/6NCrl female mice (Charles River Laboratories) mated with F1 (C57BL/6J X DBA/2J) males (The Jackson Laboratory). Microinjected blastocysts were transferred to pseudopregnant recipient females the day of injection by using standard methods (40). Chimeras were bred to C57BL/6J females (The Jackson Laboratory) to test for germ-line transmission. Germ-line transmission was confirmed by Southern blot and PCR analyses.
Generation and Maintenance of Rbx+/Gt ES Cell Lines.
Blastocysts were isolated from Rbx1Gt/+ females mated with Rbx1Gt/+ males. Blastocysts were placed in culture on irradiated mouse embryonic feeder cells in high glucose DMEM (Invitrogen) supplemented with 15% FBS (Harlan), 0.1 mM β-mercaptoethanol (Sigma), 50 IU/mL penicillin, 50 μg/mL streptomycin, 103u/mL LIF (Chemicon), and 12.5 μM PD 98,059 (Sigma). Inner cell mass outgrowths were trypsinized and passaged sequentially until ES cell lines were established in 35-mm cell culture dishes. Subsequent culture continued in the absence of antibiotics and PD098059. ES cells were passaged on gelatin coated dishes twice to eliminate feeder cells before genotyping as described (41).
PCR-Based Genotyping.
Genomic DNA was isolated from ES cells or mouse tail tips and was genotyped using primer set of ROCsi09: 5′-ACGAGATGGCTACTAGTTGTCAC-3′ and ROCsi16: 5′-GAACTCAAAATCTGCATAAGCAC-3′ for WT (809 bp) and primer set of ROCsi09 and BaygenR01: 5′-CACTCCAACCTCCGCAAACTC-3′ for mutant (622 bp). OIMR0947: 5′-GATGGACGCCAGACAAGC-3′ and OIMR0948: 5′-CTCCTGCCATTCGTATCTGC-3′ for p27 WT (180 bp). N1: 5′-CCTTCTATGGCCTTCTTGACG-3′ and K3: 5′-TGGAACCCTGTGCCATCTCTAT-3′ for p27 mutant (500 bp).
Southern Analysis.
Genomic DNA was isolated from mouse tail biopsies according to University of Michigan Transgenic Animal Model Core protocol (www.med.umich.edu/tamc/tDNA.html) and subjected to BamHI digestion. The 5′ end probe (611 bp) was generated by PCR amplification using the primer set ROC5a-01: 5′-TCTATTGACTCCGAGGGCCTGGC-3′ and ROC5a-02: 5′-AGACAAGGCACCCTTTTCAAGGAC-3′. The β-Gal probe (680 bp) was generated by PCR amplification using the primer set, β-gal01: 5′-TTATCGATGAGCGTGGTGGTTATGC-3′, and β-gal02: 5′-GCGCGTACATCGGGCAAATAATATC-3′. All of the probed were confirmed by DNA sequencing.
Immunohistochemistry.
Mouse embryos were fixed in 4% paraformaldehyde (PFA)-PBS and embedded in paraffin; 5-μm-thick sections were cut for H&E staining or for immuno-staining with ABC kits (Vector Labs). The primary antibodies used include RBX1, made against C-terminal peptide of human RBX1, LDNREWEFQKYGH, with affinity purification; p27 (BD PharMingen); p21 (BD PharMingen); cyclin E (Santa Cruz); and Gli2 (Santa Cruz). Normal goat serum was used as negative controls.
BrdU Incorporation Assay.
Pregnant mice were injected i.p with BrdU labeling reagent (2 mL/100g body weight) 2 h before killing. Embryos were dissected and fixed in 4% PFA-PBS. The assay was performed with a 5-Bromo-2′-deoxy-uridine Labeling and Detection Kit II (Roche). Slides were developed with NBT/BCIP and counterstained with Eosin (Richard-Allan Scientific).
TUNEL Assay.
Mouse embryos were dissected and fixed in 4% PFA-PBS, and embedded in paraffin; 5-μm thick sections were cut, and deparaffinized slides were prepared for the TUNEL assay by using in situ Cell Death Detection Kit (Roche). The slides were stained with the TUNEL reaction mixture, along with Label Solution for the negative control, and counterstained with propidium iodide counterstaining solution. The slides were mounted and analyzed under a fluorescence microscope.
Generation and Maintenance of MEFs.
MEFs were isolated from day E10.5–13.5 embryos. The embryos were washed once with DMEM supplemented with 25 mM Hepes buffer and 3 times with PBS. The tissue was then minced with a scalpel and digested with 0.05% trypsin solution containing 0.53 mM EDTA (Invitrogen Life Technologies) for 20 min at 37 °C. The tissue was shaken vigorously every 3 min during the incubation, and the mixture was then passed 3 times through a ½ −18G needle to further dissociate any remaining clumps. Trypsin was inactivated by addition of a 15-fold excess of MEF media. The cells from each embryo were put into 100-mm dish and incubated at 37 °C in a 5% CO2 humidified containing 15% FBS, 2 mM l-Glutamine, 0.1 mM MEM nonessential amino acids, and 10 μg/mL gentamycin.
Western Blot Analysis.
MEF or ES cells were harvested, lysed in a Titron X-100 lysis buffer, and subjected to Western blot analysis as described (42), using antibodies against RBX1, p27(BD PharMingen), p21 (BD PharMingen), cyclin E (Santa Cruz), E2F1 (Cell signaling), IκBα (Santa Cruz), β-catenine (Santa Cruz), Cyclin D1 (Santa Cruz), and p53 (Roche), as well as ß-actin (Sigma), as the loading control.
FACS Analysis.
Primary MEF cells were seeded into 60-mm dish and subjected to serum starvation (0.5%) for 24 h. The cells were harvested at 0, 8, 16, and 24 h post addition of 15% serum. Single suspended cells were fixed in 70% EtOH at 4 °C for at least 4 h, stained with 1X Propidium Iodide solution containing 400 μg/mL RNase (Roche), and analyzed in the Flow Cytometry Lab facility at the University of Michigan.
Acknowledgments.
We would like to thank Dr. Qingyang Gu for his help in immunohistochemistry staining of embryos and Dr. K. Sue O'Shea for her help in dissection of early stage embryos. This work was supported by National Cancer Institute Grants CA111554, CA116982, and CA118762 (to Y.S.). Transgenic Core support was provided by the University of Michigan Cancer Center, National Institutes of Health Grant CA46592.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Bai C, et al. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 2.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 3.Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 4.Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan P, et al. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IkBa. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 6.Seol JH, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willems AR, Schwab M, Tyers M. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 8.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 9.Wu K, et al. The SCF(HOS/beta-TRCP)-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol Cell Biol. 2000;20:1382–1393. doi: 10.1128/mcb.20.4.1382-1393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol. 2005;16:323–333. doi: 10.1016/j.semcdb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 11.Nakayama KI, Nakayama K. Ubiquitin ligases: Cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 12.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 13.Morimoto M, Nishida T, Nagayama Y, Yasuda H. Nedd8-modification of Cul1 is promoted by Roc1 as a Nedd8–E3 ligase and regulates its stability. Biochem Biophys Res Commun. 2003;301:392–398. doi: 10.1016/s0006-291x(02)03051-6. [DOI] [PubMed] [Google Scholar]
- 14.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: Building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y, Tan M, Duan H, Swaroop M. SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: Molecular cloning, biochemical properties, and biological functions. Antioxid Redox Signal. 2001;3:635–650. doi: 10.1089/15230860152542989. [DOI] [PubMed] [Google Scholar]
- 16.Gu Q, Tan M, Sun Y. SAG/ROC2/Rbx2 is a novel activator protein-1 target that promotes c-Jun degradation and inhibits 12-O-tetradecanoylphorbol-13-acetate-induced neoplastic transformation. Cancer Res. 2007;67:3616–3625. doi: 10.1158/0008-5472.CAN-06-4020. [DOI] [PubMed] [Google Scholar]
- 17.Kamura T, et al. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakem R, et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 19.Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 20.Tsvetkov LM, Yeh K-H, Lee S-J, Sun H, Zhang H. p27kip1ubiquitination and degradation is regulated by the SCFskp2 complex through phosphorylated Thr187 in p27. Cur Biol. 1999;9:661–664. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- 21.Sutterluty H, et al. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat Cell Biol. 1999;1:207–214. doi: 10.1038/12027. [DOI] [PubMed] [Google Scholar]
- 22.Noureddine MA, Donaldson TD, Thacker SA, Duronio RJ. Drosophila Roc1a encodes a RING-H2 protein with a unique function in processing the Hh signal transducer Ci by the SCF E3 ubiquitin ligase. Dev Cell. 2002;2:757–770. doi: 10.1016/s1534-5807(02)00164-8. [DOI] [PubMed] [Google Scholar]
- 23.Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 24.Fero ML, et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 25.Kiyokawa H, et al. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1) Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama K, et al. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 1996;85:707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 27.Perin JP, et al. Genomic organization and expression of the ubiquitin-proteasome complex-associated protein Rbx1/ROC1/Hrt1. Cell Mol Biol. 1999;45:1131–1137. [PubMed] [Google Scholar]
- 28.Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 29.Ciemerych MA, Yu Q, Szczepanska K, Sicinski P. CDK4 activity in mouse embryos expressing a single D-type cyclin. Int J Dev Biol. 2008;52:299–305. doi: 10.1387/ijdb.072336mc. [DOI] [PubMed] [Google Scholar]
- 30.Nagahama H, Hatakeyama S, Nakayama K, Nagata M, Tomita K. Spatial and temporal expression patterns of the cyclin-dependent kinase (CDK) inhibitors p27Kip1 and p57Kip2 during mouse development. Anat Embryol. 2001;203:77–87. doi: 10.1007/s004290000146. [DOI] [PubMed] [Google Scholar]
- 31.Swaroop M, et al. Yeast homolog of human SAG/ROC2/Rbx2/Hrt2 is essential for cell growth, but not for germination: Chip profiling implicates its role in cell cycle regulation. Oncogene. 2000;19:2855–2866. doi: 10.1038/sj.onc.1203635. [DOI] [PubMed] [Google Scholar]
- 32.Sasagawa Y, Urano T, Kohara Y, Takahashi H, Higashitani A. Caenorhabditis elegans RBX1 is essential for meiosis, mitotic chromosomal condensation and segregation, and cytokinesis. Genes Cells. 2003;8:857–872. doi: 10.1046/j.1365-2443.2003.00682.x. [DOI] [PubMed] [Google Scholar]
- 33.Dealy MJ, et al. Loss of Cul1 results in early embryonic lethality and dysregulation of cyclin E. Nat Genet. 1999;23:245–248. doi: 10.1038/13886. [DOI] [PubMed] [Google Scholar]
- 34.Singer JD, Gurian-West M, Clurman B, Roberts JM. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 1999;13:2375–2387. doi: 10.1101/gad.13.18.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li B, Ruiz JC, Chun KT. CUL-4A is critical for early embryonic development. Mol Cell Biol. 2002;22:4997–5005. doi: 10.1128/MCB.22.14.4997-5005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arai T, et al. Targeted disruption of p185/Cul7 gene results in abnormal vascular morphogenesis. Proc Natl Acad Sci USA. 2003;100:9855–9860. doi: 10.1073/pnas.1733908100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakayama K, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 2000;19:2069–2081. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hakem R, de la Pompa JL, Elia A, Potter J, Mak TW. Partial rescue of Brca1 (5–6) early embryonic lethality by p53 or p21 null mutation. Nat Genet. 1997;16:298–302. doi: 10.1038/ng0797-298. [DOI] [PubMed] [Google Scholar]
- 39.Donaldson TD, Noureddine MA, Reynolds PJ, Bradford W, Duronio RJ. (2004) Targeted disruption of Drosophila Roc1b reveals functional differences in the Roc subunit of Cullin-dependent E3 ubiquitin ligases. Mol Biol Cell. 2004;15:4892–4903. doi: 10.1091/mbc.E04-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. New York: Cold Spring Harbor Lab Press; 2003. [Google Scholar]
- 41.Hughes ED, et al. Genetic variation in C57BL/6 ES cell lines and genetic instability in the Bruce4 C57BL/6 ES cell line. Mamm Genome. 2007;18:549–558. doi: 10.1007/s00335-007-9054-0. [DOI] [PubMed] [Google Scholar]
- 42.Duan H, et al. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol. 1999;19:3145–3155. doi: 10.1128/mcb.19.4.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]