Abstract
Preclinical animal models have largely ignored the immune-suppressive mechanisms that are important in human cancers. The identification and use of such models should allow better predictions of successful human responses to immunotherapy. As a model for changes induced in nonmalignant cells by cancer, we examined T-cell function in the chronic lymphocytic leukemia (CLL) Eμ-TCL1 transgenic mouse model. With development of leukemia, Eμ-TCL1 transgenic mice developed functional T-cell defects and alteration of gene and protein expression closely resembling changes seen in CLL human patients. Furthermore, infusion of CLL cells into young Eμ-TCL1 mice induced defects comparable to those seen in mice with developed leukemia, demonstrating a causal relationship between leukemia and the T-cell defects. Altered pathways involved genes regulating actin remodeling, and T cells exhibited dysfunctional immunological synapse formation and T-cell signaling, which was reversed by the immunomodulatory drug lenalidomide. These results further demonstrate the utility of this animal model of CLL and define a versatile model to investigate both the molecular mechanisms of cancer-induced immune suppression and immunotherapeutic repair strategies.
Cancer cells induce changes in the tumor microenvironment. The mechanisms whereby this occurs are poorly understood, but include direct cancer cell-T cell interactions, production of immune-suppressive factors by cancer cells, and induction of regulatory T-cell subsets (1, 2). Understanding the elusive mechanisms of tumor-driven immune evasion will aid the refinement of existing cancer immunotherapy strategies and identify novel treatments. To date, preclinical animal models that closely model human cancer and, in particular, include examination of the immune-suppressive mechanisms used by cancer cells have been under-characterized. The identification and use of such models should allow better predictions of successful human responses to immunotherapy.
Chronic lymphocytic leukemia (CLL) is an ideal model cancer to study immune T cells that have been exposed to circulating tumor B cells and is associated with immune dysfunction. CLL is the most common adult leukemia in North America and Europe and is currently incurable. Immunotherapeutic strategies are believed to represent a promising treatment modality for this disease if immune suppression can be controlled (3). Extensive research has been carried out using CLL human patient cells, including the characterization of altered gene and protein-expression profiles (4) and suppressed functional responses in patient T cells compared to healthy-donor T cells (1, 5).
We sought to determine whether development of leukemia in the well-established Eμ-TCL1 transgenic murine model of CLL (6) would induce changes in nonmalignant T cells. To examine this, we examined changes in gene-expression profile, protein expression, and function in T cells from Eμ-TCL1 transgenic mice as they developed CLL. We demonstrate that development of CLL in these transgenic mice is associated with similar impairment of T-cell function and alteration of gene expression in CD4 and CD8 T cells to that observed in human patients with this disease. The causal relationship between leukemia and the induction of T-cell changes is demonstrated in vivo by the finding that infusion of CLL B cells into nonleukemia-bearing Eμ-TCL1 mice rapidly induces these changes, demonstrating a causal relationship. This model allows dissection of the molecular changes induced in CD4 and CD8 T cells by interaction with leukemia cells, and further supports the concept that cancer results in complex abnormalities in the immune microenvironment. The similarities with human CLL, including reversible immunological synapse dysfunction with an immunomodulating drug (1), validates the use of Eμ-TCL1 mice as a model for further analyses of ways to prevent and reverse cancer-induced immune dysfunction.
Results
Dysfunctional T-Cell Function in Eμ-TCL1 Mice That Have Developed Leukemia.
To test the hypothesis that development of leukemia would induce changes in the nonmalignant cells of the immune system, we examined changes in T-cell function in Eμ-TCL1 transgenic mice (6, 7) as they developed leukemia from 12-months of age [see supporting information (SI) Fig. S1]. Transgenic expression of TCL1 in B cells has no demonstrable effect on T cells, because before development of leukemia (tested at 3–4 months) transgenic mice had normal T-cell numbers, CD4/CD8 ratio, cytokine production on mitogen, and antigenic stimulation (data not shown), and as described below, similar gene-expression profiles to WT mice.
However, once the mice developed CLL they had decreased in vivo antigen-specific T-cell activation, suppressed T-cell mitogenic proliferation, and impaired induction of idiotype-specific CD8 T cells capable of killing CLL cells (Fig. 1) compared to age-matched control WT mice or Eμ-TCL1 transgenic mice without CLL. Leukemic mice had dysfunctional T-cell lymphokine production (Th2-preponderant) with increased IL1, IL4, and IL6 but decreased IL2, IFN-γ, and IL-12β production. These mice also had impaired proliferation and IL-2 production following cross-linking of CD3/CD28 (data not shown). These data identify defective T-cell function with the development of CLL by using this mouse model that is not a result of aging or the expression of the transgene alone.
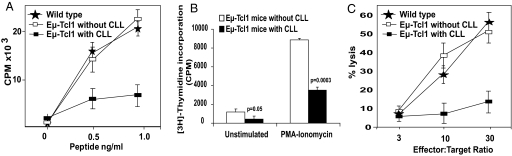
Fig. 1.
Development of CLL in Eμ-TCL1 mice induces suppressed T-cell function. (A) Decreased in vivo gp33 antigen-specific T-cell activation in mice with CLL. WT mice and mice with and without CLL were immunized with gp33 peptide. On day 8, spleen cells from immunized mice (n = 3 in each group) were stimulated with indicated concentrations of KAVYNFATM peptide in triplicate for 3 days before [3H]-Thymidine incorporation was measured. (B) Decreased response to mitogen activators in mononuclear cells from EμTCL1 mice with CLL compared to those without CLL. Mononuclear cells were stimulated with 100-ng/ml PMA and 4-μg/ml ionomycin for 3 days. Proliferation was measured using [3H]-Thymidine. (C) Decreased idiotype-specific cytotoxicity against CLL cells by CD8 T cells. CD8 cells were stimulated in vitro with dendritic cells (DCs) pulsed with Ig heavy chain (IgVH)-derived peptide GSAINYAPSI weekly for 4 weeks. Killing of CLL cells expressing this IgVH by the resulting T-cell lines was assessed at the effector:target ratios shown from wild type and Eμ-TCL1 mice with and without CLL. (A–C) Results are the mean ± SD and are representative of at least 3 independent experiments.
Molecular Basis for Defective T-Cell Function in the Eμ-TCL1 Mouse Model.
To understand the molecular basis for the observed functional defects and to compare changes seen in mice and patients with CLL (4), we performed gene-expression profiling. Unsupervised analysis of highly purified CD4 and CD8 T cells in CLL mice demonstrated altered gene-expression profiles compared to WT mice or to young Eμ-TCL1 mice without disease (Fig. S2). These findings again support the hypothesis that the altered T-cell gene-expression is not a result of aging or expression of a transgene in B cells. Because phenotypic and functional changes were induced in healthy allogeneic human T cells following short-term ex vivo coculture with CLL B cells, we next examined whether this also occurred in vivo. We therefore infused CLL B cells into 3-month-old Eμ-TCL1 mice that had not developed leukemia and after 7 days analyzed gene expression in highly purified CD4 and CD8 T cells. Infusion of CLL cells into young Eμ-TCL1 mice induced gene expression changes comparable to those seen in mice with de novo development of leukemia (see Fig. S2), demonstrating a causal relationship between leukemia and the induction of T-cell defects. Supervised analysis identified 153 differentially expressed genes (P ≤ 0.05) in CD4 T cells, with expression increased in 15 genes and decreased in 138 genes, while in CD8 T cells there were 158 differentially expressed genes (P ≤ 0.05), including 92 increased and 66 decreased genes in Eμ-TCL1 mice with CLL or infused with CLL cells compared to WT mice and Eμ-TCL1 mice without CLL (Tables S1–S3). The higher the tumor burden in these mice, the more clearly the impact on T-cell gene expression was evident. This correlates with elevated tumor counts being associated with enhanced functional T-cell defects in the human disease.
Gene-expression profile changes were validated using quantitative RT-PCR and assessment of protein expression (Fig. S3). Immunophenotyping confirmed altered expression of T-cell antigen receptor (TCR)-dependent proteins, including decreased CD28 and the negative regulator B- and T-lymphocyte attenuator (BTLA), with increased expression of cytotoxic T lymphocyte-associated antigen 4 (CTLA4). In mice with CLL there was an increase in the CD4+CD25+Foxp3+/CTLA4+ regulatory subset. Changes in expression of chemokines and chemokine receptors in CLL mice were similar to those in CLL patients (4).
Comparison of T-Cell Defects Between Eμ-TCL1 Mouse Model and Human Patients.
An important question was whether the changes in gene-expression profiles observed in the mice with CLL were similar to those observed in human patients with this disease. To examine this, analysis of gene-expression changes in T cells in CLL mice compared with those in patients (4) was performed using RESOURCERER (8, 9), a database for annotating and linking microarray resources within and across species. We identified 50 overlapping genes in CD4+ T cells (Fig. 2 A and B) and 45 overlapping genes in CD8+ T cells (Fig. 2 C and D). There was less heterogeneity of gene expression in transgenic mice than in CLL patients, making it easier to identify altered gene expression in components of specific pathways in T cells.
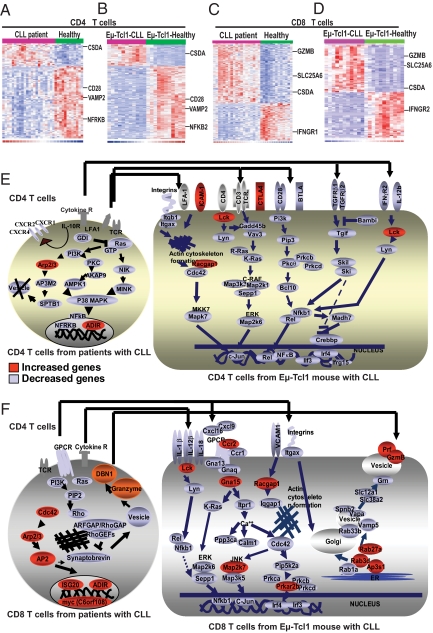
Fig. 2.
Eμ-TCL1 mouse model showing similar T-cell defects with development of CLL when compared to patients with CLL. Heat maps of differentially expressed genes by supervised analysis (P < 0.05) in (A) CD4 T cells or (C) CD8 T cells from patients with CLL compared with healthy donors, in comparison with (B) CD4 T cells or (D) CD8 T cells from Eμ-TCL1 mice with or without CLL. The heat maps represent selected genes that were expressed in both patients and Eμ-TCL1 mice with CLL. In CD4 T cells, out of 90 differentially expressed genes in patients with CLL and 152 differentially expressed genes in Eμ-TCL1 mice, 50 overlapping genes were detected using the RESOURCERER database (P ≤ 0.05), whereas in CD8 T cells, out of 273 differentially expressed genes in patients with CLL and 159 differentially expressed genes in Eμ-TCL1 mice, 45 overlapping genes were identified. Comparison of T cells from CLL with healthy T cells identified genes whose expression was above the mean in all samples shown in red, genes with expression below the mean shown in blue, and the mean expression shown in white. (E and F) T-cell pathways and functions in human patients and Eμ-TCL1 mice with CLL in (E) CD4 and (F) CD8 T cells. In CD4 cells, differentially expressed genes were involved in MAPK-dependent cell proliferation, ras-dependent Th cell differentiation, and cytokine/chemokine response pathways. In CD8 cells, differentially expressed genes were involved in vesicle trafficking, cytoskeleton organization, intracellular transport, cell motility, and cytokine-chemokine regulation pathways. Genes with increased expression are shown in red, while genes with decreased expression are shown in blue. The arrows indicate the similar pathways between patients with CLL and Eμ-TCL1 mice with CLL.
The majority of differentially expressed genes in CD4 T cells from both mice and patients with CLL were involved in cell proliferation and activation pathways with increase in Lck and the negative regulator CTLA4, and decreased expression of Nfrkb, CD28, BTLA, Pi3k, Pkc, Nfkb and Rel and Ras family members Gadd45b and Vav3 (Fig. 2 E). Within CD8 T cells there was increased RNA expression of ranzyme B (Gzmb) and perforin (Prf) in both murine and human CD8 cells in CLL, but decreased expression of granzyme and perforin in vesicles, suggestive of decreased ability to package and traffic proteins within the cell (4). In keeping with this, multiple defects within the actin cytoskeletal formation pathways were identified in both CD4 and CD8 T cells (Fig. 2 E and F). Integrity of the T-cell cytoskeleton is essential to regulate the dynamic signaling required for T-cell activation and effector function in response to immunological recognition of antigen-presenting cells (APCs). This cytoskeletal-dependent assembly of signaling molecules results in the formation of the F-actin rich immunological synapse (10). We observed impaired F-actin polymerization at the immune synapse in T cells from patients with CLL that could be reversed in vivo with the immunomodulatory drug lenalidomide (1).
Defective T-Cell Immunological Synapse Formation and Early T-Cell Signaling Can Be Repaired in Eμ-TCL1 Mice with Lenalidomide.
T-cell conjugates from mice with leukemia had suppressed antigen-dependent F-actin accumulation and early T-cell signaling at the immune synapse with CLL cells (APCs) compared to WT-mice conjugates. Moreover, we have demonstrated that infusion of CLL cells into young mice induces this T-cell defect, demonstrating an in vivo immunomodulating mechanism used by tumor cells. Treatment of both CLL cells and autologous T cells from leukemic mice with lenalidomide (0.5 μM for 24 h) enhanced the formation of the F-actin immune synapse and recruitment of tyrosine-phosphorylated proteins irrespective of the presence of exogenous antigen (Fig. 3 A and B). In addition, autologous CD8 T-cell effector function was enhanced following treatment with this agent (data not shown). Of note, the capacity to repair immunological recognition with this agent was associated with increased recruitment of the cytoskeletal-signaling molecules Lck and Cdc42 to the immunological synapse, regardless of whether the gene was increased or decreased on gene-expression profiling (Fig. 3 C and D). In addition, we have identified increased activation of Rho GTPases, including Cdc42, following drug treatment (data not shown) that is in agreement with enhanced early T-cell signaling as measured by increased tyrosine-phosphorylated activity at the synapse site. Thus, lenalidomide appears to be acting on the actin cytoskeletal signaling pathways that are dysregulated in T cells from CLL patients.
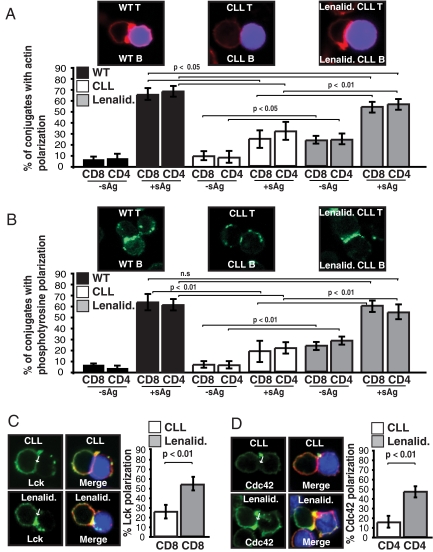
Fig. 3.
Treatment of Eμ-TCL1 mouse cells with lenalidomide significantly enhances immunological synapse formation. Autologous T-cell conjugates (with or without sAg-pulsed CLL B cells, stained blue) from untreated (CLL) or lenalidomide- (Lenalid.) treated (0.5 μM for 24h) T and CLL B cells from Eμ-TCL1 mice with CLL were scored for accumulation of (A) F-actin (red) or (B) phosphotyrosine (green) at the immune synapse. As controls, autologous age-matched WT mice T and B cells were used. Data are the mean ± SD from 3 independent experiments with 50 conjugates analyzed per experiment. (C and D) Autologous T cell-sAg-pulsed CLL B-cell conjugates (CLL B cells, stained blue) from untreated or lenalidomide-treated T and CLL cells from Eμ-TCL1 mice with CLL were stained for (C) Lck (green) or (D) Cdc42 (green), and F-actin (red). Arrows indicate protein localization at the T-cell-APC synapse site. Colocalization of proteins in the merged images is shown in yellow. Images and quantifications are representative of evaluation of 150 conjugates from 3 independent experiments.
Discussion
Our analysis demonstrates that this murine model of CLL induces similar T-cell defects to that seen in patients with CLL, confirming that the Eμ-TCL1 transgenic mouse is an appropriate animal model to study the mechanisms of immune dysregulation in cancer. The induction of these changes by infusion of CLL cells into young mice demonstrates the causal relationship of the CLL cells with the subsequent development of T-cell dysfunction. The altered gene-expression profiles in CD4 and CD8 T cells in mice with CLL compared to age-matched WT mice or to young Eμ-TCL1 mice that had not developed CLL suggest that the observed changes in gene-expression profiles did not result from aging or from transgenic expression of TCL1 alone. Additionally, because there was less heterogeneity of gene expression among the transgenic mice than that observed in patients with CLL, it was easier to identify alteration in gene expression in components of specific pathways of T-cell activation and function.
We observed that the majority of the altered genes in CD4 T cells were involved in cell proliferation, differentiation, and cytokine/chemokine-response pathways. Cross-linking of CD3/CD28 induces downstream MAPK-dependent signaling to promote T-cell proliferation and production of cytokines including IL-2 (11). CD28-dependent costimulation signaling occurs by stimulation of Pi3k, Pkc and induces transcription activity of Nfkb, Rel, and c-Jun, promoting cell proliferation and cytokine production (11–13). Most of the gene products that were down-regulated in CD4 T cells were involved in cell proliferation and differentiation pathways and can be phosphorylated by TCR stimulation (14), including Vav3, Nfkb, Rel, and c-Jun. VAV3 can induce MAPK pathway signaling through Ras family members (R-Ras, K-Ras). We showed altered gene and protein expression of molecules whose functions are dependent on TCR signals (15), including decreased expression of CD28, increased expression of the negative regulator CTLA4, and decreased expression of the BTLA in CD4 T cells. In CD8 T cells in mice with CLL, we also observed decreased expression of CD28 and increased expression of CD25 and changes in expression of chemokines and chemokine receptors, similar to those observed in human patients with CLL (4).
Functionally, these changes in gene and protein expression would be expected to result in the decreased proliferative and cytotoxic T-cell responses observed. Taken together, these results demonstrate that the leukemia cells induce changes in gene and protein expression of multiple pathways that regulate antigen recognition, immune response, T helper (Th) differentiation, and cytotoxicity, and are in keeping with the observed functional abnormalities in the T cells of mice with CLL and in CLL patients (4, 5, 16). Major defects were identified within the actin cytoskeleton-formation pathway, resulting in defective immunological synapse formation (1). The immune synapse is thought to function as a signaling platform or signalosome to translate controlled cytoskeletal signaling into appropriate T-cell functional responses in response to APCs. This synapse defect in CLL can be repaired in part by treatment with the immunomodulatory drug lenalidomide. Lenalidomide has clinical activity in CLL (17, 18) and also improves CLL B-cell APC function (19). The demonstration that treatment of Eμ-TCL1 transgenic leukemic and T cells with this agent improves the defective immune synapse formation in CLL lends further support that this mouse model can be used to examine unique agents in CLL (20).
Our results demonstrate that leukemia cells induce changes in multiple T-cell pathways regulating antigen recognition and effector function with a reversible immunological synapse dysfunction. These findings validate the use of Eμ-TCL1 mice as a model for further analyses of ways to prevent and reverse cancer-induced immune dysfunction. Now that dysfunctional intracellular targets and an immune suppressive mechanism co-opted by tumor cells have been identified, ongoing experiments are addressing ways to repair these defects using this animal model. Experimental approaches include the use of genetically manipulated T cells, monoclonal antibodies, and immunomodulatory drugs, including lenalidomide. The use of this model to understand and reverse the molecular changes in T cells induced by leukemia will likely have broad applications to maximize immune responses in patients.
Materials and Methods
Animals.
B6C3 mice were purchased from Jackson Laboratory. TCL1 gene transgenic B6C3 mice (Eμ-TCL1) were provided by C. Croce (Ohio State University, Columbus, OH). Protocols were approved by the Animal Care and Use Committee of the Dana-Farber Cancer Institute and Barts and The London School of Medicine, and mice maintained according to institute regulations. Ethical approval and a project license from the Animal Procedures Committee of the Home Office were obtained for all animal experiments. Additional methodology is described in the SI Materials and Methods.
Antibodies and Reagents.
Antibodies and reagents are listed in the SI Materials and Methods.
Cell Isolation, RNA Extraction, and Gene-Expression Profiling.
Splenic mononuclear cells were separated by ficoll-hypaque density gradient centrifugation and CD4 and CD8 T cells selected by negative depletion (Miltenyi Biotech). Purity was assessed using fluorescein-conjugated anti-CD4 and anti-CD8 antibodies. Cells were lysed in TRIzol for total RNA isolation (Life Tech) and 2 to 15 μg of total RNA used for gene-chip array. Gene-expression profiling was performed as outlined in the SI Materials and Methods. Data were normalized using DNA-Chip Analyzer (dChip) (21). To identify the genes whose expression patterns best distinguished groups, the permutation distribution of the maximum t-statistic was analyzed using Permax (22). Differentially expressed genes (filtered by P ≤ 0.05 and 1.2 < fold ≥1.2) from supervised analyses by their involvement in specific cellular pathways were determined using gene microarray pathway profiler 2.0 (GenMapp) software (23). CEL files have been deposited into the Gene Expression Omnibus and are available with accession numbers GSE8835 for human data (http://www.ncbi.nlm.nih.gov/geo/info/linking.html) GSE8836 for murine data (http://www.ncbi.nlm.nih.gov/geo/info/linking.html).
Quantitative RT-PCR and Western Immunoblot.
Representative genes were analyzed for expression by real time, quantitative PCR using FAM-TAMRA-labeled probes (Applied Biosystems) and protein expression measured by Western immunoblot and immunophenotyping. Additional methodology is described in the SI Materials and Methods.
Immune Assessment.
CTL effector assays (24), in vivo antigen specific T-cell activation assays (25, 26), and proliferation assays are described in the SI Materials and Methods.
Cell Conjugation Assays.
Cell conjugates were formed as previously described (1) and are summarized in the SI Materials and Methods.
Immunofluorescence and Confocal Microscopy Image Acquisition.
Immunofluorescent labelings were done as previously described (1) and are summarized in the SI Materials and Methods.
Quantitative Image Analysis of Protein Polymerization.
Quantification of F-actin, phosphotyrosine, Lck, and Cdc42 polarization at the immune synapse was based on our previously described method (1) by random selection of at least 50 conjugate images containing a CellTracker Blue-stained healthy or CLL B cell contacting a T cell. Polarization of proteins at the T-cell contact site was scored independently. These findings were verified using ImageJ software to calculate the relative recruitment index of proteins to the T cell-B cell contact site.
Lenalidomide Treatment.
CLL B cells and autologous T cells were treated separately ex vivo with 0.5 μM lenalidomide in acidic PBS for 24 h in full culture medium (10% serum) at 37 °C before cell-conjugation assays. Lenalidomide was prepared as previously described (1). Untreated autologous cells were also cultured separately using the acidic PBS alone without drug for 24 h in full culture medium at 37 °C before conjugation assays.
Statistics.
Statistical differences between experimental groups were evaluated by 2-tailed Student's t test. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments.
We thank Peter Varney for editorial assistance, Dr. Edward Alvin Fox and the Staff of the Dana-Farber Cancer Institute Microarray Core Facility. This work was supported by the CLL Research Consortium Grant PO1 CA 81538 from the National Cancer Institute (to J.G.G. and C.M.C.) and by grants from the CLL Global Research Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE8836).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901166106/DCSupplemental.
References
- 1.Ramsay AG, et al. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Invest. 2008;118:2427–2437. doi: 10.1172/JCI35017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyer M, et al. Reduced frequencies and suppressive function of CD4+CD25hi regulatory T cells in patients with chronic lymphocytic leukemia after therapy with fludarabine. Blood. 2005;106:2018–2025. doi: 10.1182/blood-2005-02-0642. [DOI] [PubMed] [Google Scholar]
- 3.Kater AP, van Oers MH, Kipps TJ. Cellular immune therapy for chronic lymphocytic leukemia. Blood. 2007;110:2811–2818. doi: 10.1182/blood-2007-01-068932. [DOI] [PubMed] [Google Scholar]
- 4.Gorgun G, et al. Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells. J Clin Invest. 2005;115:1797–1805. doi: 10.1172/JCI24176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krackhardt AM, et al. T-cell responses against chronic lymphocytic leukemia cells: implications for immunotherapy. Blood. 2002;100:167–173. doi: 10.1182/blood.v100.1.167. [DOI] [PubMed] [Google Scholar]
- 6.Bichi R, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA. 2002;99:6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan XJ, et al. B cell receptors in TCL1 transgenic mice resemble those of aggressive, treatment-resistant human chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2006;103:11713–11718. doi: 10.1073/pnas.0604564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai J, et al. RESOURCERER: a database for annotating and linking microarray resources within and across species. Genome Biol. 2001;2(11):SOFTWARE0002. doi: 10.1186/gb-2001-2-11-software0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, et al. Cross-referencing eukaryotic genomes: TIGR Orthologous Gene Alignments (TOGA) Genome Res. 2002;12:493–502. doi: 10.1101/gr.212002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Billadeau DD, Nolz JC, Gomez TS. Regulation of T-cell activation by the cytoskeleton. Nat Rev Immunol. 2007;7:131–143. doi: 10.1038/nri2021. [DOI] [PubMed] [Google Scholar]
- 11.Kane LP, et al. Akt provides the CD28 costimulatory signal for up-regulation of IL-2 and IFN-gamma but not TH2 cytokines. Nat Immunol. 2001;2:37–44. doi: 10.1038/83144. [DOI] [PubMed] [Google Scholar]
- 12.Coyle AJ, Gutierrez-Ramos JC. The expanding B7 superfamily: increasing complexity in costimulatory signals regulating T cell function. Nat Immunol. 2001;2:203–209. doi: 10.1038/85251. [DOI] [PubMed] [Google Scholar]
- 13.Holdorf AD, et al. Regulation of Lck activity by CD4 and CD28 in the immunological synapse. Nat Immunol. 2002;3:259–264. doi: 10.1038/ni761. [DOI] [PubMed] [Google Scholar]
- 14.Faccio R, et al. Vav3 regulates osteoclast function and bone mass. Nat Med. 2005;11:284–290. doi: 10.1038/nm1194. [DOI] [PubMed] [Google Scholar]
- 15.Wulfing C, et al. Costimulation and endogenous MHC ligands contribute to T cell recognition. Nat Immunol. 2002;3:42–47. doi: 10.1038/ni741. [DOI] [PubMed] [Google Scholar]
- 16.Buhmann R, et al. CD40-activated B-cell chronic lymphocytic leukemia cells for tumor immunotherapy: stimulation of allogeneic versus autologous T cells generates different types of effector cells. Blood. 1999;93:1992–2002. [PubMed] [Google Scholar]
- 17.Chanan-Khan A, et al. Clinical efficacy of lenalidomide in patients with relapsed or refractory chronic lymphocytic leukemia: results of a phase II study. J Clin Oncol. 2006;24:5343–5349. doi: 10.1200/JCO.2005.05.0401. [DOI] [PubMed] [Google Scholar]
- 18.Ferrajoli A, et al. Lenalidomide induces complete and partial remissions in patients with relapsed and refractory chronic lymphocytic leukemia. Blood. 2008;111:5291–5297. doi: 10.1182/blood-2007-12-130120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andritsos LA, et al. Higher doses of lenalidomide are associated with unacceptable toxicity including life-threatening tumor flare in patients with chronic lymphocytic leukemia. J Clin Oncol. 2008;26:2519–2525. doi: 10.1200/JCO.2007.13.9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson AJ, et al. Characterization of the TCL-1 transgenic mouse as a preclinical drug development tool for human chronic lymphocytic leukemia. Blood. 2006;108:1334–1338. doi: 10.1182/blood-2005-12-011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mutter GL, et al. Global expression changes of constitutive and hormonally regulated genes during endometrial neoplastic transformation. Gynecol Oncol. 2001;83:177–185. doi: 10.1006/gyno.2001.6352. [DOI] [PubMed] [Google Scholar]
- 23.Doniger SW, et al. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;R7(1):1–12. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trojan A, et al. Immunoglobulin framework-derived peptides function as cytotoxic T-cell epitopes commonly expressed in B-cell malignancies. Nat Med. 2000;6:667–672. doi: 10.1038/76243. [DOI] [PubMed] [Google Scholar]
- 25.Aichele P, et al. T cell priming versus T cell tolerance induced by synthetic peptides. J Exp Med. 1995;182:261–266. doi: 10.1084/jem.182.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pircher H, et al. Viral escape by selection of cytotoxic T cell-resistant virus variants in vivo. Nature. 1990;346:629–633. doi: 10.1038/346629a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.