Abstract
Phenotypic mutations (errors occurring during protein synthesis) are orders of magnitude more frequent than genetic mutations. Consequently, the sequences of individual protein molecules transcribed and translated from the same gene can differ. To test the effects of such mutations, we established a bacterial system in which an antibiotic resistance gene (TEM-1 β-lactamase) was transcribed by either a high-fidelity RNA polymerase or its error-prone mutant. This setup enabled the analysis of individual mRNA transcripts that were synthesized under normal or error-prone conditions. We found that an increase of ≈20-fold in the frequency of transcription errors promoted the evolution of higher TEM-1 expression levels and of more stable enzyme variants. The stabilized variants exhibited a distinct advantage under error-prone transcription, although under normal transcription they conferred resistance similar to wild-type TEM-1. They did so, primarily, by increasing TEM-1's tolerance to destabilizing deleterious mutations that arise from transcriptional errors. The stabilized TEM-1 variants also showed increased tolerance to genetic mutations. Thus, although phenotypic mutations are not individually subjected to inheritance and natural selection, as are genetic mutations, they collectively exert a direct and immediate effect on protein fitness. They may therefore play a role in shaping protein traits such as expression levels, stability, and tolerance to genetic mutations.
Keywords: protein evolution, protein stability, transcriptional errors, mutational robustness
A wealth of error-prevention and correction mechanisms have evolved to lower the mutation rates of DNA, resulting in rates of genomic mutations that are in the order of 5 × 10−10 nucleotide exchanges per position per generation (1). In contrast, the mutation rates of mRNA transcription and translation, leading to protein synthesis, are ≈5 and 6 orders of magnitude higher respectively (2–5). Because the average length of proteins in prokaryotes is ≈350 aa (6), synthesis errors would introduce, on average, a phenotypic mutation in 1 of 10 protein copies. For the largest Escherichia coli ORF (7,200 bases) errors might be found in more than half of the copies (7). In general, the generation of multiple mRNA copies from the same gene ensures a sufficient amount of wild-type mRNA and protein molecules. This may explain why the rates of phenotypic mutations have remained relatively high (8). However, in cases where the number of protein copies in the cell is very low, phenotypic mutations may significantly affect fitness (8). Indeed, >80% of E. coli genes express fewer than a hundred protein copies, and many are produced at ≤20 copies per cell (9). At high expression rates, the fraction of mutated and misfolded proteins can lead to accelerated aggregation and reduced fitness (10–12). Thus, abnormal proteins resulting from phenotypic mutations can burden cells by reducing the amount of functional protein, wasting metabolic resources, engaging folding and degradation pathways, and creating toxic aggregates. Indeed, under specific circumstances, phenotypic mutations have been found to promote adverse physiological conditions such as senescence (13, 14), neurodegeneration (15, 16), and cancer (17).
The responses of organisms, and their protein components, to genetic mutations have been the subject of numerous studies. However, the evolutionary implications of phenotypic mutations have been mostly explored by theoretical means (5, 8, 11, 18). Here, we examined the effects of phenotypic mutations in an experimental model system based on the expression of a plasmid-encoded ampicillin degrading enzyme, TEM-1 β-lactamase. TEM-1 confers resistance to ampicillin on bacterial cells, and changes in the expression levels and activity of the enzyme have a direct and quantifiable effect on the survival of E. coli in the presence of different concentrations of ampicillin. Using a T7 DNA-dependent RNA polymerase (T7 RNAP), and an error-prone mutant thereof (19), we studied the responses to an increased rate of phenotypic mutations. Reverse transcription of TEM-1 transcripts, and cloning of the resulting cDNAs, enabled the analysis of individual transcript molecules that stemmed from expression of the same gene. In this way, the sequence variations caused by transcription errors, and their effects on TEM-1's stability and function, could be analyzed. We could thus ask the question: Does an increased load of phenotypic mutations exert a selection pressure, and if so, how would TEM-1 evolve to address this pressure?
Results
Error-Prone Transcription and Reduced Ampicillin Resistance.
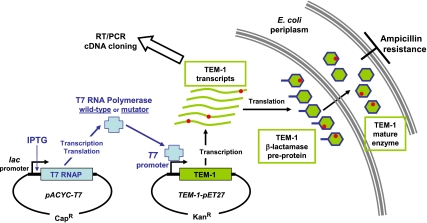
The TEM-1 gene was placed under a T7 promoter using a vector dubbed TEM-1-pET27, so that its expression, and ampicillin resistance, became dependent on the expression of a T7 RNAP (Fig. 1). The T7 RNAP, which is not an endogenous E. coli enzyme, was expressed under the lac promoter using the pACYC-T7 vector (Scheme S1d). We also used a previously described error-prone mutant, mutator-T7 RNAP, that increases the rate of transcriptional errors by 20-fold (19).
Fig. 1.
A schematic description of the experimental system. TEM-1 β-lactamase was transcribed from plasmid TEM-1-pET27 under a T7 promoter. To enable the transcription of TEM-1, wild-type T7 RNA polymerase, or a mutant that exhibits ≈20-fold higher rate of transcriptional errors (mutator T7 RNAP) were expressed from plasmid pACYC-T7. TEM-1 transcripts carrying mutations resulted in mutated enzyme molecules, and reduced levels of ampicillin resistance. The reproduction of single transcripts and their encoded TEM-1 molecules was achieved by reverse transcription and PCR amplification (RT/PCR) to obtain cDNAs that were subsequently cloned and individually analyzed.
We first examined the differences in ampicillin resistance of cells expressing TEM-1 by either wild-type, or the mutator, T7-RNAPs. For each T7-RNAP we randomly picked 50 genetically identical colonies harboring both the TEM-1-pET27 plasmid and the T7 RNAP expression plasmid. These were grown to mid-log phase and plated on a series of agar plates containing increasing concentrations of ampicillin. The fraction of surviving colonies at each ampicillin concentration was determined (Fig. S1). The mid inhibitory concentration (IC50)—i.e., the concentration of ampicillin under which 50% of the colonies survived, was found to be ≈2,700 μg/mL for wild-type T7 RNAP, and only 1,200 μg/mL for the mutator-T7-RNAP. Approximately 2/3 of this decrease in colony survival was attributed to the reduction in transcription efficiency by mutator-T7-RNAP (19) leading to a reduction in TEM-1 levels. The remaining third related to the incorporation of deleterious mutations into TEM-1 mRNAs during error-prone transcription, as the following comparison of TEM-1 cDNAs generated from both types of cells—those containing wild-type T7 RNAP, and those containing the mutator RNAP—had indicated.
Total RNA was isolated from cultures of both cell types, followed by reverse transcription of mRNA molecules using a TEM-1 specific primer. The resulting cDNAs were PCR amplified and cloned into the pET27 vector that was previously used for the expression of wild-type TEM-1. Representative plasmids from the resulting cDNA libraries were sequenced, expressed, and tested for their ability to confer ampicillin resistance relative to wild-type TEM-1.
The observed mutation frequency in all cDNA libraries included contributions from reverse transcription and cDNA amplification errors. Upon reduction of these contributions from the mutation rate in cDNAs derived from wild-type T7 RNAP (0.29 × 10−3) (Table 1) we calculated that its “net” mutation rate was 0.06 × 10−3 as originally reported in refs. 19 and 20. This rate is similar to that of native E.coli RNAP (≈0.01 × 10−3) (2, 4).
Table 1.
Mutation frequencies in cDNAs libraries
| T7 RNA polymerase | Transcribed TEM-1 gene | Number of base-pairs sequenced | Observed mutation frequency* × 10−3 | Estimated net mutation frequency† × 10−3 |
|---|---|---|---|---|
| Wild-type | Wild-type | 24,276 | 0.29 ± 0.05 | 0.06 ± 0.05 |
| Mutator | Wild-type | 24,276 | 1.7 ± 0.13 | 1.47 ± 0.13 |
| Mutator | Met182Thr-Glu147Lys | 13,872 | 1.6 ± 0.26 | 1.43 ± 0.26 |
| Mutator‡ | Met182Thr-Arg120Gly | 17,340 | 1.04 ± 0.2 c | 0.81 ± 0.2‡ |
*The total number of mutations identified in cDNA clones divided by the number of base-pairs sequenced. The errors were estimated from the comparison of the mutation frequencies in two independently prepared cDNA libraries.
†The estimated ″net″ mutation frequency was obtained by removing the expected contributions of reverse transcription (3 × 10−5) (44), and PCR amplification (≈2 × 10−4) (45), from the observed mutation frequency.
‡The observed mutation frequency in this cDNA library was lower than in the two other libraries obtained from error-prone transcription, probably due to residual amouns of plasmid DNA that were not destroyed by the DNase treatment, and contaminated the PCR amplification.
The observed mutation frequency in cDNAs derived from the mutator-T7-RNAP was ≈6-fold higher. The net mutation rate due to error-prone transcription (after subtracting the expected contributions of reverse transcription and cDNA amplification) was therefore ≈1.4 × 10−3, which is 24-fold higher than wild-type T7 RNAP. These values are in agreement with the original report of mutator-T7-RNAP (1.25 × 10−3) (19). The mutator's error frequency resulted in, on average, 1.3 mutations per TEM-1 transcript, as opposed to ≈0.05 for wild-type T7 RNAP. This rate is 10-fold greater than the estimated normal rate of protein synthesis errors in E.coli (2–4, 21), which normally results from both transcription and translation errors. Notably, despite the experimental noise of mutations related to the production of cDNAs, in transcripts generated by the mutator T7 RNAP, the majority of mutations [(1.47/1.7) × 10−3 = 85%] resulted from error prone transcription and not from the process of generating the cDNAs.
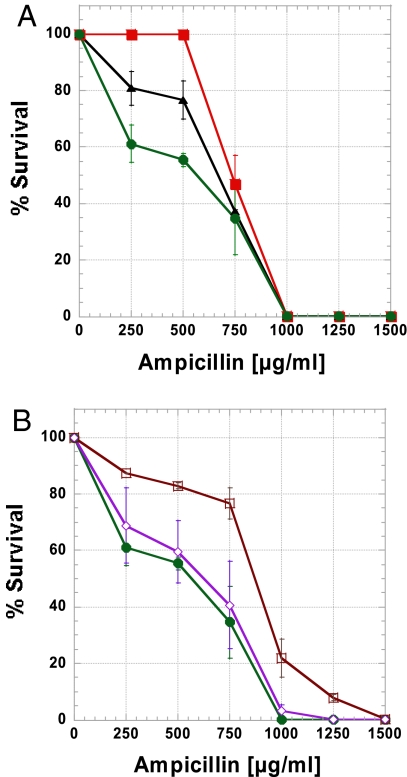
A marked difference in the percentage of surviving cells at various ampicillin concentrations was observed upon transformation of the different cDNA libraries (Fig. 2A). Because of errors incorporated during their production, TEM-1 cDNAs derived from wild-type T7 RNAP transcription were compromised by ≈20% relative to the control. However, TEM-1 cDNAs derived from mutator-T7 RNAP transcription were compromised by ≈40%. This difference in survival was the outcome of the ≈6-fold higher mutation rate observed in the cDNA library from mutator-T7-RNAP (Table 1).
Fig. 2.
Ampicillin resistance of cells expressing TEM-1 cDNA libraries. From each library, 80 randomly chosen cDNA clones were assayed for their ability to confer ampicillin resistance. Each line represents an average of 2 independently generated and assayed cDNA libraries. (A) A comparison of wild-type TEM-1 gene as control (not a cDNA library) (red squares) and cDNA libraries generated from the same TEM-1 gene using either wild-type T7 RNAP (black triangles) or mutator-T7 RNAP (green circles). (B) A comparison of cDNA libraries of wild-type TEM-1 (green circles) and cDNAs of evolved TEM-1 variants R120G-M182T (brown squares) and E147K-M182T (purple diamonds) all transcribed by the mutator-T7 RNAP.
Deleterious Nature of Transcriptional Errors.
To unravel the link between reduced ampicillin resistance and the incorporation of transcriptional errors, we sequenced 40 cDNA clones derived from transcription of wild-type TEM-1 by the mutator-T7-RNAP. As shown in Table 2, clones that exhibited severely compromised ampicillin resistance levels, carried deleterious mutations of various sorts: nonsense mutations (frameshifts due to single nucleotide insertions and deletions, and stop codons, comprising 25% of the mutations) and missense mutations including mutations in TEM-1's active-site, and in its scaffold. The severity of the latter was estimated by predicting their destabilizing effects (ΔΔG values) using FoldX (22, 23). Indeed, these mutations were predicted to be more destabilizing (average ΔΔG value ≈ 2.3 kcal/mol) than the average of all possible single nucleotide mutations in TEM-1 (1.4 kcal/mol) (23). Thus, the loss of ampicillin resistance in cDNA clones of TEM-1 was related to the incorporation of deleterious mutations, often by compromising protein stability. As expected, clones from the same cDNA library that exhibited wild-type-like resistance carried fewer mutations overall (44% carried no nonsynonymous mutations), did not incorporate nonsense mutations, and exhibited lower predicted ΔΔG values (≈1.5 kcal/mol, on average; Table S1).
Table 2.
Mutational analysis of cDNA clones exhibiting low, or no, ampicillin resistance (IC50 ≤ 500 μg/ml)
| cDNA library | wt-TEM1 | E147K + M182T | R120G + M182T | |
|---|---|---|---|---|
| Average number of mutations per clone* | 1.4 ± 0.5 | 2.2 ± 0.8 | 1.3 ± 0.5 | |
| Nonsense mutations | % clones carrying deletions† | 8.3% | 19.2% | 17.4% |
| % clones carrying insertions† | 8.3% | 23.1% | 17.4% | |
| % clones carrying stop codons | 8.3% | 15.4% | 13% | |
| Total % clones carrying nonsense mutations | 25% | 57.7% | 47.8% | |
| Missense mutations | % clones carrying active site mutations | 8.3% | 7.7% | 13% |
| % clones carrying only destabilizing mutations | 66.7% | 34.6% | 39.2% | |
| Average predicted ΔΔG of missense mutations per clone, kcal/mol | 2.33 ± 1.78 | 3.6 ± 2.3 | 3.53 ± 2.61 | |
*Mutations identified in clones sequenced from each library (23–26 clones per library) see also Table S5.
†All contain single base deletions or insertions that were found to cause frameshifts.
Selection for Increased Ampicillin Resistance Under Error-Prone Transcription.
Next, we attempted to evolve TEM-1 to regain ampicillin resistance under error-prone transcription by cloning a random library of TEM-1 mutants into pET27 and selecting for increased ampicillin resistance in cells that express mutator-T7 RNAP. The library, taken from a genetic drift described in ref. 24, was generated by error-prone PCR of wild-type TEM-1. It was subjected to a purifying selection to purge inactive TEM-1 variants by plating on low levels of ampicillin, and contained, on average, 2.2 ± 1.5 nonsynonymous mutations per gene. Initially, cells transformed with this library (≥106) were plated on agar plates containing 250 μg/mL ampicillin. Surviving colonies were pooled and replated on selection plates. This protocol was repeated while increasing the ampicillin concentration by 50 μg/mL for each round of selection. After 4 rounds of selection, plasmids of surviving variants were subjected to another round of mutagenesis by error-prone PCR, recloned, transformed and reselected on increasing levels of ampicillin. The cycle of 4 selection rounds and mutagenesis was repeated 3 times to increase the diversity of selected clones. Random samples of surviving clones were sequenced and their levels of ampicillin resistance measured (SI Materials and Methods).
Elevated Resistance by Increased TEM-1 Doses.
The first isolated variants that had evolved improved ampicillin resistance showed enhanced expression, or export, of TEM-1. The fixed mutations occurred in the N-terminal signal-peptide of TEM-1 that directs its export to the periplasm and is subsequently cleaved to generate the mature enzyme. Variants showing high levels of ampicillin resistance carried either the Q6R, or the H7R, signal peptide mutations. These mutations are likely to increase antibiotic resistance by rendering the signal peptide more basic, and facilitating export by increasing the affinity of the protein to the negatively charged lipid membrane (25). A test of active TEM-1 levels using a chromogenic substrate and Western blots confirmed that the increased levels of ampicillin resistance of these mutants correlate with a 2-fold increase in the periplasmic levels of active TEM-1 (Fig. 3 and Fig. S2c).
Fig. 3.
Active enzyme levels of wild type TEM-1 and evolved variants. Wild-type TEM-1 β-lactamase and its evolved variants harboring either signal peptide mutations (Q6R, H7R) or promoter deletion mutations (ΔC-117, ΔG-123) were expressed in cells using either wild-type T7 RNAP (black bars), or mutator T7 RNAP (gray bars). After IPTG induction, the cells were incubated with nitrocefin (a synthetic, chromogenic, β-lactamase substrate) and the initial rates of nitrocefin breakdown, proportional to the periplasmic levels of active β-lactamase were monitored. TEM-1 levels were confirmed by a Western blot analysis (Fig. S2c).
Two other mutations were observed in selected variants with enhanced resistance. These 2 single base deletions (del C -117 and G -123) occurred in the lacO operator sequence downstream to the T7 promoter that controls TEM-1's expression. They increased ampicillin resistance levels under mutator-T7-RNAP transcription to 3,000 (μg/mL) by alleviating the lacI-mediated repression of transcription. Although the promoter region was not mutated during library making, these mutations rose spontaneously and were subsequently selected. Their role was confirmed by recloning wild-type TEM-1 into the mutated plasmids, and validating the increased resistance levels. As expected, the enzyme levels of these promoter mutants were 2.5-fold higher than those produced by the normal promoter (Fig. 3 and Fig. S2c).
Both types of mutations, signal peptide and promoter, were particularly beneficial under our selection and increased ampicillin resistance levels by ≥2.5-fold (Fig. S2). However, their emergence is most likely related to the low processivity of mutator-T7-RNAP (19), which leads to reduced expression levels, because they provided a similar advantage under high-fidelity transcription (Fig. 3). Nevertheless, their elevated expression levels were also compensating for the increased fraction of inactive enzyme molecules carrying phenotypic mutations. In fact, similar mutations in TEM-1's signal peptide (26) and promoter region (27) were found in clinically isolated variants with extended spectrum β-lactamase activities.
Elevated Resistance by Mutations in the Mature Protein.
To explore additional solutions to the deleterious effects of transcriptional errors, we amplified the TEM-1 variants library with a primer that restored the sequence of the wild-type's signal peptide, thus restricting mutations to the mature enzyme. The reamplified library was cloned to pET27 and selected for increased levels of ampicillin resistance under error-prone transcription as described above. We subsequently analyzed and sequenced a total of 106 variants isolated from various rounds of selection (Table S2).
On average, the analyzed variants contained 5 ± 2.5 mutations per clone, of which 53% were nonsynonymous, and exhibited under error-prone transcription significantly higher ampicillin resistance levels than wild-type TEM-1. The 3 most frequent nonsynonymous mutations were M182T (in 22% of all analyzed clones), R120G, and E147K (15%, and 13%, of the clones, respectively) (Table S3a). M182T and R120G were present in various mutational backgrounds, but E147K was found exclusively in combination with M182T (Table S3c). These 2 mutations were the most frequent pair of nonsynonymous mutations, found in 13% of the selected variants. The combination of M182T and R120G was the second most prevalent combination (7.5%).
We then analyzed the individual and pairwise contributions of these 3 mutations to resistance. The corresponding TEM-1 mutants were engineered by site-directed mutagenesis and their ampicillin resistance measured. Apart from the single E147K mutation, all mutants tested exhibited improved resistance values relative to wild-type TEM-1. The double-mutants (E147K-M182T and R120G-M182T) conferred ampicillin resistance levels that were higher than M182T on its own (Fig. 4A). Thus, E147K was only advantageous at the background of M182T, explaining why this mutation never appeared on its own. Notably, these mutants exhibited comparable resistance levels to wild-type TEM-1 (or even lower in the case of E147K) when transcribed by wild-type T7 RNAP (Fig. 4B). Thus, the selected mutations confer an advantage only under high rates of phenotypic mutations.
Fig. 4.
Ampicillin resistance of cells expressing TEM-1 variants. For each TEM-1 variant, 80 randomly chosen clones were assayed for their ability to confer resistance at different ampicillin concentrations. Each line represents the average for 2 independent measurements. (A) Wild-type TEM-1, and its double mutants E147K-M182T and R120G-M182T, transcribed by mutator-T7 RNAP. Single TEM-1 mutants: E147K, R120G and M182T similarly transcribed, are shown for comparison. (B) The same genes transcribed by wild-type T7 RNAP.
We also tested 2 representative synonymous mutations that occurred at high frequency in the selected variants: R259R (CGT to CGG; 15.1%) and F151F (TTT to TTC; 10.4%) (Table S3b). These mutations contributed subtly but reproducibly to resistance under error-prone transcription, on their own, and together with the stabilizing mutation M182T (Fig. S3). Their accumulation may be due to a beneficial effect on translational fidelity, similar to the selection of synonymous mutations that increase translational fidelity (11).
Evolved Variants Exhibit Increase Stability.
To analyze the contributions of the mutations identified here (i.e., E147K, R120G, M182T) we determined the stability and enzymatic parameters of the purified protein mutants (Table S4). The kinetic parameters of these mutants were very similar to those of wild-type TEM-1. However, the observed increases in thermodynamic stability of these mutants (ΔTm values) were largely in agreement with their increased ampicillin resistance (e.g. ΔTm for M182T was 5 °C, and for R120G-M182T, 7.3 °C). The double-mutant E147K-M182T, comprised an exception whereby in contrast to its higher resistance level relative to M182T (Fig. 4A), its Tm value was slightly lower (3.7 °C).
It appears that the increased fitness of the variants evolved under error-prone transcription is related to their increased stability. Two of the 3 mutations (M182T and R120G) were shown to increase TEM-1 stability (both thermodynamic and kinetic stability (28–30)), and the third one (E147K) seems related to a stabilizing mutation in the same position (E147G) (29, 31). The first 2 mutations were also shown to act as global suppressors toward a wide range of destabilizing genetic mutations in TEM-1 (29).
Evolved Variants Buffer Deleterious Transcription Mutations.
Bacteria-expressing cDNAs, derived under error-prone transcription from cells that expressed the double-mutants (E147K-M182T and R120G-M182T), showed increased levels of ampicillin resistance compared with bacteria containing cDNAs from wild-type TEM-1 derived under the same conditions (Fig. 2B). An examination of cDNA sequences revealed that highly destabilizing mutations were much more frequent in cDNAs derived from the double-mutants than in cDNAs from wild-type TEM-1. Specifically, the cDNAs of double-mutants showing severely compromised resistance (including cDNAs showing no resistance) (Table 2) carried a higher fraction of nonsense mutations (≥48%) than wild-type cDNAs (≈25%). The average ΔΔG values of mutations (as predicted by FoldX) were up to 1.3 kcal/mol more destabilizing in cDNAs derived from the double-mutants than those observed in cDNAs derived from wild-type TEM-1 (2.3 kcal/mol vs. 3.6 and 3.5 kcal/mol) (Table 2). The cDNAs from both wild-type and double-mutants that enabled bacterial survival at high ampicillin concentrations exhibited less differences in the severity of deleterious mutations because at these resistance levels, tolerance to mutations was very low (Table S1).
Discussion
Similar to certain pathological manifestations (15, 32–34), our experimental system introduced phenotypic mutations at a rate much higher than normal. The normal rate of phenotypic mutations probably results from a balance between the fitness cost imposed by their deleterious effects, and the cost of decreasing their incidence. The latter is considerable—increasing translational fidelity by ribosomes, for example, resulted in slower growth rates (35). Loss of protein function due to phenotypic mutations sets an upper limit to their frequency, but there might be little pressure to reduce their occurrence beyond the current frequency (8). Nonetheless, our results suggest that phenotypic mutations impose a considerable fitness cost and that they may have had a role in the evolution of certain traits. However, our experimental system and the theoretical model (8), were both limited to the effects of phenotypic mutations at the protein level (loss of function due to deleterious mutations), and did not address possible effects on the organism such as the toxicity of aggregates (10, 11, 15).
Our experiment, using TEM-1 as a model protein, indicated an immediate effect of phenotypic mutations on protein dose and stability. Both effects were quantified based on the characteristics of the variants evolved toward higher fitness under error-prone transcription. Increasing the rate of transcription mutations by a factor of 24, to a rate of ≈1 mutation per copy, resulted in the evolution of TEM-1 variants exhibiting 2-fold higher expression levels and variants with the ability to buffer destabilizing mutations with ΔΔG values that are ≥1.3 kcal/mol higher than wild-type TEM-1 (Table 2). Although higher expression levels may have evolved partly in compensation for the low processivity of mutator-T7 RNAP, which leads to lower expression levels, the evolved tolerance to destabilizing mutations is consistent (or perhaps even an underestimation) with the stabilizing contribution of M182T (2.7 kcal/mol) (28), and the additional stabilizing mutations seen here. Moreover, stabilizing mutations conferred an advantage only under error-prone transcription. Hence they have evolved specifically in response to the introduction of transcription mutations by mutator-T7 RNAP, and not in response to its reduced transcription levels. Increased expression and stability can have an additive effect under error-prone transcription as can be seen by comparing the separate and combined gains in ampicillin resistance levels they produced (Figs. S2a and S4a). However, although increased expression provides an immediate solution, it imposes a fitness cost due to the cost of expression (36), and the potential deleterious effects of aggregates that become considerably more adverse at high expression levels (11, 37). Thus, an increase in protein stability may better serve as a long-term solution for phenotypic mutations because it does not carry with it the penalties of increased expression levels. Phenotypic mutations originating from transcription errors differ from those that originate in translation errors by being more random. Translation errors are highly codon biased (21) and can promote a selection for high-fidelity codons (11). However, they are ≈10-fold more frequent than transcription errors (3), perhaps indicating the limits of codon optimization for error-reduction. An increase in protein stability may thus provide a general strategy for dealing with errors that originate from both stages of protein synthesis.
There are fundamental differences between the selections for robustness against genetic and phenotypic mutations. The increase in tolerance to phenotypic mutations is classified as environmental robustness—namely as robustness against nonheritable perturbations such as temperature, salinity, developmental noise or transcription errors (38, 39). However, heritability is essential for the evolution of buffering mechanisms that endow cells with robustness against such perturbations. There is, therefore, a major difference in the benefit for cells that inherit mutational buffers such as protein stabilizing mutations. Although such mutations present an immediate advantage to the cell by buffering phenotypic mutations, they only present a potential future advantage to the cell's progeny by subsequently buffering genomic mutations. Thus, the acquisition of robustness against phenotypic mutations constitutes a form of “first order adaptation,” which might be observed in a wide range of organisms (11). The acquisition of robustness against genetic mutations is a “second-order adaptation” that may only be dominant under very high genetic mutation rates (38, 39). Indeed, we have previously subjected TEM-1 to a greatly increased rate of genetic mutations (29). The emergence of R120G and M182T under an intense load of genetic mutations was a more subtle phenomenon. After 18 rounds of mutation and selection, these mutations were enriched in the population, but not fixed (29). Unlike the variants isolated from the selections for phenotypic robustness described here, variants selected under an increased load of genetic mutations also carried a load of deleterious mutations such that, on average, their overall stability was lower than wild-type TEM-1 (29). In contrast, under an increased rate of phenotypic mutations, both R120G and M182T were enriched within 4–5 rounds and fixed by the 8th round (Table S3). Our results, therefore, suggest that the emergence of mutational robustness in natural proteins could be the result of the direct selection pressure imposed by phenotypic mutations rather than a second-order adaptation to genetic mutations (38).
As previously mentioned, some of the stabilizing mutations that evolved in response to protein synthesis errors and genetic mutations were similar. Two of the 3 most prevalent mutations that were selected under error-prone transcription (R120G and M182T) were shown to act as global suppressors and increase TEM-1's tolerance to genetic mutations (29, 40, 41). Interestingly, both suppressors also bring TEM-1's sequence closer to its predicted ancestor and family consensus (29). In contrast, the selected mutation E147K, which provides increased fitness under error-prone transcription only when combined with M182T (Fig. 4A), and the selected synonymous mutations, do not correlate with increased thermodynamic stability. It is possible that E147K contributes to TEM-1's kinetic stability, as observed for other mutations (29), and that the synonymous mutations help reduce translation errors (11). Thus, the requirements for TEM-1's tolerance to genetic and phenotypic mutations appear to be overlapping, but not necessarily identical.
The mutational robustness of proteins can be described as a margin of stability beyond what is required to maintain the protein's structure and function (39, 41). This margin endows proteins with the ability to temporarily buffer the deleterious effects of destabilizing mutations, and thus evolve faster (24, 41). Likewise, the expression of proteins can often reach levels that are higher than needed for survival (39, 42), and this excess can buffer various environmental and genetic perturbations. Among other factors, these extra margins of stability and expression may have resulted from the need to address phenotypic rather than genetic mutations.
Materials and Methods
A detailed description of materials and methods is available in SI Materials and Methods.
IC50 Values.
Freshly transformed colonies were picked from agar plates into 96-deep-well plates and grown overnight in liquid medium. These cultures were diluted into a new set of 96-deep-well plates, grown to mid-log phase, rediluted, and replica plated using a 96-pin replicator on a set of agar plates containing varying concentrations of ampicillin (0–3,000 μg/mL). The surviving colonies were counted after overnight growth. IC50 was defined as the concentration of ampicillin under which 50% of the colonies carrying a given TEM-1 variant and T7 RNAP, survived.
Directed Evolution.
The TEM-1 library was taken from a genetic drift described in ref. 24. Specifically, we used the 5th round of Lib250, that was subjected to purifying selection on 250 μg/mL ampicillin. The library was recloned into pET-27, transformed to JM109 cells containing the mutator-pACYC-T7 plasmid (≥106 transformants), and plated on agar plates. Overnight colonies were collected, pooled, and grown to mid-log phase in liquid medium. Cultures were diluted (1:100) and plated on agar plates with ampicillin (250 μg/mL). Surviving colonies were pooled, diluted and grown for the next round. Ampicillin concentrations were increased by 50 μg/mL for each round of selection. After 4 selection rounds (i.e., 1 selection cycle) plasmids of surviving variants were extracted and their TEM-1 genes were mutagenized using error-prone PCR. Overall, a total of 3 selection cycles, resulting in 12 selection rounds and 2 mutagenesis steps, were performed.
Engineered Mutants.
TEM-1 mutants were constructed by all-around PCR on TEM-1-pET27. Their sequence was verified, and IC50 values were determined. Purified proteins were obtained after recloning each TEM-1 variant to pET24 containing an ompA signal peptide region, expression, and purification of the secreted protein (29, 43). Apparent midpoint temperatures of melting (Tm) and kinetic parameters were determined as described in ref. 29.
β-lactamase Levels by Nitrocefin.
Individual overnight colonies expressing either wild-type TEM-1, or its variants, were picked into 96-deep-well plates, grown as in IC50 assays, and induced by IPTG. Cultures were diluted in potassium phosphate buffer pH 7.25 inside ELISA plates, and nitrocefin (50 μg/mL) was added. Product release was measured as absorption in 486 nm, and enzyme levels were determined by comparing the linear phases of nitrocefin hydrolysis (v0).
Supplementary Material
Acknowledgments.
We thank Dan Hartl for an inspirational discussion of the possible role of phenotypic mutations; Allan Drummond and the reviewers of this article for their insightful comments; Joelle Pelletier (Université de Montréal, Montréal) for the TEM-1 expression plasmid; W.T. McAllister (SUNY Downstate Medical Center, Brooklyn, NY) for the T7 expression plasmid; and Shimon Bershtein, Nobuhiko Tokuriki, and Shalev Itzkovitz for their invaluable support. This work was supported by the European Union MiFEM network and the Wolgin prize.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809506106/DCSupplemental.
References
- 1.Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blank A, Gallant JA, Burgess RR, Loeb LA. An RNA polymerase mutant with reduced accuracy of chain elongation. Biochemistry. 1986;25:5920–5928. doi: 10.1021/bi00368a013. [DOI] [PubMed] [Google Scholar]
- 3.Ellis N, Gallant J. An estimate of the global error frequency in translation. Mol Gen Genet. 1982;188:169–172. doi: 10.1007/BF00332670. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberger RF, Hilton J. The frequency of transcriptional and translational errors at nonsense codons in the lacZ gene of Escherichia coli. Mol Gen Genet. 1983;191:207–212. doi: 10.1007/BF00334815. [DOI] [PubMed] [Google Scholar]
- 5.Willensdorfer M, Burger R, Nowak MA. Phenotypic mutation rates and the abundance of abnormal proteins in yeast. PLoS Comput Biol. 2007;3:e203. doi: 10.1371/journal.pcbi.0030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skovgaard M, Jensen LJ, Brunak S, Ussery D, Krogh A. On the total number of genes and their length distribution in complete microbial genomes. Trends Genet. 2001;17:425–428. doi: 10.1016/s0168-9525(01)02372-1. [DOI] [PubMed] [Google Scholar]
- 7.Poole AM, Logan DT. Modern mRNA proofreading and repair: Clues that the last universal common ancestor possessed an RNA genome? Mol Biol Evol. 2005;22:1444–1455. doi: 10.1093/molbev/msi132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burger R, Willensdorfer M, Nowak MA. Why are phenotypic mutation rates much higher than genotypic mutation rates? Genetics. 2006;172:197–206. doi: 10.1534/genetics.105.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guptasarma P. Does replication-induced transcription regulate synthesis of the myriad low copy number proteins of Escherichia coli? Bioessays. 1995;17:987–997. doi: 10.1002/bies.950171112. [DOI] [PubMed] [Google Scholar]
- 10.Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci USA. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tartaglia GG, Pechmann S, Dobson CM, Vendruscolo M. Life on the edge: A link between gene expression levels and aggregation rates of human proteins. Trends Biochem Sci. 2007;32:204–206. doi: 10.1016/j.tibs.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Ballesteros M, Fredriksson A, Henriksson J, Nystrom T. Bacterial senescence: Protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 2001;20:5280–5289. doi: 10.1093/emboj/20.18.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bacher JM, Schimmel P. An editing-defective aminoacyl-tRNA synthetase is mutagenic in aging bacteria via the SOS response. Proc Natl Acad Sci USA. 2007;104:1907–1912. doi: 10.1073/pnas.0610835104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 16.van Leeuwen FW, et al. Molecular misreading: A new type of transcript mutation expressed during aging. Neurobiol Aging. 2000;21:879–891. doi: 10.1016/s0197-4580(00)00151-2. [DOI] [PubMed] [Google Scholar]
- 17.Brulliard M, et al. Nonrandom variations in human cancer ESTs indicate that mRNA heterogeneity increases during carcinogenesis. Proc Natl Acad Sci USA. 2007;104:7522–7527. doi: 10.1073/pnas.0611076104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitehead DJ, Wilke CO, Vernazobres D, Bornberg-Bauer E. The look-ahead effect of phenotypic mutations. Biol Direct. 2008;3:18. doi: 10.1186/1745-6150-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brakmann S, Grzeszik S. An error-prone T7 RNA polymerase mutant generated by directed evolution. Chembiochem. 2001;2:212–219. doi: 10.1002/1439-7633(20010302)2:3<212::AID-CBIC212>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 20.Remington KM, Bennett SE, Harris CM, Harris TM, Bebenek K. Highly mutagenic bypass synthesis by T7 RNA polymerase of site-specific benzopyrene diol epoxide-adducted template DNA. J Biol Chem. 1998;273:13170–13176. doi: 10.1074/jbc.273.21.13170. [DOI] [PubMed] [Google Scholar]
- 21.Kramer EB, Farabaugh PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. Rna. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: A study of more than 1000 mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 23.Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS. The stability effects of protein mutations appear to be universally distributed. J Mol Biol. 2007;369:1318–1332. doi: 10.1016/j.jmb.2007.03.069. [DOI] [PubMed] [Google Scholar]
- 24.Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- 25.Inouye S, et al. Role of positive charge on the amino-terminal region of the signal peptide in protein secretion across the membrane. Proc Natl Acad Sci USA. 1982;79:3438–3441. doi: 10.1073/pnas.79.11.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Champs C, et al. New TEM variant (TEM-92) produced by Proteus mirabilis and Providencia stuartii isolates. Antimicrob Agents Chemother. 2001;45:1278–1280. doi: 10.1128/AAC.45.4.1278-1280.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robin F, et al. Evolution of TEM-type enzymes: Biochemical and genetic characterization of two new complex mutant TEM enzymes, TEM-151 and TEM-152, from a single patient. Antimicrob Agents Chemother. 2007;51:1304–1309. doi: 10.1128/AAC.01058-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sideraki V, Huang W, Palzkill T, Gilbert HF. A secondary drug resistance mutation of TEM-1 beta-lactamase that suppresses misfolding and aggregation. Proc Natl Acad Sci USA. 2001;98:283–288. doi: 10.1073/pnas.011454198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bershtein S, Goldin K, Tawfik DS. Intense neutral drifts yield robust and evolvable consensus proteins. J Mol Biol. 2008;379:1029–1044. doi: 10.1016/j.jmb.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 30.Baker D, Agard DA. Kinetics versus thermodynamics in protein folding. Biochemistry. 1994;33:7505–7509. doi: 10.1021/bi00190a002. [DOI] [PubMed] [Google Scholar]
- 31.Kather I, Jakob R, Dobbek H, Schmid FX. Increased folding stability of TEM-1 beta-lactamase by in vitro selection. J Mol Biol. 2008 doi: 10.1016/j.jmb.2008.07.082. [DOI] [PubMed] [Google Scholar]
- 32.van Leeuwen FW, Kros JM, Kamphorst W, van Schravendijk C, de Vos RA. Molecular misreading: The occurrence of frameshift proteins in different diseases. Biochem Soc Trans. 2006;34(Pt 5):738–742. doi: 10.1042/BST0340738. [DOI] [PubMed] [Google Scholar]
- 33.Nangle LA, Motta CM, Schimmel P. Global effects of mistranslation from an editing defect in mammalian cells. Chem Biol. 2006;13:1091–1100. doi: 10.1016/j.chembiol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Laxminarayana D, Kammer GM. mRNA mutations of type I protein kinase A regulatory subunit alpha in T lymphocytes of a subject with systemic lupus erythematosus. Int Immunol. 2000;12:1521–1529. doi: 10.1093/intimm/12.11.1521. [DOI] [PubMed] [Google Scholar]
- 35.Kurland CG. Translational accuracy and the fitness of bacteria. Annu Rev Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 36.Stoebel DM, Dean AM, Dykhuizen DE. The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics. 2008;178:1653–1660. doi: 10.1534/genetics.107.085399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci USA. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Visser JA, et al. Perspective: Evolution and detection of genetic robustness. Evolution. 2003;57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 39.Wagner A. Robustness and Evolvability in Living Systems. xii. Princeton: Princeton Univ Press; 2007. p. 366. [Google Scholar]
- 40.Huang W, Palzkill T. A natural polymorphism in beta-lactamase is a global suppressor. Proc Natl Acad Sci USA. 1997;94:8801–8806. doi: 10.1073/pnas.94.16.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bloom JD, et al. Thermodynamic prediction of protein neutrality. Proc Natl Acad Sci USA. 2005;102:606–611. doi: 10.1073/pnas.0406744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bar-Even A, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 43.Sosa-Peinado A, Mustafi D, Makinen MW. Overexpression and biosynthetic deuterium enrichment of TEM-1 beta-lactamase for structural characterization by magnetic resonance methods. Protein Expr Purif. 2000;19:235–245. doi: 10.1006/prep.2000.1243. [DOI] [PubMed] [Google Scholar]
- 44.Ji JP, Loeb LA. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry. 1992;31:954–958. doi: 10.1021/bi00119a002. [DOI] [PubMed] [Google Scholar]
- 45.Bracho MA, Moya A, Barrio E. Contribution of Taq polymerase-induced errors to the estimation of RNA virus diversity. J Gen Virol. 1998;79(Pt 12):2921–2928. doi: 10.1099/0022-1317-79-12-2921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.