Abstract
Recombineering technology allows the modification of large DNA constructs without using restriction enzymes, enabling the use of bacterial artificial chromosomes (BACs) in genetic engineering of animals and plants as well as in the studies of structures and functions of chromosomal elements in DNA replication and transcription. Here, we report a new selection scheme of BAC recombineering. A dual kanamycin and streptomycin selection marker was constructed using the kanamycin resistance gene and bacterial rpsL+ gene. Recombination cassettes generated using this dual marker were used to make precise modifications in BAC constructs in a two-step procedure without leaving behind any unwanted sequences. The dual marker was first inserted into the site of modifications by positive selection of kanamycin resistance. In the second step, the counter-selection of streptomycin sensitivity resulted in the replacement of the dual marker with intended modified sequences. This method of BAC modification worked as efficiently as the previously reported galK method and provided a faster and more cost-effective alternative to the galK method.
Keywords: Bacterial artificial chromosome, recombineering, homologous recombination, kanamycin and streptomycin selection
Introduction
The completion of the Human Genome Project has not only yielded an extraordinary amount of primary sequence data but also generated a huge selection of well-mapped constructs containing large genomic DNA inserts in the forms of bacterial artificial chromosomes (BACs), P1 artificial chromosomes (PACs), and yeast artificial chromosomes (YACs). These large constructs carry sufficient DNA sequences to encompass most eukaryotic genes, including all cis-acting regulatory elements and many eukaryotic gene clusters. Such constructs, BACs in particular, have become indispensable tools for the mapping of genomic sequences, genetic engineering of animals and plants, and analysis of structures and functions of genomic elements essential for DNA replication, recombination, and transcription (1–4). However, conventional molecular cloning methods using restriction enzymes are unsuitable for modifications of these constructs, due to their large sizes. The development of phage-based Escherichia coli homologous recombination systems, also called recombination-mediated engineering or recombineering, has enabled a wide variety of modifications of large DNA constructs that were virtually impossible in the past (5,6). The two widely used recombineering systems are based on bacterial phage-encoded resombinases, one uses episomal plasmids to supply RecE/RecT of the Rac phage (5,7), and the other utilizes a temperature-sensitive λ repressor to control the expression of recombinases from a λ prophage (7–9). In addition, multiple variations of the technique have been developed (10–12).
In the λ-prophage-based Red recombination system, gam, bet and exo are expressed from a λ-prophage in E. coli host strain DY380, and their expressions are under tight control of the temperature-sensitive λ-cI857 repressor. At 32°C, the recombination system is inactive because of the active λ repressor. Upon shifting to 42°C, the λ repressor becomes inactivated and the recombinases are coordinately expressed from the pL promoter, allowing homologous recombination to occur. Because homologous recombination is an infrequent event even in the presence of recombinases, a selectable marker would be necessary for introducing mutations or other modifications to BAC constructs. Previously, selection markers were often flanked by site-specific recombination target sites (SSRTs), such as loxP or FRT sites. These sites were then removed in a subsequent recombination by inducing the expression of Cre or Flp recombinase with arabinose in modified DY380 cells, EL250 or EL350, respectively (8). While this strategy was efficient, the leftover single loxP or FRT sites might prohibit additional modifications using the same sites. The unwanted sequences might also complicate the interpretation of experimental data. To solve this problem, two-step positive/negative selection schemes were developed, allowing precise modifications of BAC constructs. Muyrers et al. reported a method using neomycin and SacB as positive and negative selection markers, respectively (7). However, the vector backbones (pBACe3.6 and pTARBAC series) in most currently available BAC libraries contain a SacB gene, making it unsuitable for modifying many BAC constructs. Recently, the Copeland laboratory have developed a galK positive/negative selection method, involving a positive selection in minimal medium and a negative selection in 2-deoxy-galactose (DOG) in special bacterial hosts with a deletion at the galK locus (13).
With the need of making precise BAC modifications for studying telomerase gene regulation, we have also developed a new selection scheme for a two-step recombineering procedure, using a positive kanamycin-resistance marker and a negative streptomycin-sensitive rpsL marker. This strategy, which was developed before the galK method was published and has been used routinely in our laboratory for more than five years, conferred several advantages over the galK selection scheme. Because the selections are carried out in the regular Luria–Bertani (LB) medium and do not require the use of minimal medium and DOG, the kanamycin/streptomycin selection strategy is a faster and more cost-effective alternative to the galK selection scheme.
Materials and Methods
Bacterial strain and BAC clones
DY380, which contains a defective λ prophage with cI857 repressor, was generously provided by the Copeland laboratory at the National Cancer Institute. BAC clones, RP24-183M22, RP23-412H3, RP11-117B23, and RP11-478M20, were purchased from Research Genetics, Inc.
Generation of a positive/negative selection marker
The selectable marker, rpsL+-kana, was created by placing the E.coli ribosomal S12 gene (rpsL) in front of the aminoglycoside phosphotransferase gene (aph). The RpsL expression results in streptomycin sensitivity and is referred to as rpsL+, whereas the Aph expression results in kanamycin resistance and is referred to as kana. To generate the dual marker, the rpsL+ gene was first isolated from DH5α genomic DNA by PCR amplification using primers 5'-GTTGCCATTAAATAGCTCCTGGTAGATCTAGG-3' and 5'-GAAGCGTCCTAAGGCTTAATGGTAGATCTAG-3’, followed by direct PCR cloning into pCR4Blunt-TOPO (Invitrogen) and sequencing confirmation. The rpsL+ gene was then inserted into the Bgl II site of pREP4 (Qiagen) that contained kana. To ensure the appropriate transcription and translation of both kana and rpsL+, rpsL+ was inserted into the Bgl II site upstream of the kana start codon but downstream of its transcriptional start site, generating pREP4-rpsL+. Finally, the 1.74-kb (rpsL+-kana) cassette was excised from pREP4-rpsL+ with Hind III and Nru I and inserted between Sma I and Hind III sites of pBluescript SK+ (Stratagene), resulting in pSK+rpsL+-kana (Figure 1B).
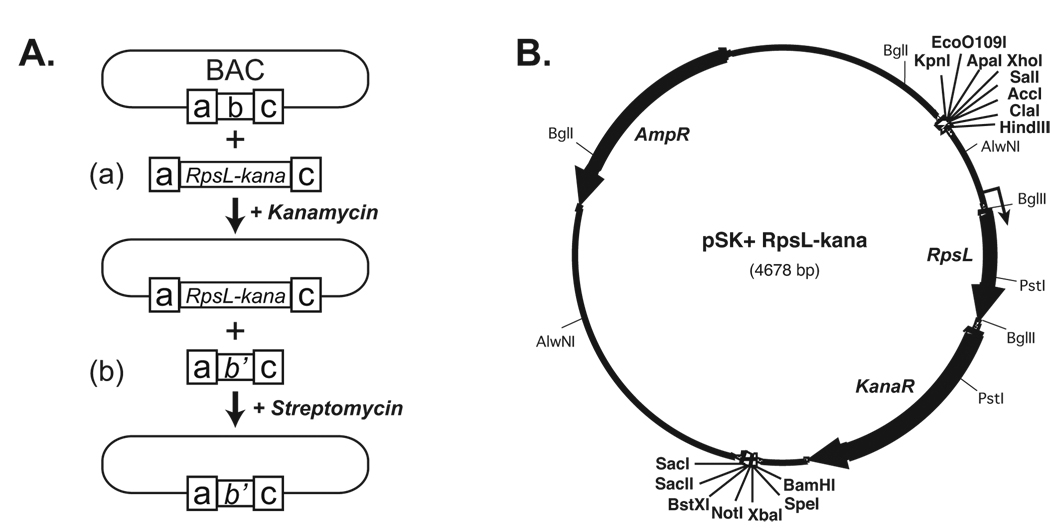
Figure 1. Diagrams of the experimental design.
A. A positive and negative selection strategy for BAC recombineering. a & c, homology arms; b & b’, original and modified BAC sequences, respectively. B. The map of pSK+rpsL+-kana plasmid. Three open reading frames are indicated by solid black arrows. The thin arrow indicates the position of transcription start site of rpsL+-kana bicistronic transcript. Unique restriction sites are shown in bold type.
Construction of recombination cassettes
The mTERT promoter was amplified from BAC clone RP24-183M22 and subcloned into pBluescript SK+. PCR-based mutagenesis was performed to create Hind III and Sal I sites in place of the ATG start codon, resulting in pSK+mTERTmATG. To place the rpsL+-kana cassette at the mTERT start codon, the 1.74-kb fragment was excised from pSK+rpsL+-kana with Hind III/Sal I and inserted into pSK+mTERTmATG through Hind III/Sal I sites, resulting in pSK+mTERTrpsL+-kana. pSK+mTERT-Fluc and pSK+mTERT-Rluc were generated by inserting the Firefly and Renilla luciferase expression cassettes from pGL3-basic and pRL-GL3 into Hind III/Sal I sites of pSK+mTERTmATG. To place rpsL+-kana at the hTERT start codon, the 1.74-kb selection cassette was excised from pSK+rpsL+-kana with Xho I/BamH I and replaced the Firefly expression cassette in pYF10-Fluc, a hTERT promoter reporter routinely used in the laboratory, resulting in pYF10-rpsL+-kana. The Firefly and Renilla luciferase expression cassettes for hTERT BAC recombination were excised from plasmids pYF10-Fluc and pYF10-Rluc, respectively. The recombination cassettes for modifications of human p16Ink4a and p14Arf loci were similar constructed. The DNA fragments used for recombineering were isolated from plasmid DNAs following restriction enzyme digestion and agarose gel electrophoresis, using QiaExII DNA isolation kit (Qiagen).
Recombineering
Each step of the experimental procedure of recombineering was performed as described by Lee at al. (8). The first step of our BAC modification process was to place the rpsL+-kana cassette at the locus of interest via positive kana selection (Figure 1A(a)). One milliliter overnight culture of DY380 bacteria harboring a BAC construct was transferred to a 50 ml culture and allowed to grow at 32°C until the bacteria were in log-phase growth (A600≈0.5). At this point, the recombinases were induced by incubating the culture at 42°C for 15 minutes. Immediately following the 15-min incubation, the bacteria were chilled to 4°C, washed three times in 10 ml ice-cold 10% glycerol, and resuspended in 400 µl ice-cold 10% glycerol and used subsequently as electrocompetent cells. For recombineering, 100–200 ng of expression cassette in a volume of 1–2 µlwas added to each 50 µl electrocompetent DY380 bacteria, followed by electroporation at 1.75 kV and 186 Ω with a capacitance of 25 µF, using a 0.1 cm gap cuvette and a BTX ECM-600 electroporator (Harvard Apparatus). The bacteria were then recovered in 1 ml SOC medium for 1 hour at 32°C with shaking, plated on LB/agar plates supplemented with 12.5 µg/ml chloramphenicol and 20 µg/ml kanamycin, and incubated for 16 hours at 32°C to allow colony formation. DNA minipreps were performed from cultures of ten kana resistant colonies for each BAC using a standard alkaline lysis method (14), and the integrity of each intermediate BAC construct was verified by restriction enzyme digestions followed by agarose gel electrophoresis.
The second step was to replace the rpsL+-kana cassette in the BAC intermediate construct with a luciferase cassette containing the same homology arms via negative selection of loss of rpsL+ (Figure 1A(b)). Induction of recombination and electroporation were performed as in the first step. Following electroporation, 100 µl of a 1:25 diluted DY380 were plated on LB/agar plates supplemented with 12.5 µg/ml chloramphenicol and 1 mg/ml streptomycin to select for bacteria harboring BACs that have lost the rpsL+-kana cassette. To identify those colonies that underwent homologous recombinations, resulting in the replacement of rpsL+-kana cassette with the luciferase cassette, single-colony PCRs were performed with primer pairs specific for the desired homologous recombination events. The PCR mixture was gently mixed up and down two to three times with a standard 200 µl pipet tip that had picked a colony and the tip was subsequently ejected into a 0.5 ml LB medium which was allowed to grow at 32°C. The PCR primers are 5’-CCTCCGCCTACCTAACCTTC-3’ and 5’-AACAGTACCGGAATGCCAAG-3’ for the mouse BAC recombinant 183M22-Fluc; 5’-GGGAGGTGTGGGAGGTTTT-3’ and 5’-CACACAGAAACCACGGTCAC-3’ for the human BAC recombinant 117B23-Fluc. Following amplification, PCR products were analyzed on 1.2–2.0% agarose gel depending on the sizes of products. The cultures corresponding to colonies with positive PCR products were brought up to total a volume of 5 ml LB with chloramphenicol and streptomycin, and grown overnight at 32°C. These pre-screened clones were further tested by restriction enzyme digestion analysis.
Restriction enzyme digestion analysis
For each step of BAC modification, single enzyme digestions of BAC minipreps with up to four restriction enzymes were used to ensure accurate assessment of desired arrangements. Due to the large sizes of BACs, we utilized Gene Construction Kit 2.5 (Textco, Inc.) to predict digestion profiles, which was subsequently compared to the real patterns on a 0.7% agarose gel.
Results and Discussion
To make precise BAC modifications without leaving behind any unwanted sequences, we developed a two-step recombineering strategy as shown in Figure 1A. The strategy utilizes a dual antibiotic marker for a positive selection of kanamycin resistance and a negative selection of the bacterial ribosomal protein S12 (rpsL+) gene, which confers streptomycin sensitivity (15). The dual marker was generated by inserting the rpsL+ gene between the transcription start site and the initiation codon of kanamycin resistance gene in pREP4, resulting in pSK+rpsL+-kana (Figure 1B).
To generate reporters for studying transcriptional regulation of the TERT (telomerase reverse transcriptase) genes (16), we inserted luciferase expression cassettes into the initiation codons of the TERT genes in BAC constructs, using the two-step procedure described as follows. To make use of the recombineering method reported by Lee at al. (8), BAC constructs of interest were introduced into bacterial strain DY380. For mTERT modifications, the mouse BAC containing DY380 cells, upon activation of the Red recombination system at 42°C, were transformed with the recombination cassette isolated from pSK+mTERT-rpsL+-kana. Following electroporation, the bacteria were plated onto LB/agar plates containing 20 µg/ml kanamycin. Using two different BACs containing the mTERT locus, RP24-183M22 and RP23-412H3, we obtained comparable recombination efficiencies, a total of 5,000–10,000 colonies or 70–140 colonies per nanogram of DNA (Table 1). Likewise, the hTERT locus in RP11-117B23 was recombined with the hTERT recombination cassette from pYF10-rpsL+-kana with a similar efficiency. The resultant modified BACs, which we called BAC intermediates, were verified by analyzing the restriction enzyme digestion patterns on a 0.7% agarose gel, to ensure that only intended recombinations occurred (Figure 2).
Table 1.
Efficiencies of recombineering using positive and negative selection markers
| Kanamycin selection | ||||||
|---|---|---|---|---|---|---|
| BAC constructs | Recombination fragments | DNA volumes | DNA concentrations | Total # of colonies | Efficiencies | |
| 183M22 | mTERT-Kana/rpsL | 2 µl | 70 ng/µl | ~10000 | ~70/ng DNA | |
| 412H3 | mTERT-Kana/rpsL | 0.5 µl | 70 ng/µl | ~5000 | ~140/ng DNA | |
| 412H3 | mTERT-Kana/rpsL | 1 µl | 70 ng/µl | ~10000 | ~140/ng DNA | |
| 117B23 | pYF10-Kana/rpsL | 1 µl | 250 ng/µl | 12400 | 50/ng DNA | |
| 478M20 | p16-Kana/rpsL | 1 µl | 50 ng/µl | ~4000 | ~80/ng DNA | |
| 478M20 | p14-Kana/rpsL | 1 µl | 160 ng/µl | ~38400 | ~240/ng DNA | |
| Streptomycin selection | ||||||
| BAC constructs | Recombination fragments | DNA volumes | DNA concentrations | Total # colonies examined | # PCR positive | # correctly modified BACs with no unwanted rearrangement |
| 183M22 | mTERT-Fluc | 1 µl | 200 ng/µl | 28 | 8 (29%) | 5 (63%) |
| 183M22 | mTERT-Fluc | 1 µl | 276 ng/µl | 43 | 4 (9%) | 2 (50%) |
| 117B23 | pYF10-Fluc | 1 µl | 270 ng/µl | 68 | 8 (12%) | 7 (88%) |
| 117B23 | pYF10-hRluc | 1 µl | 240 µg/µl | 110 | 14 (13%) | 11 (79%) |
| 478M20 | p16-hRluc | 1 µl | 370 µg/µl | 120 | 5 (4.2%) | 1 (20%) |
| 478M20 | p14-Fluc | 1 µl | 120 µg/µl | 160 | 2 (1.3%) | 2 (100%) |
| Overall | 529 | 41 (7.8%) | 28 (68%) | |||
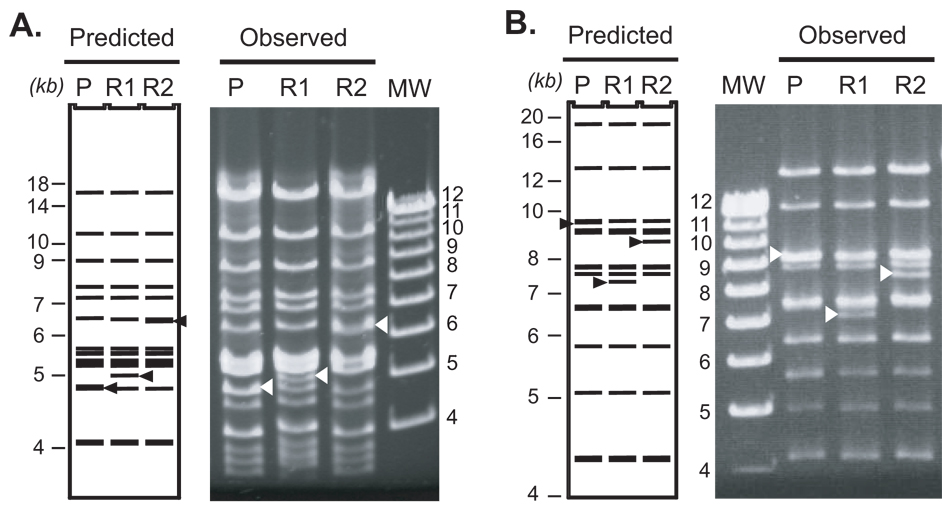
Figure 2. Restriction enzyme digestion patterns of recombineering products of the mTERT and hTERT loci in BAC constructs.
Mouse 183M22 and human 117B23 BACs were subjected to the two-step recombineering. Shown here are (A) the mouse 183M22 and (B) the human 117B23 BAC clones digested with BglII and HincII, respectively. Left panels are the computer-predicted digestion patterns, whereas right panels show the gel patterns of digested BAC clones. P, R1, and R2 represent parental, rpsL+-kana intermediate, and luciferase-containing recombinant BACs, respectively. MW, molecular weight marker in kb. Triangles indicate restriction fragments that change size upon recombineering.
The second step of recombineering involved a negative selection against the rpsL+-kana cassette, i.e. the loss of streptomycin sensitivity. Following the verification of BAC intermediates, the rpsL+-kana fragment was replaced with a luciferase cassette. Figure 2 shows examples of this second step modification for both mTERT BAC and hTERT BAC constructs. Recombination fragments for mouse and human loci were isolated from pSK+mTERT-Fluc and pYF10-Fluc, respectively, and electroporated into bacteria containing the mouse and human BAC intermediates. The bacteria were selected in LB/agar plates containing 1 mg/ml streptomycin. Although streptomycin did not completely inhibit bacterial growth, streptomycin resistant colonies were easily identifiable above a thin film of bacteria. The candidates of homologous recombination were identified by single-colony PCR using a pair of primers specific for the recombined DNA (Figure 3). For mTERT and hTERT BACs and their mutant derivatives, about 14% of randomly picked colonies were found to be PCR positives (Table 1 and data not shown). To demonstrate the general utility of the approach, we also modified BAC clone RP11-478M20, which contains 163 kb human genomic sequence including the p16Ink4a and p14Arf loci. A Renilla and a Firefly luciferase cassette were inserted into the translational initiation codons of the p16Ink4a and p14Arf, respectively. The efficiencies of these recombinations were somewhat lower (2.5%, Table 1), likely due to higher basal rearrangements of this BAC clone. While the reason for this difference is unknown, it is very likely that this BAC is prone to intra-molecular recombination due to the presence of larger amounts of repetitive sequences.
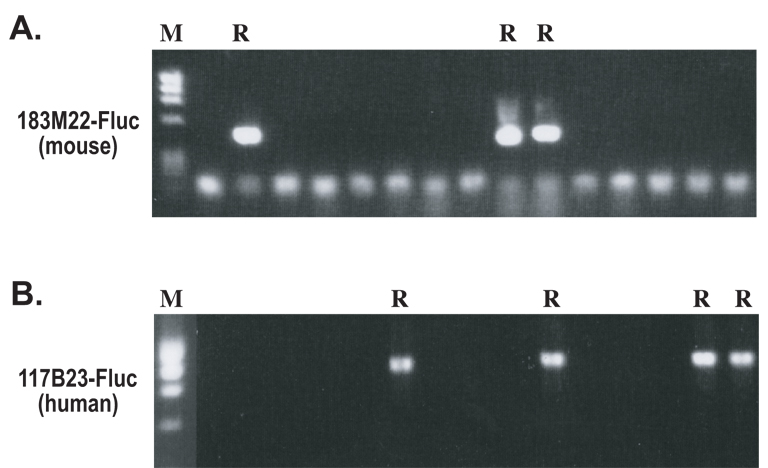
Figure 3. Single-colony PCR screening experiments to identify potential recombinants.
Colonies from the second recombination step for either the mouse BAC 183M22 (A) and human BAC 117B23 (B) were screened by single-colony PCR, using one primer specific for either the mouse or human sequences and the other primer for the Fluc gene. R represents potential recombinants.
The candidate colonies were expanded in culture and BAC DNA minipreps were extracted and digested with restriction enzymes, followed by analysis on a 0.7% agarose gel. Although the sequences of a BAC could be readily assembled using available human and mouse genomic sequence databases, the patterns of a restriction enzyme digested BAC construct did not always perfectly match the predicted patterns by Gene Construction Kit™ based on available sequences (Figure 2). We believe that this discrepancy was due to the presence of repetitive sequences, especially mini-satellites and micro-satellites, which were often polymorphic. Therefore, we emphasized the consistency of band patterns before and after each recombination step. To ensure that a modified BAC construct contained only intended recombination and no other rearrangements, the BAC construct, before and after recombination, was always verified with single enzyme digestion using multiple restriction enzymes. A correctly recombined product was confirmed when all the band patterns were the same as the parental BAC except for the intended changes (Figure 2). Among the colonies identified by single-colony PCR analysis, an overall 68% were confirmed as the products of intended recombinations with no unwanted rearrangements (Table 1).
In our laboratory, we have found that the overall efficiencies of this kanamycin/streptomycin selection scheme were very similar to the galK method (13). For introduction of point mutations and deletions into the hTERT BAC 117B23 with substrate fragments containing 50 bp homology arms, we observed an average of 70 colonies per nanogram input fragment upon galK positive selection. In the second step of galK counter-selection, about 16% of randomly picked colonies contained intended recombination as determined by colony PCR analysis (data not shown). Similar rates were obtained using the kanamycin/streptomycin method for the same BAC constructs, as shown in Table 1.
Overall, the first positive selection step was relatively easy to perform. The majority (>95%) of the kanamycin resistant colonies contained recombineering products, because no background colonies would form without the incorporation of the kanamycin. The second counter-selection step of the rpsL+ marker required higher recombineering efficiency. Even without recombination fragments, we consistently observed 20,000 to 50,000 colonies upon induction of the recombination system. We encountered the same problem with counter-selecting the galK marker. It is likely that intra-molecular recombinations, possibly through repetitive sequences, resulted in the loss of rpsL+ or galK marker. From our experience, if we could obtain more than 5,000 kanamycin resistant colonies in the first step, at least 10% of all the colonies in the second step (using the same homology arms) would turn out to be intended recombineering products.
Since the development of this two-step BAC recombination method, we have successfully modified a number of BAC constructs, including various reporter insertions and deletions. This selection scheme has several advantages compared to the galK method. First, the experiments can be performed in the original DY380 strain and its derivatives and do not require E.coli strains with galK deletions. Second, kanamycin and streptomycin resistant colonies form overnight in regular LB medium, without the need of special minimal medium. In contrast, selections for and against galK are performed in minimum media. In each step, colony growth takes at least three days under the recommended experimental conditions. Thus, the kanamycin/streptomycin selection scheme is a more timesaving approach. Finally, this modification procedure is more cost-effective since kanamycin and streptomycin are very inexpensive compared to minimal medium and DOG. Therefore, while the galK method is an efficient and user-friendly approach, the kanamycin/streptomycin technique is a faster and more cost-effective alternative.
Acknowledgements
DY380 recombineering system was kindly provided by the Copeland laboratory at National Cancer Institute, Frederick, MD. We thank Dr. Renjith Mathew for critical review of the manuscript. The study was supported in part by NIH grant GM071725. JZ is a Research Scholar of American Cancer Society. The authors declare no competing interests.
References
- 1.Heintz N. Nat Rev Neurosci. 2001;2(12):861–870. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- 2.Muyrers JP, Zhang Y, Stewart AF. Trends Biochem Sci. 2001;26(5):325–331. doi: 10.1016/s0968-0004(00)01757-6. [DOI] [PubMed] [Google Scholar]
- 3.Tsyrulnyk A, Moriggl R. Methods Mol Biol. 2008;430:269–293. doi: 10.1007/978-1-59745-182-6_19. [DOI] [PubMed] [Google Scholar]
- 4.Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, Pelletier L, Kittler R, Hua S, Naumann R, Augsburg M, Sykora MM, Hofemeister H, Zhang Y, Nasmyth K, White KP, Dietzel S, Mechtler K, Durbin R, Stewart AF, Peters JM, Buchholz F, Hyman AA. Nat Methods. 2008;5(5):409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. Nature Genetics. 1998;20(2):123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 6.Copeland NG, Jenkins NA, Court DL. Nat Rev Genet. 2001;2(10):769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 7.Muyrers JP, Zhang Y, Benes V, Testa G, Ansorge W, Stewart AF. EMBO Rep. 2000;1(3):239–243. doi: 10.1093/embo-reports/kvd049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. Genomics. 2001;73(1):56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 9.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(11):5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hegde S, Paulson RF. Biotechniques. 2004;36(6):936–938. doi: 10.2144/04366BM03. 940. [DOI] [PubMed] [Google Scholar]
- 11.Sopher BL, La Spada AR. Gene. 2006;371(1):136–143. doi: 10.1016/j.gene.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 12.Rivero-Muller A, Lajic S, Huhtaniemi I. Nucleic Acids Res. 2007;35(10):e78. doi: 10.1093/nar/gkm250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Nucleic Acids Res. 2005;33(4):e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Russell DW. Molecular Cloning, A Laboratory Mannual. Third Ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 15.Stavropoulos TA, Strathdee CA. Genomics. 2001;72(1):99–104. doi: 10.1006/geno.2000.6481. [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Hu C, Zhu J. Molecular Biology of the Cell. 2007;18(2):669–677. doi: 10.1091/mbc.E06-09-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]