Abstract
Brain tumors are horrific diseases with almost universally fatal outcomes; new therapeutics are desperately needed and will come from improved understandings of glioma biology. Exosomes are endosomally derived 30–100 nm membranous vesicles released from many cell types into the extracellular milieu; surprisingly, exosomes are virtually unstudied in neuro-oncology. These microvesicles were used as vaccines in other tumor settings, but their immunological significance is unevaluated in brain tumors. Our purpose here is to report the initial biochemical, proteomic, and immunological studies on murine brain tumor exosomes, following known procedures to isolate exosomes. Our findings show that these vesicles have biophysical characteristics and proteomic profiles similar to exosomes from other cell types but that brain tumor exosomes have unique features (e.g., very basic isoelectric points, expressing the mutated tumor antigen EGFRvIII and the putatively immunosuppressive cytokine TGF-β). Administration of such exosomes into syngeneic animals produced both humoral and cellular immune responses in immunized hosts capable of rejecting subsequent tumor challenges but failed to prolong survival in established orthotopic models. Control animals received saline or cell lysate vaccines and showed no antitumor responses. Exosomes and microvesicles isolated from sera of patients with brain tumors also possess EGFR, EGFRvIII, and TGF-β. We conclude that exosomes released from brain tumor cells are biochemically/biophysically like other exosomes and have immune-modulating properties. They can escape the blood-brain barrier, with potential systemic and distal signaling and immune consequences.—Graner, M. W., Alzate, O., Dechkovskaia, A. M., Keene, J. D., Sampson, J. H., Mitchell, D. A., Bigner, D. D. Proteomic and immunologic analyses of brain tumor exosomes.
Keywords: microvesicles, heat shock proteins, antibody, T cells, EGFRvIII, patient sera
Brain tumors such as malignant gliomas are devastating diseases with abysmal prognoses. For patients with glioblastoma multiforme (GBM), the most common form of high-grade malignant gliomas, median survival times are <15 mo (1), and the tumors are almost uniformly fatal. Despite maximal surgical, radiological, and chemotherapeutic interventions, these figures have changed little in the past two decades (2). New therapeutic strategies will likely evolve from a better understanding of brain tumor biology. One unstudied area in neuro-oncology is that of extracellular vesicular trafficking in the form of exosomes.
Exosomes are membrane-enclosed vesicles 30–100 nm in diameter that are derived from endosomes during the formation of multivesicular bodies (MVBs) (3). The fate of the MVBs and their contents is often lysosomal degradation; however, some MVBs fuse with the plasma membrane, releasing the vesicular contents into the extracellular space, begetting exosomes. Exosome release has implications for extracellular (in trans) signaling (4), deportation of toxic substances, including chemotherapeutic agents (5), viral passage (6), and horizontal mRNA/microRNA transfer (7). The role of exosomes in cell biology is not clear; in reticulocyte maturation (where exosomes were perhaps first described), exosomes are disposal mechanisms for unnecessary proteins and membrane components (8). Most exosome research has been in hematopoietic and immune system cells (9), focusing on the potential of exosomes as vehicles of immunity (10). There have also been several studies on immunity and tumor cell exosomes (11,12,13), but none on brain tumor exosomes.
We recently identified exosomes from brain tumor cell lines (14) and characterized their heat shock protein content as potential sources of extracellular heat shock proteins. This implies that brain tumor exosomes may be immunogenic and capable of driving antitumor immunity, perhaps mediated by their heat shock proteome; however, there are many indications that tumor exosomes may be immunosuppressive (13, 15, 16). This may be especially significant for high-grade gliomas, as those patients are often profoundly immunosuppressed (17, 18). In that regard, we have conducted further characterization of exosomes from SMA560vIII, a syngeneic murine (VM/Dk) anaplastic astrocytoma model (19). This particular cell line is stably transduced to express murine mutant epidermal growth factor receptor (EGFR) variant III (EGFRvIII). A splice site mutation truncates the EGFRvIII extracellular domain, resulting in its constitutive activation. Approximately 50–60% of all high-grade gliomas express EGFRvIII, and it is a bona fide tumor-specific antigen with potent oncogenic properties (20). With this aggressive tumor model, we can test in vivo the potential immunostimulatory or immunosuppressive aspects of exosomes coupled with in vitro mechanistic characterizations. Herein, we show that SMA560vIII exosomes possess interesting similarities to known characteristics of exosomes from other cell types but that there are notable differences, likely related to cell type. We performed a proteomic analysis of these exosomes and identified proteins that appear unique to brain tumor-derived exosomes. We also show that administration of SMA560vIII exosomes prophylactically protected mice against subcutaneous tumor challenge but failed to prolong survival in an orthotopic, preestablished tumor setting. In vitro studies indicated that the immunity has both humoral and T-cell components, which we believe is a novel finding for tumor exosomes. Like their murine counterparts, exosomes/microvesicles collected from the sera of patients with GBM possess EGFRvIII, a potent tumor-specific antigen, as well as wild-type EGFR, but the vesicles also display transforming growth factor beta 1 (TGF-β1), a potentially immunosuppressive cytokine. This work describes the physical and biochemical characteristics of novel brain tumor exosomes, demonstrates their existence in patient sera, and explores their complicated immune modulation.
MATERIALS AND METHODS
Cell lines and culture: lysate preparations
Cell lines D247MG (21), D283MED (22), SMA560 (19), and SMA560vIII (a gift from Dr. Glenn Dranoff, Dana Farber Cancer Institute, Boston, MA, USA; ref. 23) were grown as described previously (14). H2159MG is a pediatric glioma that was grown initially as a xenograft; it was harvested, disaggregated, and grown in neural stem cell medium. D54MG is from an adult glioma sublime of A-172, and D456MG is from a pediatric glioma. X43,T is from an adult glioma and is a gift from C. David James (University of California, San Francisco, CA, USA). These cells were grown in stem sell medium as described previously (24). Cell lysates were prepared as described previously (14).
Exosome preparations and analyses
Exosomes were prepared as described previously (14). Exosomes were further purified by density gradient centrifugation through an OptiPrep (Axis-Shield; Greiner Bio One Inc., Monroe, NC, USA) step gradient prepared in 20 mM HEPES buffer, pH 7.4, spun at 100,000 g for 18 h in a Beckman SW-41 rotor (Beckman Instruments, Fullerton, CA, USA). The gradient consisted of steps of 0–60% OptiPrep. One-milliliter fractions were collected, fraction densities were determined, and other analyses, including electron microscopy, LDS-PAGE, and Western blot analysis, were as described before (14).
Acetylcholinesterase (AChE) activities were assessed following a modified procedure from Khan et al. (25); a kinetic assay was performed using a Tecan microplate reader (Phoenix Research Products, Hayward, CA, USA) with a recording absorbance at 412 nm. AChE activity is reported in arbitrary units (μmol substrate hydrolyzed/min) relative to the blank.
Western blot analysis was performed as described (14). In general, lysates were loaded at 5–20 μg, while exosome samples were loaded with as much volume in gel wells as possible, so comparisons are only qualitative. The following antibodies were used: anti-ALIX (AL2-interacting protein X), mAb clone 2H12 (Santa Cruz Biotechnologies, Santa Cruz, CA, USA); anti-PDI (protein disulfide isomerase), rabbit polyclonal SPA-890 (Assay Designs/Stressgen, Ann Arbor, MI, USA); anti-CRT (calreticulin), rabbit polyclonal SPA-600 (Stressgen); anti-transferrin, mAb clone 2A2 (Fitzgerald, Concord, MA, USA); anti-alpha-1 antitrypsin, chicken (IgY) polyclonal (Affinity Bioreagents, Golden, CO, USA); anti-GAPDH (glyceraldehyde 3-phosphate dehydrogenase), rabbit polyclonal (Abcam, Cambridge, MA, USA); anti-CD9, mAb clone MM2.57 (Millipore, Billerica, MA, USA); chicken anti-TGF-β (transforming growth factor beta), AF-101-NA, R&D Systems, Minneapolis, MN, USA), anti-actin (Sigma-Aldrich, St. Louis, MO, USA), anti-L1/NCAM (CD171) (UJ127.11; Santa Cruz); anti-EGFR (extracellular domain, Ab-5, LabVision/Thermo Scientific, Fremont, CA, USA). Antibodies to EGFRvIII (mAb L8A4) and to GPNMB (glycoprotein nonmetastatic B, rabbit polyclonal 2640) have been described previously (26, 27). Secondary antibodies were HRP (horseradish peroxidase)-labeled donkey anti-rabbit IgG, or sheep anti-mouse IgG, both from GE Healthcare (Piscataway, NJ, USA). Donkey anti-chicken HRP-labeled secondary antibody was from Affinity Bioreagents. Native TGF-β1 (purified from human platelets) was from R&D Systems.
Free-solution isoelectric focusing was performed using a Rotofor device (Bio-Rad Laboratories, Hercules, CA, USA). OptiPrep fractions containing exosomes were diluted to 30 ml in water that contained 0.25% (v/v) each of Fluka High-Resolution carrier ampholytes, pH ranges 3–10, 3–6, 4–6, 5–7 (Sigma-Aldrich). The sample was loaded into the focusing chamber (0.1 M H3PO, anode; 0.1M NaOH, cathode). Isoelectric focusing was conducted at 15-W constant power for 4 h. Twenty fractions were harvested, and the pH of each fraction was measured. AChE assays were performed on each fraction, and the proteins in the fractions were separated on LDS-PAGE followed by electroblotting; Western blots were probed with anti-ALIX antibodies as described above.
Exosome bead fluorescence-activated cell sorting (FACS)
Exosomes were attached to 4 μm aldehyde/sulfate latex beads (Invitrogen, Carlsbad, CA, USA) by mixing ∼30 μg exosomes in a 100 μl volume of beads for 2 h at room temperature. This suspension was diluted to 1 ml with PBS, and the reaction was stopped with 100 mM glycine. Exosome-bound beads were washed in PBS/1% BSA, blocked with 10% normal goat serum and Fc-blocker (Invitrogen), and stained for FACS with FITC-labeled antibodies L8A4 (for EGFRvIII) and antibodies against HSP27 and HSP70, as described previously (14).
Sample preparation for two-dimensional (2-D) gel electrophoresis
Exosomes (from OptiPrep gradient fractions) were diluted in PBS and pelleted using a Beckman Coulter TLA 100.3 rotor (500,000 g, 20 min). Pellets were diluted in 100 μl PBS and then were supplemented with 100 μl phenol and vortexed. The mixture was heated to 70°C for 10 min in a safety hood, and transferred to ice for 5 min, followed by centrifugation (5000 g, 10 min). The aqueous phase was discarded, and these steps were repeated by adding 100 μl diH2O. After spinning, the aqueous phase was discarded, 200 μl of acetone was added, vortexed, and spun as before. The supernatant was discarded, the previous step was repeated, and the resulting pellet was air-dried. The pellet was dissolved in solubilization buffer (8 M urea, 4% CHAPS, 30 mM Tris, pH 8.5). Protein concentration was determined with the 2D-Quant kit (GE Healthcare), and 360 μg of total protein was used for 2-D gel electrophoresis. We should point out that brain tumor exosomes have proven exceptionally resistant to lysis by detergent-based extraction methods.
2-D gel electrophoresis, 2-D DIGE, imaging, and protein identification
2-D gel electrophoresis, 2-D DIGE (difference gel electrophoresis), and imaging were performed as described (28). For protein identification, particular protein spots were chosen on the basis of both similarities and differences in migration compared to proteins from human brain tumor exosomes in 2-D DIGE. Some spots had coordinates identical to those of the human samples; others appeared as charge variants, and still others were unique. Protein spots were processed for protein identification by tandem mass spectrometry at the Duke-UNC Michael Hooker Proteomics Center of the University of North Carolina, Chapel Hill, as described (29) (30). Protein subcellular localizations were determined from literature searches and confirmed using the TargetP program (www.cbs.dtu.dk/services/TargetP/) (31) and Ingenuity Systems software (Redwood City, CA, USA; http://www.ingenuity.com/index.html). Pathway analyses and network constructions were assembled using the Ingenuity software.
Vaccination studies
All animal studies were done under the auspices of the Duke University Institutional Animal Care and Use Committee following approved protocols. VM/Dk mice (6–12 wk of age) were subcutaneously immunized on days −14 and −7 with 50 μg SMA560vIII exosomes (diluted in PBS—no adjuvants were used). Control mice received either PBS or 50 μg SMA560vIII lysate. On day 0, mice were implanted subcutaneously with 1 × 106 SMA560vIII cells on the flank opposite the vaccine injections. Tumor cells were harvested from disaggregated in vivo grown SMA560vIII tumors to avoid the potential for carry-over of fetal calf serum antigens from cells grown in tissue culture. Tumor growth was subsequently monitored as described (14). Experiments were performed on three independent occasions. Mice that survived the initial tumor challenge were subcutaneously rechallenged 90–135 days after the primary tumor, again with 1 × 106 SMA560vIII cells on the flank opposite the first tumor. Age-matched naive mice received the same tumor inoculations to verify tumor take, and tumor volumes were determined as above. In an orthotopic, preestablished tumor setting, mice were intracranially injected with 5000 SMA560vIII cells (from in vivo-grown tumors) as described (32). Those experiments were performed twice.
Immunoassays: ELISPOTs and ELISAs
ELISPOT assays were performed as described previously (14) with mouse interferon-gamma kits from R&D Systems (Minneapolis, MN, USA). Splenocytes were restimulated with 10 μg/ml of EGFRvIII “Peptide 3,” a 13-amino acid peptide with a terminal cysteine (LEEKKGNYVVTDHC) that spans the EGFRvIII mutation (AnaSpec, San Jose, CA, USA). Other restimulants include 10 μg/ml of SMA560vIII lysate and 10 μg/ml of SMA560vIII exosomes. Assays were performed in quadruplicate. ELISAs were used to assess serum antibody titers from mice. Sera were obtained from sedated mice (either naive mice, control mice immunized with PBS, control tumor-bearing mice, mice immunized with exosomes, or exosome-immunized mice that survived tumor challenges). ELISA plates were prepared by overnight incubation with SMA560vIII exosomes in PBS at 10 μg/well. Sera were serially diluted and incubated with antigen overnight; bound antibody was detected with biotinylated goat anti-mouse secondary antibodies specific for murine isotypes IgG, IgA, and IgM (Invitrogen/Zymed) followed by streptavidin-HRP (Sigma). Color was developed with 3,3′,5,5′-tetramethylbenzidine (Sigma); development was stopped with sulfuric acid, and absorbance was read at 492 nm on a Tecan microplate reader.
Isolation of exosomes from patient sera
Exosomes were isolated from the sera of patients with high-grade gliomas by differential centrifugation. Deidentified serum samples (collected from patients prior to surgery for high-grade gliomas following appropriate consenting as required and approved by Duke University Internal Review Board) were separated from whole blood and stored frozen until used. Samples from 12 patients were apportioned into 4 pools and diluted with PBS leaving ∼2 ml total volume for each pool. An equal volume of human AB serum (Sigma) was used as a control. Samples were centrifuged at 10,000 g for 30 min; supernatants were saved and centrifuged at 100,000 g for 1 h in a Beckman Optima TL-100 centrifuge using a TLA-100.3 rotor. Pellets were collected, allowed to drain, and submitted for electron microscopy, as described above, or resuspended in Laemmli sample buffer (Bio-Rad) with 2-mercaptoethanol and 1% saponin (Sigma). Samples were boiled for 15 min, and maximum loading volumes (50 μl) were loaded onto 10% PAGE gels or 4–20% gradient gels (Bio-Rad). Gels were electroblotted and probed with antibodies as described above.
Statistical analyses
Where meaningful, Student’s t test was used to determine statistical significance between measurements. In some cases, Kaplan-Meier curves were also generated and analyzed by log-rank statistics to determine survival differences between groups.
RESULTS
Biochemical characterization of brain tumor exosomes
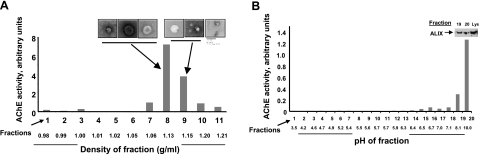
We previously demonstrated the presence of brain tumor exosomes in the spent media from human and murine glioma lines (14). Those vesicles were determined to be exosomal microparticles by electron microscopy and Western blot analysis for canonical heat shock protein content. Here, we have purified and characterized exosomes from the media of the murine brain tumor line SMA560vIII (SMA560 parental cells transduced with the murine homologue of the mutant EGFRvIII). These exosomes were collected by differential centrifugation and were further purified by density gradient centrifugation through an OptiPrep medium step gradient. OptiPrep was chosen as the medium due to its ready solubility, lack of endotoxin contamination, and putative preservation of biological function. Fractions of increasing density were collected, and those presumably enriched in exosomes were identified by their AChE activity (Fig. 1). The AChE activity was present in fractions whose densities are consistent with those reported for exosomes derived from other cell types (33, 34). Uranyl acetate-negative stain electron microscopy (Fig. 1A, insets) confirmed the presence of membrane vesicles with 50- to 100-nm diameters characteristic of exosomes. We have performed similar isolations and characterizations of exosomes from A20 murine lymphoma to verify our techniques (data not shown).
Figure 1.
Characterization of murine brain tumor exosomes by density gradient centrifugation, electron microscopy, free-solution isoelectric focusing and marker (AChE, ALIX) content. A) SMA560vIII exosomes were harvested from spent media by differential centrifugation, followed by density gradient centrifugation. Fractions containing exosomes were identified by AChE activity and negative-stain electron microscopy. Scale bar = 100 nm. B) Exosomes isolated from the appropriate fractions in A were subjected to solution phase isoelectric focusing in a pH 3–10 gradient. AChE activity was used again to identify exosome-containing fractions, and their presence was verified by Western blot analysis with antibodies to ALIX (shown for fractions 19 and 20, with lysate (Lys, 5 μg) as a positive control.
Because the net overall charge of exosomes may influence their biological properties (e.g., interactions with cell surfaces, stroma, serum components, etc.), we examined SMA560vIII exosome migration in a solution-based isoelectric focusing gradient. Despite the presence of many proteins with acidic isoelectric points (pIs, see below), these lipid-protein complexes focused to the far basic end of the pH gradient (Fig. 1B). The presence of exosomes was again demonstrated by AChE activity and by Western blot analysis for ALIX (ALG-2-interacting protein X, a molecular marker of exosomes; ref. 35) in the high-pH fractions. This exosome property may indicate that certain lipids are preferentially surface exposed or that potent counter ion effects lead to an overall alkaline pI.
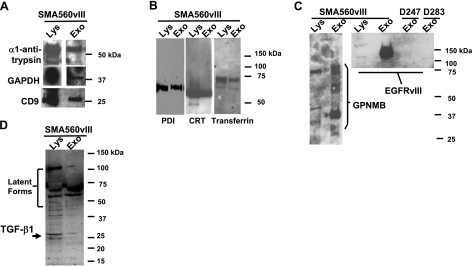
Previously, we have demonstrated the presence of canonical heat shock proteins (HSPs 90, 70, 60, and 27) in brain tumor exosomes (14). Here, we characterized the murine brain tumor exosomes by probing exosome protein blots with antibodies that specifically recognize known exosome constituents alpha 1-antitrypsin, GAPDH, and one of the prototypical tetraspanins found in exosomes, CD9, which is abundantly expressed in brain (36) (Fig. 2A). The alpha 1-antitrypsin antibody is specific for human and rodent proteins. We also identified the metal-binding proteins and chaperones PDI, CRT, and transferrin (Fig. 2B). PDI was present in murine mast cell exosomes (7); CRT and PDI have been identified in MCF-7 exosomes and microvesicles (Steve Griffiths, Atlantic Cancer Research Institute, Moncton, NB, Canada; personal communication, May 2008), and in plasma microparticles (37), so there is precedent for these endoplasmic reticulum residents to appear in exosomes. By Western blot analysis, we demonstrated the presence of known brain tumor antigens, such as GPNMB (26), and the well-known brain tumor antigen EGFRvIII (38, 39) (Fig. 2C). Note that exosomes from cells that do not express EGFRvIII (i.e., D247MG and D283MED) do not react with the EGFRvIII specific monoclonal antibody (mAb L8A4) (40). While Fig. 2C indicates that tumor antigens are present in exosomes, the immunomodulatory protein transforming growth factor β1 is also in brain tumor-derived exosomes (Fig. 2D).
Figure 2.
Western blot characterization of SMA560vIII exosomes. Exosome proteins (Exo) and lysate proteins (Lys) of parent cells were separated on LDS-PAGE and electroblotted to nitrocellulose. Blots were probed with antibodies against proteins listed. A) Typical proteins found in exosomes: α1-antitrypsin, GAPDH, and the tetraspanin CD9; 20 μg of lysate was loaded. B) Metal binding proteins and chaperones: PDI, CRT, and transferrin; 5 μg of lysate was loaded. C) Known brain tumor antigens: GPNMB and EGFRvIII; 10 μg of lysate was loaded. D) Immunomodulatory cytokine TGF-β1; 10 μg of lysate was loaded. Molecular mass markers for each section are shown at right.
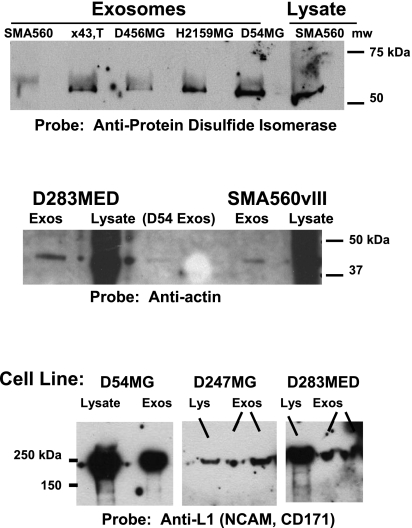
To ensure that our results were not exclusive to murine brain tumor exosomes, we probed a variety of human glioma- and medulloblastoma-derived exosomes with antibodies against PDI, actin, and L1-NCAM/(CD171), the latter of which is also considered an operationally specific brain tumor antigen (Fig. 3). Please note that the amounts of exosome proteins loaded are generally far less than those of the corresponding lysates, so only qualitative comparisons may be made.
Figure 3.
Exosomes isolated from a variety of brain tumor cell lines contain the chaperone PDI, the structural protein actin, and the cell adhesion molecule (and brain tumor antigen) L1-NCAM (CD171). Top: exosomes derived from the murine brain tumor model SMA560; from gliomas X43,T, D456MG, and H2159MG (pediatric gliomas); and D54MG all react with anti-PDI antibodies in Western blots. SMA560 lysate is shown as a positive control; 5 μg of lysate was loaded. Middle: Western blots for actin in exosome proteins (and lysates) of D283 medulloblastoma, D54MG, and SMA560vIII; 10 μg of lysate was loaded. Bottom: Western blots of lysates and exosomes from D54MG, D247MG, and D283MED are positive for L1-NCAM/CD171, which is considered an operationally specific tumor antigen; 10 μg of lysate was loaded.
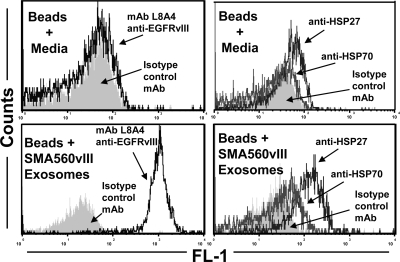
The cell surface heat shock protein content of many brain tumor cell lines and the release of HSPs into the spent media led us to search for exosomes from these cells (14). The mechanism of exosome generation as intraendosomal vesicles suggests the possibility that cell surface chaperones could emerge on the surfaces of released exosomes. We used an aldehyde sulfate-latex bead preparation to bind and enrich exosomes nonspecifically; this enabled us to stain for surface markers and analyze the exosome-coated beads by FACS (Fig. 4). As seen in the lower left panel of Fig. 4, the exosome surfaces were stained with the L8A4 mAb specific for EGFRvIII. This is considered to be a positive control for the assay, since the mAb detected EGFRvIII in the SMA50vIII exosomes by Western blot (Fig. 2); indeed, it did not stain exosomes from the parental SMA560 line (data not shown). The lower right panel shows results of exosome-bead FACS using antibodies to HSPs 27 and 70, both of which were found on the surfaces of SMA560 and SMA50vIII cells in FACS and in Western blot analysis of SMA560 exosomes (14) (data not shown). We believe this is the first report of HSP27 on exosome surfaces. The implications of these results are discussed below.
Figure 4.
Brain tumor exosome surfaces display EGFRvIII and heat shock proteins 27 and 70. Exosomes were bound to aldehyde-sulfate latex beads, were stained with fluorescently labeled antibodies to EGFRvIII (mAb L8A4; left panels) and to HSPs 27 and 70 (right panels), and were analyzed by flow cytometry. Staining of beads soaked in cell culture media as controls are shown in top panels. Isotype control staining profiles are shown as gray fill.
Proteomic characterization of brain tumor exosomes
The proteome of SMA560vIII exosomes was analyzed by 2-D polyacrylamide gel electrophoresis (2-D PAGE). We selected proteins for further analysis based on migration patterns in 2-D DIGE, as described in Materials and Methods. Thus, some SMA560vIII exosome proteins selected had virtually identical migration patterns (molecular mass and isoelectric point) with those exosome proteins from human H2159MG cells; some appeared to be species-specific isoforms; others were clearly distinct compared to the human proteins (unpublished results).
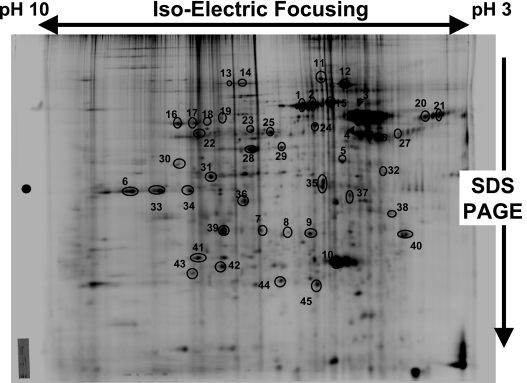
Forty-five isolated protein spots were identified by peptide mass fingerprinting using mass spectrometry (MS) and MS/MS data for de novo peptide sequencing. The Brilliant-Blue Coomassie-stained gel with the denoted spots is shown in Fig. 5. We identified 36 distinct proteins from these samples (Table 1), which we have categorized according to predicted molecular mass and isoelectric point, the functions ascribed to the proteins in the literature and by computer algorithm searches, putative intracellular or extracellular localizations, and with respect to their previous identification in exosomes. Note that many of the proteins in this sample have acidic pIs, which belies the very basic isoelectric point of the vesicles as a whole (Fig. 1B). These include the HSC70 and HSP60 proteins that we previously identified by Western blot analysis (14). Proteins that are unique to these brain tumor exosomes compared to exosomes from other cell types (based on our literature searches) include arrestin domain-containing protein 2 (#13), 3DPGH (#23), collapsing response mediator protein 2 (#24), CENP-P (#26), sialic acid synthase (#30), eIF3β (#34), PCNA (#37), MTApase (#38), EF-1β (#39), and proteasome subunit α type2-C (#41).
Figure 5.
Two-dimensional polyacrylamide gel electrophoresis of brain tumor exosomes. 2-D PAGE was performed to separate exosome proteins by isoelectric focusing in the first dimension, and by SDS-PAGE in the second dimension. Gel was stained with Brilliant-Blue Coomassie dye; circled and numbered proteins were identified by mass spectrometry (see Table 1).
TABLE 1.
Results of brain tumor exosome proteomic analyses
| Spot | Protein name | Accession no. | Mass/pI | Peptide count (match score) | Ion score | Function | Subcellular localization | Exosome reference |
|---|---|---|---|---|---|---|---|---|
| 1 | Serum albumin (precursor, bovine) | P02769 | 69.3/5.82 | 13 (255) | 189 | Ion, lipid transport; osmotic pressure | Extracellular space | 35 |
| 2 | Serum albumin (precursor, murine) | P07724 | 68.7/5.75 | 8 (91) | 67 | Ion, lipid transport; osmotic pressure | Extracellular space | 88, 89 |
| 3 | α-1-Antitrypsin (precursor, bovine) | P34955 | 46.1/6.05 | 12 (260) | 174 | Secreted extracellular protease inhibitor | Extracellular space | 66 |
| 4 | Vitamin D-binding protein (precursor, bovine) | Q3MHN5 | 53.3/5.36 | 4 (67) | 55 | Sterol transport; actin and Ig binding | Extracellular space | |
| 5 | β-Actin | P60710 | 41.7/5.29 | 10 (161) | 104 | Cytoarchitecture, motility | Cytosola | 90b |
| 6 | GAPDH | P16858 | 35.8/8.44 | 7 (95) | 63 | Step 1 of glycolytic enzyme complex | Plasma membrane | 91b |
| 7 | Gag-pro-pol polyprotien (precursor, murine) | Q1KYM2 | 194/9.05 | 12 (94) | 79 | Endogenous murine retroviral polyprotein (processed) | Plasma membrane | 35 |
| 8 | Gag polyprotein pr65 | Q1KYL8 | 60.1/8.52 | 5 (158) | 145 | Isoelectric variant of #7 | Plasma membrane | |
| 9 | Gag polyprotein pr65 | Q1KYL8 | 60.1/8.52 | 7 (133) | 114 | Isoelectric variant of #7, 8 | Plasma membrane | |
| 10 | Apolipoprotein A-1 (precursor) | JC137 (MSDB) | 30.4/5.52 | 5 (65) | 50 | Lipid binding, cholesterol metabolism | Extracellular space | 35, 67 (bovine) |
| 11 | Major vault protein | Q922X6 | 95.9/5.43 | 20 (128) | 18 | Ribonucleoprotein; “vault” subunit | Nucleusc | 90, 92 |
| 12 | Transitional ER ATPase (p97/VCP) | Q01853 | 89.3/5.14 | 23 (355) | 210 | ER vesicular trafficking | ER/Golgi | 7 |
| 13 | Arrestin domain-containing protein 2 | Q9D668 | 44.3/9.32 | 2 (33) | 27 | Scaffold/adaptor G-protein regulator | Cytosolc | |
| 14 | Transitional ER ATPase (p97/VCP) | Q01853 | 89.3/5.14 | 14 (87) | 42 | Modified variant of #12 | ER/Golgi | |
| 15 | Heat shock cognate 71 (HSC70, HSP70) | P63017 | 70.9/5.37 | 26 (495) | 289 | Chaperone, protein folding | Cytosola | 67b |
| 16 | Pyruvate kinase isozyme M1/M2 | P52480 | 57.8/7.15 | 23 (218) | 54 | Step 5 of glycolytic enzyme complex | Cytosold | 7, 90, 93 |
| 17 | Pyruvate kinase isozyme M1/M2 | P52480 | 57.8/7.15 | 20 (210) | 71 | Isoelectric variant of #16 | Cytosold | |
| 18 | Pyruvate kinase isozyme M1/M2 | P52480 | 57.8/7.15 | 10 (57) | 23 | Isoelectric variant of #16, 17 | Cytosold | |
| 19 | TCP-1ζ (HSP60) | P80317 | 58.0/6.63 | 11 (78) | 31 | Chaperonin complex | Cytosola | 92 |
| 20 | α-2-HS-glycoprotein (Fetuin-A, bovine precursor) | P12763 | 38.4/5.26 | 5 (102) | 74 | Lipid binding, promotes endocytosis; protease inhibitor | Extracellular space | 67, 94 (human) |
| 21 | α-2-HS-glycoprotein (Fetuin-A, bovine precursor) | P12763 | 38.4/5.26 | 6 (187) | 144 | Isoelectric variant of #21 | Extracellular space | |
| 22 | No significant hits | |||||||
| 23 | d-3 phosphoglycerate dehydrogenase (3PDGH, rat) | O08651 | 56.5/6.28 | 10 (90) | 27 | Enzyme of serine biosynthesis | Cytosol | |
| 24 | Collapsing response mediator protein 2 | P47942 | 62.3/5.95 | 17 (235) | 125 | Growth cone collapse; axon formation | Cytosolc | |
| 25 | TCP-1β (HSP60) | P80314 | 57.5/5.97 | 24 (317) | 122 | Chaperonin complex | Cytosola | 92, 95 |
| 26 | Centromere protein P | Q9CZ92 | 33.3/5.18 | 5 (39) | 24 | Component of centromere complex | Nucleus | |
| 27 | α-Enolase | P17182 | 47.1/6.37 | 19 (322) | 167 | Step 4 of glycolytic enzyme complex | Cytosold | 90b |
| 28 | α-Enolase | P17182 | 47.1/6.37 | 14 (124) | 34 | Isoform of #28 | Cytosold | |
| 29 | Phosphoglycerate kinase 1 | P09411 | 44.4/8.02 | 4 (36) | 26 | Step 2 of glycolytic enzyme complex | Cytosold | 7, 66, 95 |
| 30 | Sialic acid synthase | Q3TFB5 | 31.2/7.14 | 9 (183) | 130 | Sialic acid synthesis | Cytosol | |
| 31 | 40S ribosomal protein SA (34/67 kDa laminin receptor) | P14206 | 32.8/4.80 | 8 (101) | 54 | Ribosome component; on tumor surfaces, binds laminin | Plasma membrane | 7 |
| 32 | GAPDH | P16858 | 35.8/8.44 | 8 (128) | 87 | Isoelectric variant of #6 | Plasma membrane | |
| 33 | GAPDH | P16858 | 35.8/8.44 | 9 (85) | 37 | Isoelectric variant of #6 | Plasma membrane | |
| 34 | Eukaryotic translation initiation factor 3 subunit 2 (eIF3β) | Q9QZD9 | 36.5/5.38 | 13 (213) | 120 | Protein translation; binds 40S ribosome, initiator methionine tRNA | Cytosol | |
| 35 | Syntenin | O88601 | 32.3/6.66 | 2 (65) | 51 | Scaffold/adaptor Proteolypid complexes | Plasma membrane | 35b |
| 36 | Tubulin β-5 | P99024 | 49.7/4.78 | 13 (166) | 82 | Cytoskeleton, cell division, transport | Cytosol | 35 |
| 37 | Proliferating cell nuclear Ag (PCNA) | P17918 | 28.8/4.66 | 9 (123) | 150 | DNA replication | Nucleus | |
| 38 | S-methyl-5-thio-adenosine phosphorylase (MTApase) | Q9CQ65 | 31.1/6.71 | 18 (303) | 150 | Polyamine metabolism salvage pathway | Nucleus | |
| 39 | Elongation factor 1-β (EF-1β) | O70251 | 24.7/4.53 | 3 (58) | 45 | Protein synthesis, GDP/GTP exchange | Cytosol | |
| 40 | RAN (Ras family member; TC4) | Q3ULW0 | 24.7/7.71 | 10 (163) | 92 | Nucleocytoplasmic transport, GTP binding | Nucleusc | 95 |
| 41 | Proteasome subunit α type-2 component C3 | P49722 | 25.9/8.39 | 10 (165) | 91 | Protease, regulatory subunit of proteasome | Cytosol | |
| 42 | Glutathione S-transferase P1 (GST-pi) | P19157 | 23.6/7.69 | 3 (73) | 59 | Conjugates GSH in detoxification reactions | Cytosol, nucleus | 95 |
| 43 | Adenine phosphoribosyl transferase (APRT) | Q564P4 | 19.7/6.31 | 10 (338) | 258 | Purine salvage pathway, formation of AMP | Cytosol | 66 |
| 44 | Ferritin heavy chain | P09528 | 20.9/5.53 | 5 (108) | 82 | Iron binding complex | Cytosola | 7 |
| 45 | Ig γ-3 C region (IgG3 constant region) | P22436 | 36.2/8.53 | 10 (353) | 282 | Constant region of IgG heavy chain | Extracellular space | 88 |
Accession numbers are from the UniProt Knowledge base (Swiss-Prot/TrEMBL), http://us.expasy.org/sprot/(spot 10 is from MSDB; http://csc-fserve.hh.med.ic.ac.uk/msdb.html). Mass/pI refers to predicted molecular mass (kDa) and isoelectric point of the protein using the program Compute pI/Mw (http://ca.expasy.org/cgi-bin/pi_tool). Peptide count refers to number of peptides that match the theoretical digest of the primary protein identified. Match score refers to quality of the peptide-mass fingerprint match and quality of the MS/MS peptide fragment ion matches (if MS/MS data were generated). Scores ≥ 95 are considered significant. Ion score is quality of MS/MS peptide fragment ion matches only (if MS/MS data were generated). Scores ≥ 20 are significant. Functions and subcellular localizations were inferred from literature searches, from the TargetP algorithm, and from Ingenuity software; exosome reference denotes literature citations of previous proteomic analyses for the given protein.
Cytosolic protein that has been identified on tumor cell surfaces or extracellularly.
Protein has been identified in numerous exosome types; only limited reference citations are shown.
Cytosolic or nuclear protein that has known associations with intracellular membranes.
Cytosolic protein that has known alternate extracellular or membrane localizations in normal cell types.
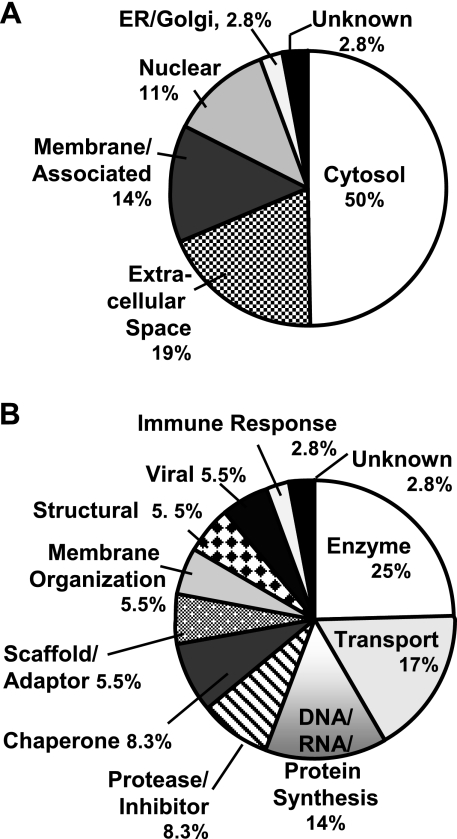
We have displayed the cellular and secreted distributions of the identified proteins based on database and literature predictions (Fig. 6A); clearly, most of the SMA560vIII exosome proteins identified were cytosolic, but many of those had known membrane associations, as well as unusual localizations in tumors (e.g., the heat shock proteins; ref. 14). The proteins identified were also classified by predicted functions (Fig. 6B), and the majority of our proteomic analysis revealed enzymes and other metabolic entities. There were also a number of proteins involved in DNA/RNA interactions and protein synthesis, extracellular transport proteins, chaperones, and proteases or protease inhibitors. Many of the molecules identified are multifunctional proteins whose activities alone or in complexes may differ from their traditionally recognized roles. Thus, these classifications are suggestive rather than definitive, and they require some flexibility in interpretation. As such, the contextual functions of these proteins could be potentially unique to intercellular exchanges via exosomal microparticles.
Figure 6.
Categorization of proteins identified in Fig. 5 and Table 1 by subcellular and extracellular localization (A) and by putative function (B). A) Percentage determinations are given: cytosolic proteins, 18 of 36 total; secreted in the extracellular space, 7 of 36; membrane and/or membrane associated (both cytoplasmic and extracellular faces) 5 of 36; nuclear, 4 of 36; ER and Golgi, and unknown, 1 of 36 for each. B) Percentage determinations are based on enzymes and metabolic proteins, 9 of 36 total; transport proteins, 6 of 36; DNA- and RNA-binding proteins and translation and protein synthesis, 5 of 36; chaperones and proteases or protease inhibitors, 3 of 36, respectively; scaffold and adaptor proteins, membrane-organizing proteins, structural proteins, and viral proteins, 2 of 36 for each category; immune response and unknown, 1 of 36 for each. In circumstances in which a protein may have multiple functions or localizations, it received assignment based on the preponderance of literature references.
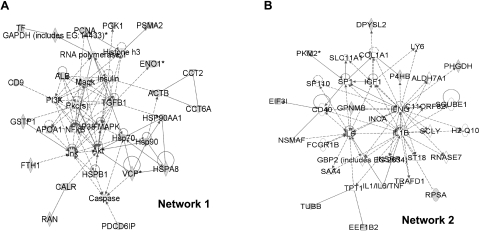
Using Ingenuity software, we performed network pathway analyses combining the proteins identified in our proteomic studies with information from the Western blot analyses (Figs. 2, 3) and from the HSPs found in exosomes from our previous work (14). The algorithms revealed 8 networks, the top two of which were described as “hematologic/immunological/respiratory disease” and “cell-cell signaling/interaction, cancer, hematological system development and function” (Table 2). Those networks are shown in Fig. 7. In network 1, note the relationships among many of the exosome proteins, as well as their ties to major signaling molecules in cancer, in general, and brain tumors, in particular (AKT, PI3K, MAPK; Fig. 7A). Pathway analyses such as these may also provide insight into roles for proteins whose functions are currently poorly understood, such as GPNMB (Fig. 7B). Network 2 demonstrates several exosome protein interactions with important immunological proteins such as CD40, interferon-gamma, interleukin-6, and interleukin-1β. The interconnectedness of the proteins in these networks highlights the multiple links that exosomes may have to various normal and pathological cellular states.
TABLE 2.
Ingenuity software pathway analyses, highest scoring networks
| Network | Molecules in network | Score | Focus molecules | Top functions |
|---|---|---|---|---|
| 1 | ACTB, Akt, ALB, APOA1, CALR, Caspase, CCT2 CCT6A, CD9, ENO1, FTH1, GAPDH, GSTP1, Histone h3, Hsp70, Hsp90, HSP90AA1, HSPA8, HSPB1, Insulin, Jnk, Mapk, NFkB, P38, MAPK, PCNA, PDCD6IP, PGK1, PI3K, Pkc(s), PSMA2 RAN, RNA polymerase II, TF, TGFB1, VCP | 55 | 22 | Hematological disease, immunological disease, respiratory disease |
| 2 | ALDH7A1, C11ORF82, CD40, COL1A1, DPYSL2, EEF1B2, EIF3I, FCGR1B, GBP2, GPNMB, H2-Q10, IFNG, IGF1, IL1/IL6/TNF, IL1B, INCA, INSRR, LY6, NSMAF, P4HB PHGDH, PMK2, RNASE7, RPSA, SAA4, SCLY, SCUBE1, SLC11A, SP1, SP110, ST18, TPT1, TRAFD1, TUBB | 17 | 9 | Cell-to-cell signaling and interaction, cancer, hematological system development, and function |
Proteins derived from brain tumor exosomes [by proteomics, Western blotting, and from our previous publication (14)] are underscored.
Figure 7.
Network analyses of brain tumor exosome proteins identified by proteomics and Western blot analysis. Eight networks were discerned using Ingenuity software (Table 2), with the two significant-scoring ones shown here. Proteins derived empirically from this study are shown as gray-filled symbols; direct protein-protein interactions are shown as solid lines; indirect interactions are shown as broken lines. A) Network 1 had pathways in “hematological, immunological, and respiratory disease.” B) Network 2 pathways were involved in “cell-cell signaling/interaction, cancer, and hematological system development and function.”
Immune responses to brain tumor exosomes
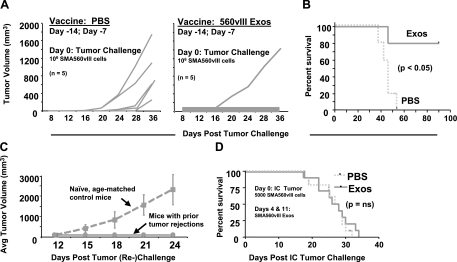
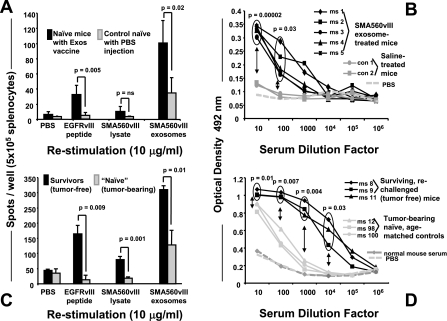
Much of the scientific attention devoted to microparticles like exosomes is in cancer immunotherapy, in which exosomes appear to be immune stimulating vehicles, whether derived from antigen-pulsed dendritic cells (“dexasomes”) or from tumors themselves (“texasomes”) (10). We immunized mice prophylactically with tumor-derived exosomes under the pretense of evaluating either the immune stimulation or potential immune suppression of the exosomes, which would be more easily measured in a subcutaneous challenge. Thus, we injected mice with 50 μg of SMA560vIII exosomes on days −14 and −7; mice received subcutaneous tumor cell challenges (1×106 SMA560vIII cells) on day 0, and we monitored tumor growth subsequently. Approximately 80% of the mice immunized with the exosomes rejected their tumors (Fig. 8A shows results typical of 3 experiments); most of those mice did develop palpable tumors that later regressed. In the control group, all mice died from their tumors by day 55 (Fig. 8B); similar results were seen for mice vaccinated with tumor lysate (data not shown). Exosome administration generated protective immunity in these animals with no evidence of immune suppression. On rechallenge 135 days after their first tumor injection, all of the surviving mice that had been originally immunized with SMA560vIII exosomes rejected their second tumor inoculation, whereas all of the age-matched control mice developed tumor (Fig. 8C). In this setting, exosome vaccination coupled with tumor rejection provided immunological memory.
Figure 8.
Antitumor responses of mice immunized with SMA560vIII exosomes. A) Groups of 5 VM/Dk mice were immunized with 50 μg of SMA560vIII exosomes (exos) or with saline (PBS) 14 and 7 days prior to subcutaneous SMA560vIII tumor challenge (106 cells). B, C) Individual tumor volumes were monitored over time, as was survival (B), which was statistically significant in a Kaplan-Meier analysis. Prophylactically immunized mice that survived their initial tumor challenge were rechallenged 135 days after the initial tumor inoculation, along with age-matched naive controls (C), and tumor volumes were monitored (shown here as averages). D) In a preestablished orthotopic model, groups of 10 mice were implanted with 5000 SMA560vIII cells intracranially. Four and 11 days later, mice were treated with 50 μg of SMA560vIII exosomes or with PBS, and survival was monitored. Survival differences between the treated and control groups were not significant (ns).
We followed the prophylactic immunization experiments with exosome vaccination in a preestablished setting using an orthotopic [intracranial (i.c.)] SMA560vIII model. VM/Dk mice were inoculated by stereotactic injections with 5000 SMA560vIII cells directly into the brain, and 4 days later were treated with subcutaneous injections of 50 μg SMA560vIII exosomes. Control animals received saline instead of exosomes. The vaccination was repeated 1 wk later. Unlike the response to the prophylactic vaccines, treating animals bearing i.c. tumors with tumor-derived exosomes well after tumor implantation demonstrated no obvious antitumor effect in terms of overall survival (Fig. 8D).
While the orthotopic, preestablished tumor model used here is quite stringent, the lack of any notable antitumor activity from exosome vaccines was puzzling, given the results of the prophylactic immunizations. We therefore attempted to identify potential mechanisms of the protective vaccine effects. We immunized naive VM/Dk mice with SMA560vIII exosomes 14 and 7 days before spleen and sera harvest. The splenocytes were used in interferon-gamma (IFN-γ) ELISPOT assays to test for cellular-based immune responses, and the sera were used in ELISA assessments for humoral immunity (with SMA560vIII exosomes as antigen targets). Splenocytes were restimulated either with SMA560vIII tumor lysate, with Peptide 3 (a 14-mer peptide from the splice site region specific to the EGFRvIII mutation), or with SMA560vIII exosomes. From the exosome-vaccinated mice, a significant number of splenocytes produced IFN-γ on restimulation with sources of tumor antigen, including responses to the EGFRvIII peptide (Fig. 9A) There were also significant serum antibody titers against exosomes, albeit at relatively low dilutions (Fig. 9B), despite only two immunizations performed in the absence of added adjuvant.
Figure 9.
ELISPOT and ELISA measurements of immune responses from exosome-immunized mice. A, B) Naive VM/Dk mice were twice immunized with 50 μg of SMA560vIII exosomes (days −14 and −7) or with PBS as controls. On day 0, spleens and sera were harvested and restimulated (as shown) in vitro in ELISPOT assays for INF-γ release from splenocytes (A) and serum antibody titers against 10 μg/well SMA560vIII exosomes in ELISA assays (B). C, D) Same assays as in A and B, except that mice either were survivors of primary and secondary subcutaneous tumor inoculations (following exosome vaccination), or were age-matched controls that were growing tumors at the time of tissue and sera harvest. Statistically significant differences in outputs are noted in the figure.
To examine responses in tumor-bearing and tumor-challenged mice (from Fig. 8C), we harvested spleens and sera from mice in the tumor rechallenge experiment 25 days after tumor rechallenge. This was the termination point, as the average tumor volume for the control (age-matched naive) mice was >2000 mm3. Again, the splenocytes and sera were used in ELISPOT and ELISA assays, respectively. Splenocytes were restimulated as in Fig. 9A. From the exosome-vaccinated mice (i.e., those that had rejected tumors both initially and on rechallenge), there was significant splenocyte production of IFN-γ on restimulation with sources of tumor antigen, including strong responses to the EGFRvIII peptide (Fig. 9C). The patterns of responses to peptide, lysate, and exosomes were similar to those of the naive mice, but the magnitudes of the outputs were 2 to 3 times higher than those from naive animals. Exosome-immunized mice that had rejected tumors also showed high-titer serum antibodies against SMA560vIII exosomes detected by ELISA (Fig. 9D). Thus, SMA560vIII exosome administration resulted in antibody production and T-cell activation. The presence of antitumor antibodies and T cells at the time of tumor inoculation apparently induced adequate tumor cell death to drive a further activated T-cell (and memory) response. In an intracranial, preestablished tumor setting, such responses were at insufficient levels and failed to impact tumor growth. Variable and generally low T-cell and antibody outputs accompanied treatments in the intracranial setting (data not shown).
Sera from patients with high-grade gliomas contain exosome-like entities
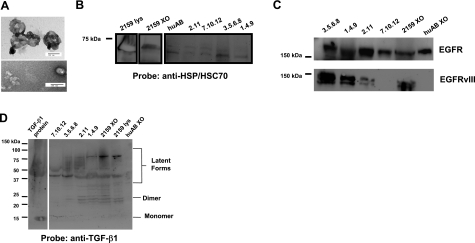
From our data here and previously (14), it is evident that brain tumor cell lines release exosomes when those cells are grown in culture. In addition, numerous studies have identified tumor-derived exosomes in malignant effusions (12), ascites (41), and sera (42) of cancer patients. Thus, it is reasonable to ask whether patients with brain tumors also have tumor-derived vesicles in their sera, as well as exosomes that are normally found there. One complicating factor in the clinical scenario of brain tumors might be the presence of the blood-brain barrier, which could theoretically prevent such vesicles from releasing into the periphery, and therefore render exosome-induced immune modulations only locally rather than systemically. By differential centrifugation, we isolated material from pooled sera of several patients with high-grade gliomas. The pellets from high-speed centrifugations bore physical characteristics of exosomes and microvesicles (e.g., membrane-enclosed vesicles seen in electron microscopy ranging from 50 nm to 200 nm in diameter, Fig. 10A) and biochemical characteristics as well (presence of HSP/HSC70, Fig. 10B). EGFR was present in the exosomes and microvesicles from both patient sera and normal serum (Fig. 10C, top) while the brain tumor-specific antigen EGFRvIII was present in exosomes and microvesicles from most of the patient sera; it was absent in vesicles from normal serum (Fig. 10C, bottom). This was also true for the immune modulatory cytokine TGF-β1 (Fig. 10D). Thus, “normal” exosomes and microvesicles contain HSP/HSC70 and EGFR, but neither EGFRvIII nor TGF-β1. These may be distinguishing features of glioma exosomes with biomarker implications, but their presence also supports the immune duality of these exosomes having both antigenic and suppressive components. Similar to previous reports (7), we did not detect DNA in our patient sera exosomes (data not shown).
Figure 10.
Tumor-derived exosomes and microvesicles are present in sera from patients with high-grade gliomas. Pooled sera from patients with high-grade gliomas were subjected to differential centrifugation as described in Materials and Methods. A) High-speed pellets were sectioned and visualized by electron microscopy (scale bar=100 nm), or were resuspended in SDS-PAGE sample buffer with 1% saponin, and boiled for 15 min. B–D) Maximum volumes of exosome samples were loaded (number-decimal samples are patient-derived; huAB refers to exosomes from human AB serum; 2159XO refers to exosomes from H2159MG cells; 2159 lys refers to lysate from those cells, 5 μg loaded). Proteins were separated on 10% Tris-glycine gels (B, C) or 4–20% gels (D) and blotted to nitrocellulose. Blots were probed with antibodies to canonical HSP/HSC70 (B), to EGFR (C, top), to EGFRvIII (mAb L8A4; C, bottom, and to TGF-β1 (D). Note that exosomes from human AB serum do not react with Abs vs. EGFRvIII or TGF-β1 but do react with Abs vs. EGFR and HSP/HSC70. Native human TGF-β1 protein (5 ng) was used as a positive control (D). Molecular mass markers are shown at left. Lines or boxes around blots indicate differences in exposure time or positioning of blot strips.
DISCUSSION
We previously isolated exosomes from several brain tumor cell lines, where their appearance in electron micrographs (membrane-bound vesicles of 30–100 nm diameter), and their heat shock protein content warranted the exosome designation (14). In this current work, we have further characterized exosomes from the murine brain tumor cell line SMA560vIII, which expresses the mutated EGFR variant 3—EGFRvIII. The mutated receptor is constitutively activated and oncogenic (38, 39), and it is also a tumor-specific antigen, and is capable of triggering specific immune responses (Fig. 9A, C). This antigen has also allowed us to identify tumor-derived vesicles harvested from the sera of patients with brain tumors (Fig. 10C), establishing that exosomes and microvesicles from gliomas access compartments external of the CNS. This is important in light of recent evidence that EGFRvIII can be transferred via microvesicles derived from tumor cells that possess the oncogenic receptor to tumor cells that lack expression of EGFRvIII, with concomitant activation of downstream signaling pathways and an increased aggressive phenotype (43). Microvesicles share similar properties with exosomes but are larger and are formed by budding from the plasma membrane. Thus, tumor-derived exosomes may elicit phenotype changes in adjacent or distant cells by horizontal transfer of membrane components, other proteins, or nucleic acids (7). Because glioma cells do not appear to exit the CNS (44), exosomes and/or other types of microvesicles may act as surrogates of cells that may influence processes and systems distant from the tumor site, which could include immune responses.
The immune impact of tumor exosomes is an ongoing controversy, and this glioma model system allowed us to examine exosome immune modulatory capacities, which have been the focus of many studies in cancer immunology (10, 45,46,47,48). This is particularly true for dendritic cell exosomes as cell-free vaccines (49, 50), including Phase I clinical trials (51, 52). There are reports of tumor-derived exosomes serving as antitumor vaccines (11, 12, 53, 54), but there has been considerable concern that tumor exosomes have immunosuppressive properties (13, 41, 55). The cellular “targets” of this immunosuppression include the innate responders such as natural killer cells (56, 57), effector T cells (58), and antigen-presenting cells (59, 60).
Our immunization results imply that brain tumor exosomes indeed provide immune stimulation involving both T-cell and antibody-based mechanisms. Immunization with SMA560vIII exosomes prior to subcutaneous tumor implantation led to tumor rejection and long-term immunological memory (Fig. 8A–C) with both T-cell activation and high-titer serum antibodies against exosomes (Fig. 9C, D). Because patients with high-grade gliomas are frequently profoundly immune suppressed (17), we considered the possibility that exosomes may play a role in that suppression. We had initially wondered whether such “vaccination” would actually induce immune suppression with reduced tumor latency and enhanced growth, but this did not appear to be the case. Analysis of twice-vaccinated mice just prior to tumor implantation revealed similar cellular and humoral responses, although somewhat muted compared to the outputs from the mice that successfully rejected their tumors (Fig. 9A, B). Reexposure to antigen in the form of injected tumor cells apparently served as a powerful boost, resulting in primary and secondary tumor rejections. However, in a more rigorous and realistic setting, with orthotopic intracranial tumor implantation followed by vaccine therapy 4 and 11 days later, we saw no effect from the exosome injections in terms of prolonged survival (Fig. 8D). Thus, the levels and/or the speed of the exosome-driven immune responses are insufficient to impede tumor progression under these conditions. Since exosome immunizations lead to antibody production (Fig. 9B, D), we speculate that antibody access to the intracranial tumor may be a limiting factor compared to subcutaneous tumors (32). Also, if there is an associated Th2 cytokine shift leading to antibody production at the expense of the Th1 cytokines necessary to generate activated cytotoxic T cells, the contribution of those lymphocytes may be minimized and ineffective against the intracranial tumor. While there has been a report of exosome immunization leading to protective antibody responses against murine tumors expressing the SARS virus S protein (61), those exosomes were derived from an engineered human cell line in the context of an antiviral vaccine. We believe our data here are the first to show that tumor exosome injections lead to antibody responses that may play a role in antitumor immunity.
Previously, we injected syngeneic mice with heat-shocked SMA560 cells and showed that those stressed tumor cells were rejected, presumably in an immune-based mechanism (14). Dying, stressed cells can provide “danger signals” leading to antitumor immune responses (62), so in this study, we wanted to ensure that our microvesicle immunizations consisted of classically defined exosomes from the SMA560vIII cells rather than cellular debris. Indeed, discerning immune responses to apoptotic bodies, membrane “blebs,” or other tumor-derived detritus could pose a challenge if the materials investigated are not well characterized. Vesicles released from these cells share biophysical and biochemical properties with exosomes derived from other cell types. Namely, the appearance, buoyant density, AChE activity, and protein content of these brain tumor-derived microvesicles are characteristic of exosomes (33, 34, 63) (Figs. 1A and 2A; Table 1). These brain tumor exosomes also exhibit a previously unreported property in that as intact particles, they possess a very alkaline isoelectric point (Fig. 1B), which contrasts with the pIs of the overall protein population as seen in 2-D gels (Fig. 5), indicating that the lipid component of vesicles may be a dominant physical property. The significance of this is unclear, but it likely influences exosome interactions with cells both proximal and distal. The strongly basic pI may prove attractive to acidic cell surface components, such as proteoglycans and glycosaminoglycans, which are frequently used as receptors by viruses (64). We suspect that, like viruses, exosomes may have profound impacts on the activities of recipient cells in terms of acquisition of new proteins and even nucleic acid information that may alter recipient cell phenotypes (7, 65). The cellular targets and downstream effects of brain tumor exosomes will be an area of great research interest.
As part of our characterization of novel brain tumor exosomes, we conducted the first proteomic analysis of them. We identified more than 40 proteins by Western blot analysis (Figs. 2, 3), by bead FACS (Fig. 4), and by 2-D PAGE and MS/MS techniques (Fig. 5, Table 1). Overall, our proteomic profile is consistent with other reports, including 4 of the 5 enzymes of the “payoff” phase of glycolysis (the 5th member, step 3 enzyme phosphoglycerate mutase, has been found in other exosomes types; refs. 66, 67). These glycolytic enzymes have been isolated as complexes from the cytosolic side of plasma membrane preparations of glioma cells (68). In general, numerous proteins that we identified are components of oligomeric complexes (major vault protein, TCP-1/HSP60, proteasome subunit α, actin, tubulin, 40S ribosomal protein SA, elongation, and translation factors, and ferritin). The formation of complexes may contribute to the accumulation of such proteins in exosomes (69) as higher-order oligomers. Interestingly, that publication demonstrated the targeting of various viral and retroviral Gag proteins to exosomes, blurring the distinction between viral particles and exosomes. We also identified a murine retroviral Gag polyprotein (pr65) and its precursor (Table 1); this particular retrovirus was implicated in progression of murine melanoma by subversion of immune surveillance (70). Intriguingly, human cytomegalovirus (HCMV) has been identified in patient gliomas and HCMV DNA found in their peripheral blood (71). One may speculate that the HCMV DNA from these patients may originate from tumor exosomes acting as viral particles; the implications of this scenario are manifold.
We also categorized the proteomic results by subcellular and extracellular localization and function (Fig. 6), and with network analyses (Fig. 7). Many of these proteins are multifunctional members of complexes and appear in various subcellular and extracellular locations, e.g., the heat shock proteins. HSPs are expressed throughout cellular organelles and on tumor cell surfaces, with diverse roles in protein folding and innate immune stimulus (72). α-enolase (a glycolytic enzyme) and actin (a structural protein) have been shown to be cell surface plasminogen and angiostatin receptors as well (73, 74). Thus, our assessments of the rigidity of these categories may necessitate more windows and fewer walls.
The networks arising from our pathway analyses provided anticipated interactions (e.g., the HSPs as major nodes surrounding signaling molecules, Fig. 7A), as well as known associations between the transcription factor SP1 and the genes of glycolysis (75). The networks also may help establish roles for proteins that lack distinct functions, such as GPNMB (intertwined with CD40, IFN-γ, IL-6, and IL-1β, Fig. 7B), or which may be involved in numerous activities depending on context. Curiously, one of the roles noted for GPNMB in mice is in the formation of melanosomes, where GPNMB mutations disrupt those vesicles and lead to pigmentary glaucoma (76). Since IL-1β is an immunocytokine that is secreted without a known signal sequence (77), it is conceivable that GPNMB plays a role in vesicular release of IL-1β via exosomes. Another indirect relationship between TGF-β and α-enolase suggests that the growth factor, which has known influences on the expression of neural-specific enolase (78, 79), may also have impacts on the non-neural glycolytic protein as well. Indeed, a number of metabolic enzymes feed into signaling molecule pathways in our network models, suggesting that exosomes may represent a mechanism for linkage of metabolism and intracellular/extracellular signaling. As the relationship between NF-κB and ApoA-1 may play roles in lipid metabolism (80, 81), transport of ApoA-1 via exosomes may work to either distribute ApoA-1 to other cells, or to expel it from the cells, depending on NF-κB activation status and the lipid biochemistry of the cells. This could also influence the production of exosomes or other vesicles. Our traditional views of the functions and territories of many proteins may be vastly oversimplified and will require network webs to fully appreciate the impact.
Brain tumor exosomes displayed surface HSPs (Fig. 4); since exosomes are topographically identical to the cell, cell surface proteins may become exosome surface proteins. We had previously reported HSPs on brain tumor cell surfaces (14), and such HSP localizations have been described before for exosomes from colon and pancreatic carcinoma cell lines that display HSP70 on their surfaces (34, 82). However, we believe this is the first report of HSP27 on exosome surfaces. The exosome surface HSPs, as well as other known tumor surface antigens (e.g., EGFRvIII), may be exploitable biomarkers if these exosomes are present in accessible fluids such as blood or cerebrospinal fluid. Indeed, we have isolated exosomes and microvesicles from sera of patients with gliomas and have shown that those vesicles possess EGFR and EGFRvIII, as well as HSP/HSC70 (typically found in exosomes) and TGF-β1. EGFRvIII and TGF-β1 may distinguish tumor-derived exosomes from those normally found in serum and may serve as tumor biomarkers (Fig. 10). Such exosome and microvesicle biomarker searches are ongoing in urine and amniotic fluid (83). There is also the intriguing possibility of using exosome-cloaked nucleic acids (e.g., mRNAs or miRNAs) as sources of biomarkers as well (7, 65). Please note that we have used the term “microvesicle” to describe the small membranous materials that we isolated from patient sera. Those vesicles were slightly larger and more amorphous in appearance in electron microscopy (Fig. 10A) than we had seen for tissue culture-derived exosomes, and thus there may be microvesicles in that population. As mentioned above, these distinctions are subtle and somewhat controversial (84), and at some point will likely become semantic.
Exosome surface HSPs may contribute to exosome reactivity with antigen-presenting cells or with natural killer cells; on the other hand, cell surface HSP27 has been associated with increased tumor aggressiveness (85), so further study of exosome surface HSPs will be of great interest. Although exosome surface HSPs may promote interactions with and/or internalization by antigen-presenting cells, the rest of the exosomal content may determine whether the outcome is immune activation or immune suppression (86). TGF-β in exosomes (Figs. 2D and 10D) may be one of the factors that leads to immune suppression, particularly if exosomes are a protected systemic reservoir of the cytokine.
The balance between immunity and immunosuppression may be intimately bound up in exosomes. As our results indicate, the nature and overall effectiveness of the immunity are likely to be influenced by a variety of extrinsic and intrinsic factors. These would include the context and timing of the tumor implantation as related to the window of immune opportunity of the exosome vaccine. The antigen content and immune-stimulatory components of the exosomes are undoubtedly crucial, but possibly countered by the immunoregulatory factors such as Fas ligand, lipids, viruses (70), or even TGF-β activity modulation (e.g., by eIF3β; ref. 87). On the basis of the antibody responses seen in our experiments, it is conceivable that exosomes released by tumors in vivo may lead to host antibody production. However, if the tumor target is difficult to access (e.g., in the brain), and/or if there is a constant source of “decoy” tumors (i.e., the exosomes) to saturate circulating antibody, such immunity would be essentially futile. The shift of immune attention to antibody production and away from cytotoxic T-cell activity would also presumably benefit the tumor. All of these considerations are currently theoretical and merit further study. Brain tumor exosomes may maintain functions inherent in exosome neurobiology, in which inflammation is tightly controlled and immune modulation works to protect the CNS. It may be these same capacities that brain tumor exosomes exploit to reduce antitumor immune responses; an improved understanding of brain tumor exosomes may provide us with more therapeutic avenues to treat these dreadful diseases.
Acknowledgments
The authors thank Dr. Sara Miller of the Duke University Cancer Center electron microscopy facility for performing the electron microscopy; R. Ian Cumming, Shelley Davis, and Nichole Satterwhite for superb technical support; and Dr. Steve Griffiths (Atlantic Cancer Research Institute, Moncton, NB, Canada) for ideas and sharing unpublished observations. This work was supported by the Duke University Brain Cancer Specialized Programs of Research Excellence 5 P50 CA108786-02; SPORE Career Development Award (to M.W.G.); National Institute of Neurological Disorders and Stroke SRC 5 P50 20023-21; the Pediatric Brain Tumor Foundation; the Brian Cless Research Foundation; and the Southeast Brain Tumor Foundation (to M.W.G.).
References
- Stupp R, Mason W P, van den Bent M J, Weller M, Fisher B, Taphoorn M J, Belanger K, Brandes A A, Marosi C, Bogdahn U, Curschmann J, Janzer R C, Ludwin S K, Gorlia T, Allgeier A, Lacombe D, Cairncross J G, Eisenhauer E, Mirimanoff R O. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64:479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- Keller S, Sanderson M P, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Calzolari A, Raggi C, Deaglio S, Sposi N M, Stafsnes M, Fecchi K, Parolini I, Malavasi F, Peschle C, Sargiacomo M, Testa U. TfR2 localizes in lipid raft domains and is released in exosomes to activate signal transduction along the MAPK pathway. J Cell Sci. 2006;119:4486–4498. doi: 10.1242/jcs.03228. [DOI] [PubMed] [Google Scholar]
- Safaei R, Larson B J, Cheng T C, Gibson M A, Otani S, Naerdemann W, Howell S B. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther. 2005;4:1595–1604. doi: 10.1158/1535-7163.MCT-05-0102. [DOI] [PubMed] [Google Scholar]
- Pelchen-Matthews A, Raposo G, Marsh M. Endosomes, exosomes and Trojan viruses. Trends Microbiol. 2004;12:310–316. doi: 10.1016/j.tim.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee J J, Lotvall J O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Johnstone R M. Exosomes biological significance: a concise review. Blood Cells Mol Dis. 2006;36:315–321. doi: 10.1016/j.bcmd.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Denzer K, Kleijmeer M J, Heijnen H F, Stoorvogel W, Geuze H J. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci. 2000;113:3365–3374. doi: 10.1242/jcs.113.19.3365. [DOI] [PubMed] [Google Scholar]
- Mignot G, Roux S, Thery C, Segura E, Zitvogel L. Prospects for exosomes in immunotherapy of cancer. J Cell Mol Med. 2006;10:376–388. doi: 10.1111/j.1582-4934.2006.tb00406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, Angevin E, Amigorena S, Zitvogel L. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7:297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- Andre F, Schartz N E, Movassagh M, Flament C, Pautier P, Morice P, Pomel C, Lhomme C, Escudier B, Le Chevalier T, Tursz T, Amigorena S, Raposo G, Angevin E, Zitvogel L. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360:295–305. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- Valenti R, Huber V, Iero M, Filipazzi P, Parmiani G, Rivoltini L. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 2007;67:2912–2915. doi: 10.1158/0008-5472.CAN-07-0520. [DOI] [PubMed] [Google Scholar]
- Graner M W, Cumming R I, Bigner D D. The heat shock response and chaperones/heat shock proteins in brain tumors: surface expression, release, and possible immune consequences. J Neurosci. 2007;27:11214–11227. doi: 10.1523/JNEUROSCI.3588-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D D, Gercel-Taylor C, Lyons K S, Stanson J, Whiteside T L. T-cell apoptosis and suppression of T-cell receptor/CD3-zeta by Fas ligand-containing membrane vesicles shed from ovarian tumors. Clin Cancer Res. 2003;9:5113–5119. [PubMed] [Google Scholar]
- Gabrilovich D. Mechanisms and functional significance of tumour-induced dendritic-cell defects. Nat Rev Immunol. 2004;4:941–952. doi: 10.1038/nri1498. [DOI] [PubMed] [Google Scholar]
- Fecci P E, Mitchell D A, Whitesides J F, Xie W, Friedman A H, Archer G E, Herndon J E, 2nd, Bigner D D, Dranoff G, Sampson J H. Increased regulatory T-cell fraction amidst a diminished CD4 compartment explains cellular immune defects in patients with malignant glioma. Cancer Res. 2006;66:3294–3302. doi: 10.1158/0008-5472.CAN-05-3773. [DOI] [PubMed] [Google Scholar]
- Gomez G G, Kruse C A. Mechanisms of malignant glioma immune resistance and sources of immunosuppression. Gene Ther Mol Biol. 2006;10:133–146. [PMC free article] [PubMed] [Google Scholar]
- Serano R D, Pegram C N, Bigner D D. Tumorigenic cell culture lines from a spontaneous VM/Dk murine astrocytoma (SMA) Acta Neuropathol (Berl) 1980;51:53–64. doi: 10.1007/BF00688850. [DOI] [PubMed] [Google Scholar]
- Pedersen M W, Meltorn M, Damstrup L, Poulsen H S. The type III epidermal growth factor receptor mutation. Biological significance and potential target for anti-cancer therapy Ann. Oncol. 2001;12:745–760. doi: 10.1023/a:1011177318162. [DOI] [PubMed] [Google Scholar]
- Garg P K, John C S, Zalutsky M R. Preparation and preliminary evaluation of 4-[211At]astato-N-piperidinoethyl benzamide. Nucl Med Biol. 1995;22:467–473. doi: 10.1016/0969-8051(94)00134-6. [DOI] [PubMed] [Google Scholar]
- Friedman H S, Burger P C, Bigner S H, Trojanowski J Q, Wikstrand C J, Halperin E C, Bigner D D. Establishment and characterization of the human medulloblastoma cell line and transplantable xenograft D283 Med. J Neuropathol Exp Neurol. 1985;44:592–605. doi: 10.1097/00005072-198511000-00005. [DOI] [PubMed] [Google Scholar]
- Sampson J H, Ashley D M, Archer G E, Fuchs H E, Dranoff G, Hale L P, Bigner D D. Characterization of a spontaneous murine astrocytoma and abrogation of its tumorigenicity by cytokine secretion. Neurosurgery. 1997;41:1365–1372. doi: 10.1097/00006123-199712000-00024. discussion 1372–1363. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland A B, Shi Q, McLendon R E, Bigner D D, Rich J N. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006;66:7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- Khan W A, Dechkovskaia A M, Herrick E A, Jones K H, Abou-Donia M B. Acute sarin exposure causes differential regulation of choline acetyltransferase, acetylcholinesterase, and acetylcholine receptors in the central nervous system of the rat. Toxicol Sci. 2000;57:112–120. doi: 10.1093/toxsci/57.1.112. [DOI] [PubMed] [Google Scholar]
- Kuan C T, Wakiya K, Dowell J M, Herndon J E, 2nd, Reardon D A, Graner M W, Riggins G J, Wikstrand C J, Bigner D D. Glycoprotein nonmetastatic melanoma protein B, a potential molecular therapeutic target in patients with glioblastoma multiforme. Clin Cancer Res. 2006;12:1970–1982. doi: 10.1158/1078-0432.CCR-05-2797. [DOI] [PubMed] [Google Scholar]
- Wikstrand C J, McLendon R E, Friedman A H, Bigner D D. Cell surface localization and density of the tumor-associated variant of the epidermal growth factor receptor, EGFRvIII. Cancer Res. 1997;57:4130–4140. [PubMed] [Google Scholar]
- Osorio C, Sullivan P M, He D N, Mace B E, Ervin J F, Strittmatter W J, Alzate O. Mortalin is regulated by APOE in hippocampus of AD patients and by human APOE in TR mice. Neurobiol Aging. 2007;28:1853–1862. doi: 10.1016/j.neurobiolaging.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Pinaud R, Osorio C, Alzate O, Jarvis E D. Profiling of experience-regulated proteins in the songbird auditory forebrain using quantitative proteomics. Eur J Neurosci. 2008;27:1409–1422. doi: 10.1111/j.1460-9568.2008.06102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C E, Warren M R, Loiselle D R, Dicheva N N, Scarlett C O, Borchers C H. Identification of components of protein complexes. Methods Mol Biol. 2005;301:117–151. doi: 10.1385/1-59259-895-1:117. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Sampson J H, Crotty L E, Lee S, Archer G E, Ashley D M, Wikstrand C J, Hale L P, Small C, Dranoff G, Friedman A H, Friedman H S, Bigner D D. Unarmed, tumor-specific monoclonal antibody effectively treats brain tumors. Proc Natl Acad Sci U S A. 2000;97:7503–7508. doi: 10.1073/pnas.130166597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gassart A, Geminard C, Fevrier B, Raposo G, Vidal M. Lipid raft-associated protein sorting in exosomes. Blood. 2003;102:4336–4344. doi: 10.1182/blood-2003-03-0871. [DOI] [PubMed] [Google Scholar]
- Gastpar R, Gehrmann M, Bausero M A, Asea A, Gross C, Schroeder J A, Multhoff G. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res. 2005;65:5238–5247. doi: 10.1158/0008-5472.CAN-04-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- Thery C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J Cell Biol. 1999;147:599–610. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Drwal G, Bourgeois T, Saltz J, Wu H M. Distinct proteome features of plasma microparticles. Proteomics. 2005;5:1940–1952. doi: 10.1002/pmic.200401057. [DOI] [PubMed] [Google Scholar]
- Moscatello D K, Ramirez G, Wong A J. A naturally occurring mutant human epidermal growth factor receptor as a target for peptide vaccine immunotherapy of tumors. Cancer Res. 1997;57:1419–1424. [PubMed] [Google Scholar]
- Kuan C T, Wikstrand C J, Bigner D D. EGFRvIII as a promising target for antibody-based brain tumor therapy. Brain Tumor Pathol. 2000;17:71–78. doi: 10.1007/BF02482738. [DOI] [PubMed] [Google Scholar]
- Wikstrand C J, Hale L P, Batra S K, Hill M L, Humphrey P A, Kurpad S N, McLendon R E, Moscatello D, Pegram C N, Reist C J, Traweek S T, Wong A J, Zalutsky M R, Bigner D D. Monoclonal antibodies against EGFRvIII are tumor specific and react with breast and lung carcinomas and malignant gliomas. Cancer Res. 1995;55:3140–3148. [PubMed] [Google Scholar]
- Taylor D D, Gercel-Taylor C. Tumour-derived exosomes and their role in cancer-associated T-cell signalling defects. Br J Cancer. 2005;92:305–311. doi: 10.1038/sj.bjc.6602316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J W, Wieckowski E, Taylor D D, Reichert T E, Watkins S, Whiteside T L. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin Cancer Res. 2005;11:1010–1020. [PubMed] [Google Scholar]
- Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- Mourad P D, Farrell L, Stamps L D, Chicoine M R, Silbergeld D L. Why are systemic glioblastoma metastases rare? Systemic and cerebral growth of mouse glioblastoma. Surg Neurol. 2005;63:511–519. doi: 10.1016/j.surneu.2004.08.062. discussion 519. [DOI] [PubMed] [Google Scholar]
- Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- Clayton A, Tabi Z. Exosomes and the MICA-NKG2D system in cancer. Blood Cells Mol Dis. 2005;34:206–213. doi: 10.1016/j.bcmd.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Kim S H, Bianco N, Menon R, Lechman E R, Shufesky W J, Morelli A E, Robbins P D. Exosomes derived from genetically modified DC expressing FasL are anti-inflammatory and immunosuppressive. Mol Ther. 2006;13:289–300. doi: 10.1016/j.ymthe.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Delcayre A, Le Pecq J B. Exosomes as novel therapeutic nanodevices. Curr Opin Mol Ther. 2006;8:31–38. [PubMed] [Google Scholar]
- Taieb J, Chaput N, Zitvogel L. Dendritic cell-derived exosomes as cell-free peptide-based vaccines. Crit Rev Immunol. 2005;25:215–223. doi: 10.1615/critrevimmunol.v25.i3.30. [DOI] [PubMed] [Google Scholar]
- Chaput N, Flament C, Viaud S, Taieb J, Roux S, Spatz A, Andre F, LePecq J B, Boussac M, Garin J, Amigorena S, Thery C, Zitvogel L. Dendritic cell derived-exosomes: biology and clinical implementations. J Leukoc Biol. 2006;80:471–478. doi: 10.1189/jlb.0206094. [DOI] [PubMed] [Google Scholar]
- Morse M A, Garst J, Osada T, Khan S, Hobeika A, Clay T M, Valente N, Shreeniwas R, Sutton M A, Delcayre A, Hsu D H, Le Pecq J B, Lyerly H K. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. [Online] J Transl Med. 2005;3:9. doi: 10.1186/1479-5876-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudier B, Dorval T, Chaput N, Andre F, Caby M P, Novault S, Flament C, Leboulaire C, Borg C, Amigorena S, Boccaccio C, Bonnerot C, Dhellin O, Movassagh M, Piperno S, Robert C, Serra V, Valente N, Le Pecq J B, Spatz A, Lantz O, Tursz T, Angevin E, Zitvogel L. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of thefirst phase I clinical trial. [Online] J Transl Med. 2005;3:10. doi: 10.1186/1479-5876-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altieri S L, Khan A N, Tomasi T B. Exosomes from plasmacytoma cells as a tumor vaccine. J Immunother. 2004;27:282–288. doi: 10.1097/00002371-200407000-00004. [DOI] [PubMed] [Google Scholar]
- Bu N, Li Q L, Feng Q, Sun B Z. Immune protection effect of exosomes against attack of L1210 tumor cells. Leuk Lymphoma. 2006;47:913–918. doi: 10.1080/10428190500376191. [DOI] [PubMed] [Google Scholar]
- Wieckowski E, Whiteside T L. Human tumor-derived vs dendritic cell-derived exosomes have distinct biologic roles and molecular profiles. Immunol Res. 2006;36:247–254. doi: 10.1385/IR:36:1:247. [DOI] [PubMed] [Google Scholar]
- Liu C, Yu S, Zinn K, Wang J, Zhang L, Jia Y, Kappes J C, Barnes S, Kimberly R P, Grizzle W E, Zhang H G. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J Immunol. 2006;176:1375–1385. doi: 10.4049/jimmunol.176.3.1375. [DOI] [PubMed] [Google Scholar]
- Clayton A, Mitchell J P, Court J, Mason M D, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 2007;67:7458–7466. doi: 10.1158/0008-5472.CAN-06-3456. [DOI] [PubMed] [Google Scholar]
- Abusamra A J, Zhong Z, Zheng X, Li M, Ichim T E, Chin J L, Min W P. Tumor exosomes expressing Fas ligand mediate CD8+ T-cell apoptosis. Blood Cells Mol Dis. 2005;35:169–173. doi: 10.1016/j.bcmd.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Valenti R, Huber V, Filipazzi P, Pilla L, Sovena G, Villa A, Corbelli A, Fais S, Parmiani G, Rivoltini L. Human tumor-released microvesicles promote the differentiation of myeloid cells with transforming growth factor-beta-mediated suppressive activity on T lymphocytes. Cancer Res. 2006;66:9290–9298. doi: 10.1158/0008-5472.CAN-06-1819. [DOI] [PubMed] [Google Scholar]
- Yu S, Liu C, Su K, Wang J, Liu Y, Zhang L, Li C, Cong Y, Kimberly R, Grizzle W E, Falkson C, Zhang H G. Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J Immunol. 2007;178:6867–6875. doi: 10.4049/jimmunol.178.11.6867. [DOI] [PubMed] [Google Scholar]
- Kuate S, Cinatl J, Doerr H W, Uberla K. Exosomal vaccines containing the S protein of the SARS coronavirus induce high levels of neutralizing antibodies. Virology. 2007;362:26–37. doi: 10.1016/j.virol.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Zeng Y, Whitesell L, Katsanis E. Stressed apoptotic tumor cells express heat shock proteins and elicit tumor-specific immunity. Blood. 2001;97:3505–3512. doi: 10.1182/blood.v97.11.3505. [DOI] [PubMed] [Google Scholar]
- Savina A, Vidal M, Colombo M I. The exosome pathway in K562 cells is regulated by Rab11. J Cell Sci. 2002;115:2505–2515. doi: 10.1242/jcs.115.12.2505. [DOI] [PubMed] [Google Scholar]
- Olofsson S, Bergstrom T. Glycoconjugate glycans as viral receptors. Ann Med. 2005;37:154–172. doi: 10.1080/07853890510007340. [DOI] [PubMed] [Google Scholar]
- Keene J D. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Pisitkun T, Shen R F, Knepper M A. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears R, Craven R A, Hanrahan S, Totty N, Upton C, Young S L, Patel P, Selby P J, Banks R E. Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics. 2004;4:4019–4031. doi: 10.1002/pmic.200400876. [DOI] [PubMed] [Google Scholar]
- Daum G, Keller K, Lange K. Association of glycolytic enzymes with the cytoplasmic side of the plasma membrane of glioma cells. Biochim Biophys Acta. 1988;939:277–281. doi: 10.1016/0005-2736(88)90071-5. [DOI] [PubMed] [Google Scholar]
- Fang Y, Wu N, Gan X, Yan W, Morrell J C, Gould S J. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. [Online] PLoS Biol. 2007;5:e158. doi: 10.1371/journal.pbio.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeney M, Pothlichet J, Renard M, Ducos B, Heidmann T. Endogenous retrovirus expression is required for murine melanoma tumor growth in vivo. Cancer Res. 2005;65:2588–2591. doi: 10.1158/0008-5472.CAN-04-4231. [DOI] [PubMed] [Google Scholar]
- Mitchell D A, Xie W, Schmittling R, Learn C, Friedman A, McLendon R E, Sampson J H. Sensitive detection of human cytomegalovirus in tumors and peripheral blood of patients diagnosed with glioblastoma. Neuro-oncol. 2007;10:10–18. doi: 10.1215/15228517-2007-035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graner M W, Bigner D D. Chaperone proteins and brain tumors: potential targets and possible therapeutics. Neuro-oncol. 2005;7:260–278. doi: 10.1215/S1152851704001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancholi V. Multifunctional alpha-enolase: its role in diseases. Cell Mol Life Sci. 2001;58:902–920. doi: 10.1007/PL00000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudani A K, Ben-Tchavtchavadze M, Porter S, Tackaberry E. Angiostatin and plasminogen share binding to endothelial cell surface actin. Biochem Cell Biol. 2005;83:28–35. doi: 10.1139/o04-109. [DOI] [PubMed] [Google Scholar]
- Girard J, Ferre P, Foufelle F. Mechanisms by which carbohydrates regulate expression of genes for glycolytic and lipogenic enzymes. Annu Rev Nutr. 1997;17:325–352. doi: 10.1146/annurev.nutr.17.1.325. [DOI] [PubMed] [Google Scholar]
- Anderson M G, Smith R S, Hawes N L, Zabaleta A, Chang B, Wiggs J L, John S W. Mutations in genes encoding melanosomal proteins cause pigmentary glaucoma in DBA/2J mice. Nat Genet. 2002;30:81–85. doi: 10.1038/ng794. [DOI] [PubMed] [Google Scholar]
- Rubartelli A, Bajetto A, Allavena G, Cozzolino F, Sitia R. Post-translational regulation of interleukin 1 beta secretion. Cytokine. 1993;5:117–124. doi: 10.1016/1043-4666(93)90050-f. [DOI] [PubMed] [Google Scholar]
- Yang G, Timme T L, Park S H, Thompson T C. Transforming growth factor beta 1 transduced mouse prostate reconstitutions: I. Induction of neuronal phenotypes Prostate. 1997;33:151–156. doi: 10.1002/(sici)1097-0045(19971101)33:3<151::aid-pros1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Burchardt T, Burchardt M, Chen M W, Cao Y, de la Taille A, Shabsigh A, Hayek O, Dorai T, Buttyan R. Transdifferentiation of prostate cancer cells to a neuroendocrine cell phenotype in vitro and in vivo. J Urol. 1999;162:1800–1805. [PubMed] [Google Scholar]
- Morishima A, Ohkubo N, Maeda N, Miki T, Mitsuda N. NF-κB regulates plasma apolipoprotein A-I and high density lipoprotein cholesterol through inhibition of peroxisome proliferator-activated receptor alpha. J Biol Chem. 2003;278:38188–38193. doi: 10.1074/jbc.M306336200. [DOI] [PubMed] [Google Scholar]
- Gerbod-Giannone M C, Li Y, Holleboom A, Han S, Hsu L C, Tabas I, Tall A R. TNFα induces ABCA1 through NF-κB in macrophages and in phagocytes ingesting apoptotic cells. Proc Natl Acad Sci U S A. 2006;103:3112–3117. doi: 10.1073/pnas.0510345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsner L, Muppala V, Gehrmann M, Lozano J, Malzahn D, Bickeboller H, Brunner E, Zientkowska M, Herrmann T, Walter L, Alves F, Multhoff G, Dressel R. The heat shock protein HSP70 promotes mouse NK cell activity against tumors that express inducible NKG2D ligands. J Immunol. 2007;179:5523–5533. doi: 10.4049/jimmunol.179.8.5523. [DOI] [PubMed] [Google Scholar]
- Keller S, Rupp C, Stoeck A, Runz S, Fogel M, Lugert S, Hager H D, Abdel-Bakky M S, Gutwein P, Altevogt P. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007;72:1095–1102. doi: 10.1038/sj.ki.5002486. [DOI] [PubMed] [Google Scholar]
- Simpson R J, Jensen S S, Lim J W. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- Bausero M A, Page D T, Osinaga E, Asea A. Surface expression of Hsp25 and Hsp72 differentially regulates tumor growth and metastasis. Tumour Biol. 2004;25:243–251. doi: 10.1159/000081387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iero M, Valenti R, Huber V, Filipazzi P, Parmiani G, Fais S, Rivoltini L. Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ. 2007;15:80–88. doi: 10.1038/sj.cdd.4402237. [DOI] [PubMed] [Google Scholar]
- Choy L, Derynck R. The type II transforming growth factor (TGF)-beta receptor-interacting protein TRIP-1 acts as a modulator of the TGF-beta response. J Biol Chem. 1998;273:31455–31462. doi: 10.1074/jbc.273.47.31455. [DOI] [PubMed] [Google Scholar]
- Bard M P, Hegmans J P, Hemmes A, Luider T M, Willemsen R, Severijnen L A, van Meerbeeck J P, Burgers S A, Hoogsteden H C, Lambrecht B N. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am J Respir Cell Mol Biol. 2004;31:114–121. doi: 10.1165/rcmb.2003-0238OC. [DOI] [PubMed] [Google Scholar]
- Hegmans J P, Bard M P, Hemmes A, Luider T M, Kleijmeer M J, Prins J B, Zitvogel L, Burgers S A, Hoogsteden H C, Lambrecht B N. Proteomic analysis of exosomes secreted by human mesothelioma cells. Am J Pathol. 2004;164:1807–1815. doi: 10.1016/S0002-9440(10)63739-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Niel G, Raposo G, Candalh C, Boussac M, Hershberg R, Cerf-Bensussan N, Heyman M. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology. 2001;121:337–349. doi: 10.1053/gast.2001.26263. [DOI] [PubMed] [Google Scholar]
- Wubbolts R, Leckie R S, Veenhuizen P T, Schwarzmann G, Mobius W, Hoernschemeyer J, Slot J W, Geuze H J, Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation J. Biol. Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- Segura E, Nicco C, Lombard B, Veron P, Raposo G, Batteux F, Amigorena S, Thery C. ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood. 2005;106:216–223. doi: 10.1182/blood-2005-01-0220. [DOI] [PubMed] [Google Scholar]
- Potolicchio I, Carven G J, Xu X, Stipp C, Riese R J, Stern L J, Santambrogio L. Proteomic analysis of microglia-derived exosomes: metabolic role of the aminopeptidase CD13 in neuropeptide catabolism. J Immunol. 2005;175:2237–2243. doi: 10.4049/jimmunol.175.4.2237. [DOI] [PubMed] [Google Scholar]
- Zhou H, Yuen P S, Pisitkun T, Gonzales P A, Yasuda H, Dear J W, Gross P, Knepper M A, Star R A. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 2006;69:1471–1476. doi: 10.1038/sj.ki.5000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertova E, Chertov O, Coren L V, Roser J D, Trubey C M, Bess J W, Jr, Sowder R C, 2nd, Barsov E, Hood B L, Fisher R J, Nagashima K, Conrads T P, Veenstra T D, Lifson J D, Ott D E. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]